Abstract
SPX domains control phosphate homeostasis in eukaryotes. Ten genes in yeast encode SPX-containing proteins, among which YDR089W is the only one of unknown function. Here, we show that YDR089W encodes a novel subunit of the vacuole transporter chaperone (VTC) complex that produces inorganic polyphosphate (polyP). The polyP synthesis transfers inorganic phosphate (Pi) from the cytosol into the acidocalcisome- and lysosome-related vacuoles of yeast, where it can be released again. It was therefore proposed for buffer changes in cytosolic Pi concentration (Thomas, M. R., and O'Shea, E. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9565–9570). Vtc5 physically interacts with the VTC complex and accelerates the accumulation of polyP synthesized by it. Deletion of VTC5 reduces polyP accumulation in vivo and in vitro. Its overexpression hyperactivates polyP production and triggers the phosphate starvation response via the PHO pathway. Because this Vtc5-induced starvation response can be reverted by shutting down polyP synthesis genetically or pharmacologically, we propose that polyP synthesis rather than Vtc5 itself is a regulator of the PHO pathway. Our observations suggest that polyP synthesis not only serves to establish a buffer for transient drops in cytosolic Pi levels but that it can actively decrease or increase the steady state of cytosolic Pi.
Keywords: homeostasis, inositol phosphate, plant physiology, vacuole, yeast, polyphosphate, acidocalcisome, phosphate homeostasis
Introduction
Phosphate is a limiting factor for the growth of living organisms. It is mainly taken up by the cells as Pi and incorporated in biological molecules, including ATP, nucleic acids, and phospholipids. Pi plays a major role in the regulation of biochemical pathways through phosphorylation, pyrophosphorylation, and polyphosphorylation. Its concentration affects the free energy liberated by the hydrolysis of ATP and other nucleoside di- and triphosphates. Therefore, Pi homeostasis (import, usage, storage, and export) is regulated (1, 2), and Pi can be accumulated and stored as a polymer of Pi units called polyphosphate (polyP).3 One mode of regulation operates on the transcriptional level. The PHO pathway, which is a Pi-dependent transcriptional program in yeast, has been extensively characterized (2). Transcription of the PHO genes is regulated by the transcription factors Pho4 and Pho2. Phosphorylated Pho2 interacts with Pho4 to induce expression of the PHO genes. Pho2 is a target of Cdc28 kinase in vitro suggesting a coordination between cell cycle progression and nutrient availability (3). Pho4 is itself regulated by the cyclin-dependent kinase Pho85 and its cyclin Pho80. Under Pi-rich conditions, the Pho80-Pho85 complex phosphorylates Pho4, thereby restricting the localization of the transcription factor to the cytosol (4, 5). Under Pi limitation, Pho80/Pho85 is inhibited, and the non-phosphorylated Pho4 is imported into the nucleus, allowing transcription of PHO genes. Pho80/85 is regulated by the cyclin-dependent kinase inhibitor Pho81. Inhibition of Pho80/85 by Pho81 is facilitated by the inositol pyrophosphate 1-IP7, which is produced by Vip1 (6). However, a recent report found that induction of the PHO pathway shows only a minor delay in vip1Δ mutants (7). In plants and yeast, many proteins involved in Pi homeostasis contain SPX domains, which have been proposed to regulate Pi metabolism by serving as receptors to inositol pyrophosphates (8).
The genome of Saccharomyces cerevisiae contains 10 open reading frames encoding SPX domain proteins (9), named after Syg1, Pho81, XPR1. Eight of these SPX proteins have functions related to Pi metabolism as follows: Pho87 and Pho90 are plasma membrane Pi importers (10, 11); Pho91 is a vacuolar Pi exporter (12); Pho81 is a cyclin-dependent kinase inhibitor (13); Gde1 is a glycerophosphocholine phosphodiesterase that hydrolyzes glycerophosphocholine, thereby releasing choline and glycerol 3-phosphate that can be used as a Pi source (14). Three other SPX domain containing proteins (Vtc2, Vtc3, and Vtc4) are part of a large complex that produces polyP, the Vacuole Transporter Chaperone (VTC) complex (15–18). AlthoughVtc4 holds the catalytic activity, Vtc2 and Vtc3 are assumed to be accessory subunits with a regulatory function (19). In line with this assumption, VTC exists in two isoforms, composed of Vtc4/Vtc3/Vtc1 or Vtc4/Vtc2/Vtc1. Vtc1 is a small protein that contributes only three transmembrane domains to the complex, which resemble the transmembrane domains of the larger VTC proteins but lack their large hydrophilic N-terminal extensions. VTC is located in the membrane of the acidocalcisome- and lysosome-like vacuole and in a peripheral location presumed to be the endoplasmic reticulum (ER), with Vtc4/Vtc3/Vtc1 being more enriched in the vacuole and Vtc4/Vtc2/Vtc1 being more prominent in the ER (19, 20). VTC couples synthesis of polyP to its translocation across the membrane, thereby sequestering polyP inside the lumen of the vacuole and avoiding its accumulation in the cytosol, where polyP can be toxic (21). PolyP is degraded when cells are shifted from Pi-rich media to Pi-starvation media (22). Therefore, polyP can influence the kinetics of changes in cytosolic Pi (1, 22). Finally, Syg1 is an SPX domain protein of unknown function. It is a homolog of the mammalian phosphate exporter Xpr1. Because both proteins localize to the plasma membrane and interact with β subunits of G-proteins (23–25), they may have similar functions in exporting Pi.
In this work, we focused on a yeast gene coding an SPX domain-containing protein of unknown function, YDR089W. We identify its product as a novel subunit of the VTC complex.
Experimental Procedures
Reagents
Creatine phosphate, creatine kinase, adenosine 5′-triphosphate, and protein G-agarose were from Roche Applied Science; N-(3-triethylammoniumpropyl)- 4-(p-diethylaminophenylhexatrienyl)-pyridinium dibromide (FM4-64) was from Invitrogen; 4′,6′-diamidino-2-phenylindole (DAPI) was from Sigma; monoclonal anti-HA antibodies (mouse ascites) were from Covance; polyclonal antibodies to GFP were from Torrey Pines Biolabs; polyclonal antibodies to β-tubulin were from Abcam (Ab15568); polyclonal antibodies against Ypt7, Vam3, Sec17, Vtc1, Vtc2, Vtc3, and Vtc4 had been raised in rabbits or goats and were prepared as described (18, 19, 26). The lytic enzyme (lyticase) (27) and recombinant Ppx1 (21) were purified as described.
Media and Growth Conditions
Cells were grown at 30 °C in standard YPD (1% yeast extract, 2% polypeptone, 2% dextrose) or in synthetic complete media (SC).
Genetic Manipulations
Yeast transformations were carried out using the lithium acetate method (25). Gene deletions and tagging were performed following established procedures (28–30). Strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Oligonucleotides used are listed in Table 3.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4741 vtc5Δ | BY4741 YDR089W::natNT2 | This study |
| BY4741 GPD1-Vtc5 | VTC5 natNT2-GPD1pr | This study |
| BY4741 GPD1-GFP-Vtc5 | VTC5 natNT2-GPD1pr-GFP | This study |
| BJ3505 | MATa pep4::HIS3 prb1-Δ1.6R lys2-208 trp1-Δ101 ura3-52 gal2 can | 60 |
| DKY6281 | MATα pho8::TRP1 leu2-3 leu2-112 lys2-801 suc2-Δ9 trp1-Δ901 ura3-52 | 61 |
| BJ3505 Vtc5-GFP | VTC5-GFP-kanMX | This study |
| BJ3505 ADH1-GFP-Vtc5 | VTC5 natNT2-ADH1pr-GFP | This study |
| DKY6281 Vtc5-GFP | VTC5-GFP-kanMX | This study |
| DKY6281 ADH1-GFP-Vtc5 | VTC5 natNT2-ADH1pr-GFP | This study |
| BJ3505 ADH1-HA-Vtc5 | VTC5 natNT2-ADH1pr-HA3 | This study |
| BJ3505 Vtc2-HA Vtc5-GFP | VTC2–3HA-kanMX Vtc5-GFP-URA3 | This study |
| BJ3505 Vtc3-HA Vtc5-GFP | VTC3–3HA-kanMX Vtc5-GFP-URA3 | This study |
| BJ3505 Vtc1-HA Vtc5-GFP | VTC1–3HA-kanMX Vtc5-GFP-URA3 | This study |
| BJ3505 Vtc4-HA Vtc5-GFP | VTC4–3HA-TRP1Vtc5-GFP-kanMX | This study |
| BJ3505 Vtc4-HA | VTC4–3HA-TRP1 | This study |
| BJ3505 Vtc4-HA vtc5Δ | VTC4–3HA-TRP1 VTC5::natNT2 | This study |
| BJ3505 Vtc5-GFP vtc2Δ | VTC5-GFP-kanMX VTC2::natNT2 | This study |
| BJ3505 Vtc5-GFP vtc3Δ | VTC5-GFP-kanMX VTC3::natNT2 | This study |
| BJ3505 Vtc5-GFP vtc4Δ | VTC5-GFP-kanMX VTC4::natNT2 | This study |
| BY4741 pho4Δ | PHO4::kanMX | Euroscarf |
| BY4741 pho81Δ | PHO81::kanMX | Euroscarf |
| BY4741 pho80Δ | PHO80::kanMX | Euroscarf |
| BY4741 pho4Δ GPD1-GFP-Vtc5 | PHO80::kanMX VTC5 natNT2-GPD1pr-GFP | This study |
| BY4741 pho81Δ GPD1-GFP-Vtc5 | PHO81::kanMX VTC5 natNT2-GPD1pr-GFP | This study |
| BY4742 Vtc4wt | VTC4::kanMX VTC4-His3 | 19 |
| BY4742 vtc4Δ | VTC4::kanMX | Euroscarf |
| BY4742 Vtc4R264A | VTC4::kanMX VTC4R264A-His3 | 19 |
| BY4742 Vtc4wt GPD1-GFP-Vtc5 | VTC4::kanMX VTC4-His3 VTC5 natNT2-GPD1pr-GFP | This study |
| BY4742 vtc4Δ GPD1-GFP-Vtc5 | VTC4::kanMX VTC5 natNT2-GPD1pr-GFP | This study |
| BY4742 Vtc4R264A GPD1-GFP-Vtc5 | VTC4::kanMX VTC4R264A-His3 VTC5 natNT2-GPD1pr-GFP | This study |
| BY4741 Pho84-GFP | Pho84-GFP-kanMX | This study |
| BY4741 Pho84-GFP vtc5Δ | Pho84-GFP-kanMX VTC5::NatNT2 | This study |
| BY4741 Pho84-GFP GPD1-Vtc5 | Pho84-GFP-kanMX VTC5 natNT2-GPD1pr | This study |
| EY1109 | MATa leu2::PHO84pr:GFP::LEU2 ADE2 | 1 |
| EY1109 vtc4Δ | VTC4::natNT2 | This study |
| EY1109 vtc5Δ | YDR089W::natNT2 | This study |
| EY1109 GPD1-Vtc5 | VTC5 natNT2-GPD1pr | This study |
| EY1109 pho80Δ | PHO80::natNT2 | This study |
| EY1109 pho81Δ | PHO81::natNT2 | This study |
TABLE 2.
Plasmids used in this study
TABLE 3.
Oligonucleotides used in this study
fwd, forward; rev, reverse.
| No. | Sequence (5′–3′) | Use |
|---|---|---|
| 1 | GTTTTTCTCAGTTACTGGAACAGTGTGCTATGGTATGATTGAAATTTTCTTTGGTGACGGTGCTGGTTTA | (fwd); tagging Vtc5 at the C terminus; templates, pKT127 or pKT209 |
| 2 | GCATATTTTATATAGACTGTACACATGAAAAACTTTTATCGTCCAAAATAATCGATGAATTCGAGCTCG | (rev); tagging Vtc5 at the C terminus; templates, pKT127 or pKT209 |
| 3 | TGGCAAGTGTATGTCTGGGA | (fwd); control C-terminal tagging of Vtc5 |
| 4 | GGG ACA ACA CCA GTG AAT | (rev); control C-terminal tagging of Vtc5 |
| 5 | TATCTCCATCTCAGCCCTAGGTAAGGATAGATACGAAGGGTTGTACAATCATGCGTACGCTGCAGGTCGAC | (fwd); tagging Vtc5 N terminus; templates, pYM-NX plasmids |
| 6 | GTAGAATTTCCATTCAGGGATTGATTTATTCAATATGCGATCTTCAAATTTCATCGATGAATTCTCTGTCG | (rev); tagging Vtc5 N terminus; templates, pYM-NX plasmids |
| 7 | AAAGTTTGTGTCTCCCCTTG | (fwd); checking VTC5 deletion and N-terminal tagging |
| 8 | GTCTGCAGCGAGGAGCCGTA | (rev); checking integration of NAT cassette |
| 9 | TATCTCCATCTCAGCCCTAGGTAAGGATAGATACGAAGGGTTGTACAATCATGCGTACGCTGCAGGTCGAC | (fwd); deleting VTC5 through NAT; template, pFA6-natNT2 |
| 10 | GCATATTTTATATAGACTGTACACATGAAAAACTTTTATCGTCCAAAATAATTAATCGATGAATTCGAGCTCG | (rev); deleting VTC5 through NAT; template, pFA6-natNT2 |
| 11 | GTGTTTCTAATTCTCTCTATTTTATTTTCCTTTTTCTGCAACCTAGTTGCTAAACGTACGCTGCAGGTCGAC | (fwd); tagging Vtc4 C terminus; template, pYM22 |
| 12 | CTAATATGATTATTACTTAATTATACAGTAAAAAAAACACGCTGTGTATTCAATCGATGAATTCGAGCTCG | (rev); tagging Vtc4 C terminus; template, pYM22 |
| 13 | AAAAGTAGAAAGAACGACTACACCTCAACATAACGACACTTTTTTGACATGCGTACGCTGCAGGTCGAC | (fwd); deleting VTC2 through NAT; template, pFA6-natNT2 |
| 14 | GGTCTCAGTAGATAGAGTACATATTCTAATCACTGCTTGGCCCCATTAATCGATGAATTCGAGCTCG | (rev); used to delete VTC2 using the NAT marker (plasmid pFA6-natNT2 as a template) |
| 15 | CTATTAGAGCGAACAGCAGAATTTGTCCTTGGT TTTCAGAGTTTGAAAATGCGTACGCTGCAGGTCGAC | (fwd); deleting VTC3 through NAT; template, pFA6-natNT2 |
| 16 | AACTGGTACTTGTGTAATATATGTGTATATAAAAAATATACATGTTCTTAATCGTATGAATTCGTAGCTCG | (rev); deleting VTC3 through NAT; template, pFA6-natNT2 |
| 17 | GGCTAACAATCAAATCGGCCAATAAAAGAGCATAACAAGGCAGGAACAGCTATGCGTACGCTGCAGGTCGAC | (fwd); deleting VTC4 through NAT; template, pFA6-natNT2 |
| 18 | GATTATTACTTAATTATACAGTAAAAAAAACACGCTGTGTATTCATTTAATCGATGAATTCGAGCTCG | (rev); deleting VTC4 through NAT; template, pFA6-natNT2 |
| 19 | CAAGAATAATGACATTGAATCTTCCAGCCCATCTCAACTTCAACATGAAGCACGGATCCCCGGGTTAATTAA | (fwd); tagging Pho84 C terminus; template, pFA6a-GFP(S65T)-kanMX6 |
| 20 | GTTTTTGTATTATTTGTTCTAGTTTACAAGTTTTAGTGCATCTTTGAGGCTTGAATTCGAGCTCGTTTAAAC | (rev); tagging Pho84 C terminus; template, pFA6a-GFP(S65T)-kanMX6 |
| 21 | CCATAACTGTGCTAGAGACG | (fwd); check C-terminal tagging of Pho84 |
| 22 | GACTAGTATGAAATTTGAAGATCGCAT | (fwd); amplifying VTC5 |
| 23 | CCGCTCGAGTTAAAAGAAAATTTCAATCATACC | (rev) amplifying VTC5 |
| 24 | CGGAATTCGGTGACGGTGCTGGTTTA | (fwd); amplifying GFP; template, pKT127 |
| 25 | CCCAAGCTTTTATTTGTACAATTCATCCAT | (rev); amplifying GFP; template, pKT127 |
| 26 | CGAGCTCTGCATTATTTAGATCGGAAAAAGTC | (fwd); amplifying PHO4 and its endogenous promoter |
| 27 | GGAATTCCGTGCTCACGTTCTGCTGTAGGTG | (rev); amplifying PHO4 and its endogenous promoter |
The plasmid used to overexpress VTC5, pRS416 GPD1-Vtc5, was obtained by cloning the VTC5 open reading frame between the XhoI and SpeI restriction sites of the pRS416 GPD1 plasmid (Euroscarf). To obtain pRS416 PPHO4-PHO4-GFP, GFP from the plasmid pKT127 was first amplified by PCR using oligo-24 and oligo-25 and inserted into pRS416 between the EcoRI and HindIII restriction sites, yielding pRS416-GFP. Then, PPHO4-PHO4 was amplified from genomic DNA using oligo-26 and oligo-27 and ligated between the SacI and EcoRI restriction sites of pRS416-GFP, yielding pRS416 PPHO4-PHO4-GFP.
Vacuole Isolation
Yeast cells were grown overnight in 2-liter flasks containing 1 liter of YPD to an OD600 nm of 0.5–1.5. Cells from l liter of culture were collected by centrifugation (1 min, 4000 × g, room temperature, JLA-10.500 rotor) and resuspended in 50 ml of 30 mm Tris-HCl buffer, pH 8.9, containing 10 mm DTT. Suspensions were incubated for 5 min at 30 °C and centrifuged as before. Cells were incubated in 15 ml of spheroplasting buffer (50 mm potassium phosphate, pH 7.5, 600 mm sorbitol in YPD with 0.2% d-glucose, and 3600 units ml−1 lyticase) for 25 min at 30 °C. Spheroplasts were collected by centrifugation (2 min, 1500 × g, 4 °C), resuspended in PS buffer (10 mm PIPES-KOH, pH 6.8, 200 mm sorbitol) containing 15% Ficoll 400, and 80 μg of DEAE-dextran was added to each tube. Samples were incubated (30 °C, 2 min) and fractionated on a discontinuous Ficoll 400 gradient (90 min, 150,000 × g, 2 °C). Vacuoles were collected from the 4-0% Ficoll 400 interface. Protein concentration in the samples was determined using the Bradford reagent, using BSA as a standard.
Immunoprecipitations
200 μg of isolated vacuoles were diluted into 1 ml of PS buffer and sedimented by centrifugation (7000 × g, 7 min, 4 °C). Pellets were resuspended in 500 μl of solubilization buffer (10 mm PIPES, pH 6.8, 200 mm sorbitol, 5% glycerol, 0.5% Triton X-100). Lysis was allowed to proceed for 20 min at 4 °C with gentle shaking. Insoluble material was removed by centrifugation (20,000 × g, 20 min, 4 °C), and supernatants were transferred to a fresh tube. Small aliquots of the supernatants were withdrawn to serve as loading controls. Anti-HA monoclonal antibodies (3 μl of serum from mouse ascites, Covance) and 30 μl of protein G-agarose beads, pre-equilibrated in solubilization buffer, were added to the remaining fraction of the solubilized samples. Tubes were placed on a rotating shaker (1 h, 4 °C). Beads were washed with solubilization buffer, and immunoprecipitated proteins were eluted by incubating the beads in 2× SDS-PAGE sample loading buffer for 10 min at 65 °C.
Polyphosphate Synthesis Assay in Vitro
PolyP synthesis by isolated yeast vacuoles was measured by DAPI-polyP fluorescence as described previously (21). Briefly, reaction master mix was prepared in the reaction buffer (10 mm PIPES/KOH, pH 6.8, 150 mm KCl, 0.5 mm MnCl2, 200 mm sorbitol) containing an ATP-regenerating system (1 mm ATP-MgCl2, 40 mm creatine phosphate and 0.25 mg/ml creatine kinase). The reactions were started by adding isolated vacuoles to a final protein concentration of 0.005 mg/ml, and the samples were incubated at 27 °C. At indicated time points, an aliquot was withdrawn and mixed with 2 volumes of stop solution (10 mm PIPES/KOH, pH 6.8, 150 mm KCl, 200 mm sorbitol, 12 mm EDTA, 0.15% Triton X-100, and 15 μm DAPI). Fluorescence of the DAPI-polyP complex was measured in a black 96-well plate using λex = 415 nm and λem = 550 nm (cutoff = 530 nm) at 27 °C. A calibration curve was produced using synthetic polyP-60 as a standard.
Determination of Polyphosphate Levels in Living Cells
Cells were grown overnight in SC medium until an OD600 nm ∼1.0 was reached. One equivalent OD of cells was collected by centrifugation (11,000 × g, 1 min, room temperature). PolyP was extracted by adding 50 μl of 2 m sulfuric acid and vortexing for 10 s. The samples were neutralized by adding 50 μl of 2 m NaOH and vortexing for 10 s. The extracts were supplemented with 100 μl of 1 m Tris malate, pH 7.4. PolyP was purified through PCR DNA purification columns (Qiagen PCR purification kit). 600 μl of binding buffer was added to the extracts, and the samples were loaded onto the columns. After two washes with washing buffer (10 mm Tris-HCl, pH 7.0, 50% ethanol, 10 mm NaCl, 0.3 mm EDTA), polyP was eluted with 110 μl of the manufacturer's elution buffer (10 mm Tris-HCl, pH 8.5). The amount of orthophosphate released after incubation with recombinant Ppx1 (0.01 μg of Ppx1 in 5 mm Tris, pH 7.0, 0.15 mm MgCl2) was determined by the Malachite Green assay.
Determination of PolyP Chain Lengths
PolyP was purified from logarithmically growing cells by glass bead lysis and phenol extraction as described (31) and dissolved in 10 mm Tris-HCl, pH 8.0, 0.5 mm EDTA. RNA concentration was determined by measuring absorbance at 260 nm. The aliquots containing 2 μg of RNA were treated with RNase A (0.04 mg/ml, 30 min at 37 °C), followed by proteinase K (0.1 mg/ml, 30 min at 50 °C). The samples were fractionated on 25% polyacrylamide gel, and polyP was visualized by negative DAPI staining (31, 32).
Quantification of Pho5 Activity
Cells were grown overnight in YPD to logarithmic phase (OD600 nm = 1–2). 50 μl of the cell suspension was mixed with 200 μl of 20 mm p-nitrophenol in 0.1 mm sodium acetate, pH 4.2. Samples were incubated at 30 °C for 15 min. The reactions were stopped by addition of 200 μl of a 10% trichloroacetic acid solution. 400 μl of 2 m sodium carbonate was added, and insoluble material was removed by centrifugation (1 min, 11,000 × g, room temperature). The absorbance of the supernatant was measured at 420 nm. Pho5 activity was expressed as OD420 nm/OD600 nm × 1000.
Fluorescence-activated Cell Sorting
Cells were grown in YPD medium to logarithmic phase. Just before measurement, 50 μl of the cell suspension was added to 950 μl of TBS. Samples were vortexed and immediately analyzed using an LSRII flow cytometer (BD Biosciences). 20,000 events were recorded for each sample. The mean fluorescence value was used to estimate protein expression levels.
Microscopy
Where indicated, vacuoles were stained with the vital dye FM4-64. Cells were grown overnight to an OD600 nm of 0.6–1.0 and diluted to OD600 nm 0.2 in fresh medium. Vacuoles were stained with 10 μm FM4-64 during 1 h at 30 °C. Cells were washed twice with dye-free medium and chased in fresh dye-free medium for 1.5 h at 30 °C. Microscopy was done on a LEICA DMI6000B inverted microscope equipped with a Hamamatsu digital CCD camera (ORCA-R2 C10600-10B), an X-Cite® series 120Q UV lamp, and a Leica 100 × 1.4 NA lens.
Protein Extraction
Two OD600 nm units of cells were harvested by centrifugation (11,000 × g, 1 min, room temperature). Pellets were resuspended in 200 μl of 0.1 m NaOH, and the suspensions were incubated for 5 min at 25 °C. Cells were collected as before and resuspended in 50 μl of 2× loading sample buffer. Samples were heated at 95 °C for 5 min, and insoluble material was removed by centrifugation (11,000 × g, 30 s, room temperature). Supernatants were loaded on SDS-polyacrylamide gels.
Statistics
Tests for significance of observed differences were performed by an unpaired Student's t test. Error bars represent the standard deviation. Differences with p values higher than 0.1 were considered as not significant.
Results
Vtc5 Is a Transmembrane Protein Localized in the Vacuolar Membrane
No significant sequence homology of YDR089W to other yeast proteins was found outside the SPX domain. Using secondary structure predictions (TMHMM) software, we noticed that YDR089W topologically resembled Vtc2, Vtc3, and Vtc4. The YDR089W ORF is of similar length as VTC2, VTC3, and VTC4, and like these genes, it encodes an SPX domain at its N terminus and three C-terminal transmembrane regions (Fig. 1A). Therefore, we explored its possible relationship to polyP metabolism and refer to YDR089W as VTC5. To determine the localization of Vtc5, the protein was genomically tagged with yeGFP at its C terminus in protease-deficient strains (BJ3505) and protease-proficient strains (DKY6281). Vtc5-yeGFP yielded a relatively weak signal around the vacuole rim in BJ3505 cells. No fluorescence signal was detected in other parts of the cell. In a protease-proficient strain (DKY6281), GFP localized mainly to the vacuole lumen (Fig. 1B). This suggests that the C-terminal end of Vtc5-yeGFP is sensitive to cleavage by vacuolar proteases and faces the vacuole lumen. When Vtc5 was tagged at the N terminus, GFP was detected on the vacuolar membrane in a protease-deficient strain as well as in a protease-proficient strain (Fig. 1B). This suggests that the N-terminal domain of Vtc5 is inaccessible to vacuolar proteases. Thus, Vtc5 is likely to be a trans-membrane protein with its C terminus in the vacuole lumen and its N terminus facing the cytosol, resembling the topology of Vtc2–4 (17).
FIGURE 1.
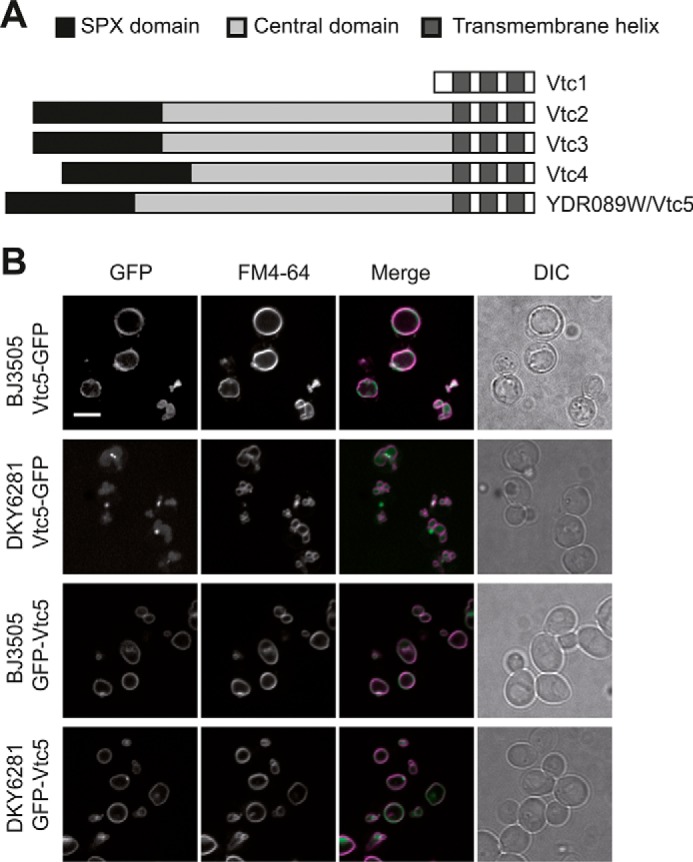
Localization and topology of Vtc5. A, schematic representation of the predicted structural features of Vtc5 and comparison with previously known VTC subunits. Three transmembrane helices were predicted using the TMHMM server. B, Vtc5-GFP and GFP-Vtc5 localization was determined by confocal fluorescence microscopy in a pep4Δ strain (BJ3505) and in a strain expressing PEP4 (DKY6281). Cells were cultivated in YPD to logarithmic phase, and vacuoles were stained with the vital dye FM4-64. The N-terminally tagged version of Vtc5 (GFP-Vtc5) was expressed from the genomically integrated construct under control of the ADH1 promoter. The ADH1 promoter results in moderate overexpression. This explains the fusion of the vacuoles into a single larger organelle, which occurs as a consequence of enhanced polyP production (52). DIC, differential interference contrast. Scale bar, 5 μm.
Vtc5 Physically Interacts with the VTC Complex
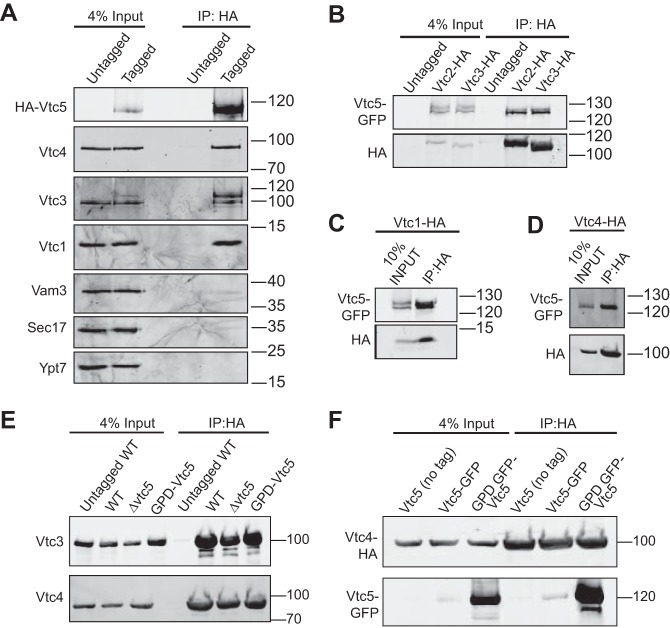
A possible interaction of Vtc5 with the VTC complex was tested by immunoprecipitation. Vacuoles were isolated from a strain expressing HA-tagged Vtc5 under control of the ADH1 promoter, solubilized in Triton X-100, and immunoprecipitated with antibodies to the HA tag (Fig. 2A). SDS-PAGE and Western blotting revealed that Vtc3, Vtc4, and Vtc1 co-precipitated with Vtc5-HA but not with untagged Vtc5. Other vacuolar proteins, such as Vam3 and Ypt7 and the peripherally associated Sec17, were not detected in the Vtc5-HA precipitate with comparable efficiency. To determine whether Vtc5 associates preferentially with the Vtc2- or Vtc3-containing isoform of VTC (19), we immunoprecipitated Vtc2-HA and Vtc3-HA from a strain expressing Vtc5-GFP. All three tagged proteins were expressed under the control of their endogenous promoters. Vtc5 was found associated with both proteins (Fig. 2B). For comparison, we performed the same experiment in strains carrying Vtc1-HA or Vtc4-HA, which are found in both VTC isoforms (Fig. 2, C and D). In all cases, the immunoprecipitates enriched Vtc5-GFP with the bait protein. This suggests that Vtc5 can associate with both Vtc2- and Vtc3-containing VTC complexes.
FIGURE 2.
Physical interaction between Vtc5 and other VTC subunits. A, co-immunoprecipitation of Vtc5 and other VTC subunits. Vacuoles were isolated from BJ3505 cells expressing Vtc5 under the control of the ADH1 promoter with or without an HA tag. The detergent-solubilized membranes were immunoprecipitated (IP) using anti-HA monoclonal antibodies. Adsorbed proteins were analyzed by SDS-PAGE and Western blotting using monoclonal mouse antibodies directed against the HA tag or polyclonal antibodies against the entire proteins. B, both VTC isoforms interact with Vtc5. Vacuoles were isolated from BJ3505 strains expressing either Vtc2-HA or Vtc3-HA and a C-terminally GFP-tagged version of Vtc5 (Vtc5-GFP), all under control of the respective endogenous promoters. HA-tagged proteins were immunoprecipitated and analyzed as in A. C and D, co-immunoprecipitation of Vtc5 with Vtc1 and Vtc4. The same experiment as in B was performed using vacuoles isolated from a strain expressing Vtc1-HA (C) or Vtc4-HA (D). E, Vtc5 does not influence the interaction between other VTC subunits. VTC5 was deleted or overexpressed from the GPD1 promoter in BJ3505 cells carrying HA-tagged or untagged Vtc4. Vacuoles were isolated and subjected to immunoprecipitation as in A, using antibodies directed against GFP or against the HA tag. F, influence of Vtc5 expression level on the Vtc4-Vtc5 interaction. VTC5 was expressed in BJ3505 VTC4-HA under the control of its endogenous promoter (Vtc5-GFP) or the strong GPD1 promoter (GPD1-GFP-Vtc5). Vtc4-HA was pulled down from isolated vacuoles as in C, and adsorbed Vtc5-GFP was detected by Western blotting for its GFP tag. The migration of relevant molecular mass markers (size in kDa) is indicated on the right-hand side of the panels.
Next, we tested whether the abundance of Vtc5 could alter the interaction between other VTC subunits. We deleted or overexpressed Vtc5 in a Vtc4-HA strain, isolated vacuoles, and immunoprecipitated Vtc4-HA. The fractions of Vtc3 associated with Vtc4 were similar in both cases (Fig. 2E). We also measured the amount of Vtc5 interacting with Vtc4 in a strain expressing genomic levels of Vtc5 and in a strain overexpressing Vtc5 under the control of the strong GPD1 promoter. The amount of Vtc5 associated with Vtc4-HA increased as the total amount of Vtc5 in the vacuole membrane increased (Fig. 2F). Because the vacuolar amounts of Vtc4-HA were not affected by VTC5 overexpression, this suggests that not all VTC complexes on the vacuoles are saturated with Vtc5 and that Vtc5 association with the complex might regulate VTC activity. This notion is supported by mass spectrometry data, which found Vtc5 to be 20× less abundant than Vtc1–4 (33).
Vtc5 Is Required for Normal Expression Levels of the VTC Complex
We asked whether Vtc5 is required for the stability of the VTC complex. Vacuoles were isolated from a strain deleted for VTC5, and the levels of the other VTC subunits were determined by Western blotting. Deletion of VTC5 decreased the levels of Vtc2, Vtc3, and Vtc4 and of the vacuolar alkaline phosphatase Pho8 by around 30% (Fig. 3, A–C) when compared with the levels of the vacuolar marker proteins Vam3 and Nyv1. No significant reduction of Vtc4 was detected in whole-cell extracts, suggesting that absence of Vtc5 interferes with sorting of the VTC complex to vacuoles. Overexpression of Vtc5 from the GPD1 promoter did not change the levels of other VTC subunits or of Pho8. We also analyzed the levels of VTC5 on vacuoles from vtc2Δ, vtc3Δ, and vtc4Δ cells but observed no significant changes (Fig. 3D). Thus, expression and sorting of Vtc5 to vacuoles do not depend on the other constituents of the VTC complex, but the presence of Vtc5 increases the abundance of the other VTC subunits on vacuoles.
FIGURE 3.
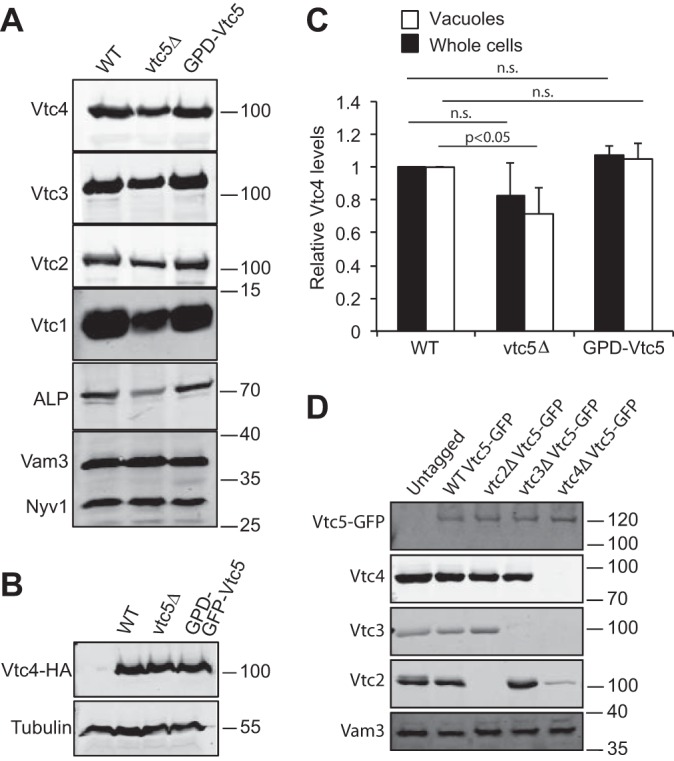
Levels of VTC subunits in vtc mutants. A, influence of Vtc5 on proteins on isolated vacuoles. 30 μg of vacuolar protein from BY4741, vtc5Δ, and BY4741 PGPD1-Vtc5 were analyzed by SDS-PAGE and Western blotting with antibodies against the indicated proteins or the HA tag. B, influence of Vtc5 on proteins in whole-cell extracts. Whole-cell extracts were prepared from cells expressing Vtc4 from its endogenous promoter with an HA tag, grown in YPD to logarithmic phase. Proteins were analyzed as in A. Note that we used Vtc4-HA in this case because Vtc4 is difficult to detect in whole-cell extracts with our antibodies. C, quantification of the Vtc4 levels from A (white bars) and B (black bars) from three independent preparations each. n.s., not significant. D, influence of Vtc2, Vtc3, and Vtc4 on the level of Vtc5. Vacuoles were isolated from the indicated BY4741 strains expressing Vtc5 with or without a GFP tag. Vacuolar proteins were analyzed as in A. Vam3 was used as a loading control. The migration of relevant molecular mass markers (size in kDa) is indicated on the right-hand side of the panels.
Vtc5 Stimulates the Accumulation of PolyP Synthesized by the VTC Complex
To test the role of Vtc5 in polyP accumulation in vivo, yeast cells were grown in SC medium to exponential phase, and polyP was extracted, purified, and quantified (34). vtc5Δ cells showed only 20% of the polyP content of wild-type cells (Fig. 4A). This phenotype was reversed by introducing a centromeric plasmid encoding VTC5 under control of the GPD1 promoter. Overexpressing VTC5 or GFP-VTC5 from the GPD1 promoter increased the polyP content of wild-type cells 3.5-fold. This increase in polyP content depended on the catalytic activity of Vtc4 because it was not observed upon overexpressing VTC5 in a vtc4R264A mutant, which leaves the VTC complex physically intact and correctly sorted but eliminates the catalytic activity of Vtc4 (Fig. 4B) (19). We determined the length of the polyP chains produced by the vtc5 mutants by PAGE and negative staining with DAPI (Fig. 4C). Yeast cells synthesize polyP chains of up to several hundred residues. Chains that differ by a single unit can be resolved up to a chain length of around 40 units, forming a ladder of individual bands. Longer chains run as a smear. Synthetic polyP of an average chain length of 60 units was used as a marker. Deletion or overexpression of Vtc5 had no impact on the length distribution of cellular polyP. This suggests that Vtc5 does not affect the processivity of the VTC complex nor does it seem to influence turnover of these chains by vacuolar endo- or exo-polyphosphatases (35–37).
FIGURE 4.

Polyphosphate accumulation in vtc5 mutants in vivo. A, polyP levels in vtc5 mutants. Wild-type (BY4741) or vtc5Δ cells were transformed with an empty vector (pRS416 PGPD1) or vectors expressing Vtc5 or GFP-Vtc5 from the strong GPD1 promoter. Cells were grown in SC-URA medium to logarithmic phase. PolyP was extracted, quantified using the Malachite Green assay, and plotted relative to the wild-type signal; n = 4. **, p < 0.01; ***, p < 0.001. B, Vtc5 does not have polyP polymerase activity. GFP-VTC5 was expressed under the control of the strong GPD1 promoter in BY4742 vtc4Δ cells producing either a wild-type version of Vtc4 (Vtc4WT) or a catalytically inactive mutant (vtc4R264A). Cells were grown in SC medium, and polyP was quantified as in A; n = 3. ***, p < 0.001. C, polyP chain length distribution. PolyP was extracted from the indicated BY4741 strains, which had been grown logarithmically in YPD. PolyP was fractionated on a 25% polyacrylamide gel and visualized by negative staining with DAPI. Equal amounts of extracted polyP were loaded for each strain. An additional sample consisting of a three times higher amount of polyP was loaded for the vtc5Δ strain (vtc5Δ 3x). Synthetic polyP of an average chain length of 60 phosphate units was loaded as a length marker.
PolyP synthesis can be assayed in vitro using isolated organelles (21). The system recapitulates the stimulation of polyP accumulation by inositol pyrophosphates, which is observed in vivo (31, 38). The inositol pyrophosphate 5-IP7 is a potent agonist and accelerates polyP accumulation by WT vacuoles over 50-fold (8). We used this in vitro system to test the influence of Vtc5 on polyP synthesis. Neither deletion nor overexpression of Vtc5 abolished the stimulation of polyP synthesis by 5-IP7 (Fig. 5, A and C). However, Vtc5 levels influenced the rates of polyP accumulation. vtc5Δ vacuoles showed 50% reduced activity in the presence of 5-IP7. In the absence of 5-IP7, wild-type vacuoles showed only very low activity (5 pmol/μg/min), which dropped to undetectable levels in vtc5Δ (<1 pmol/μg/min) (Fig. 5, B and C). Note that the activities shown have been normalized to the levels of Vtc4, which means that the observed effects cannot be explained by the reduced abundance of this catalytic subunit on vtc5Δ vacuoles. Overexpression of Vtc5 increased the rate of polyP accumulation in the absence of 5-IP7 over 10-fold, whereas the 5-IP7-stimulated activity increased only 1.2-fold. The effect of Vtc5 on polyP synthesis did not result from a change in the stability of the complex. Isolated vacuoles from strains deleted for VTC5 or overexpressing Vtc5 were incubated under the conditions used for the in vitro polyP assay, and proteins were TCA-precipitated at different time points before SDS-PAGE analysis. The levels of Vtc4 and Vtc3 were then quantified by Western blotting (Fig. 5D). Degradation of the two subunits could not be detected in either of the strains tested. These results suggest that Vtc5 can augment the catalytic activity of the VTC complex by a mechanism independent of 5-IP7.
FIGURE 5.
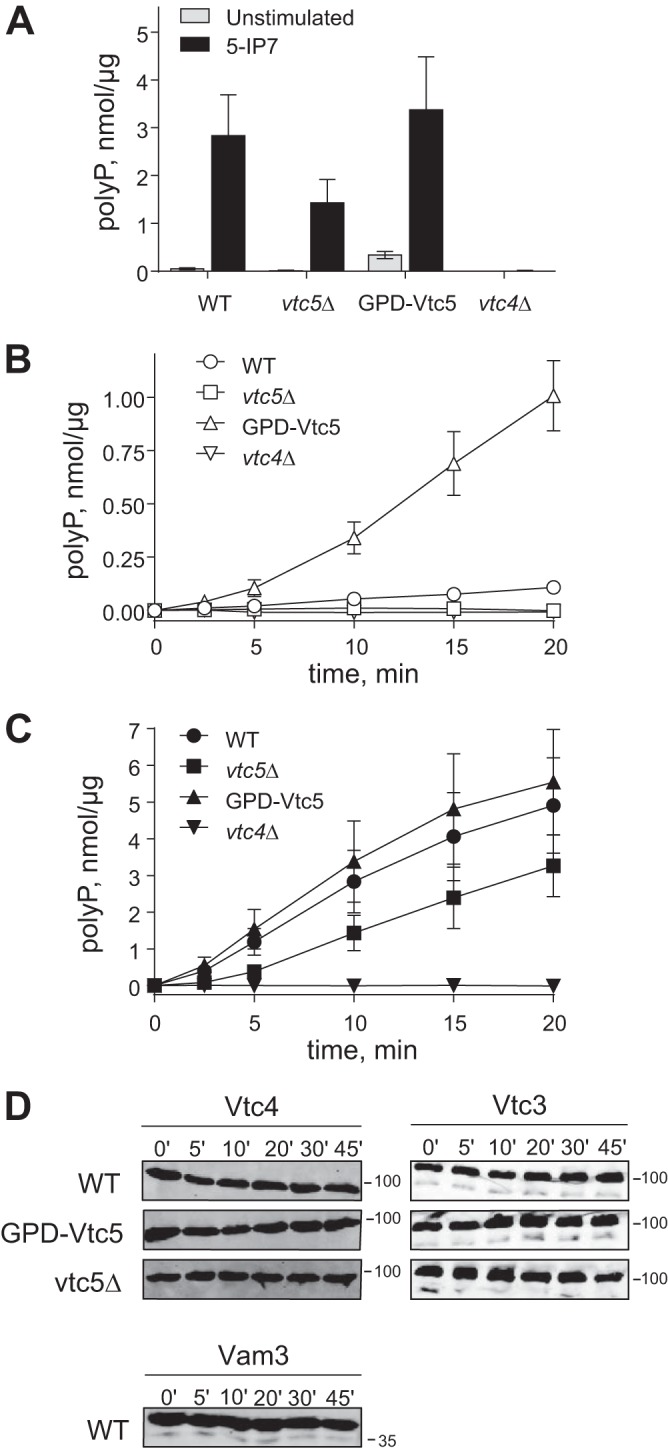
Polyphosphate synthesis of vtc5 mutants in vitro. The rate of polyP synthesis in vitro depends on Vtc5. Vacuoles were isolated from wild-type BY4741 and the indicated isogenic mutants. Isolated vacuoles were incubated with an ATP-regenerating system, and polyP thus produced was quantified using a DAPI-based assay. A, amount of polyP produced after 10 min of reaction by 0.005 μg/μl of vacuoles in the presence (5-IP7) or absence (unstimulated) of 0.5 μm 5-IP7 was plotted. B and C, time courses of polyP synthesis, measured as in A in the absence (B) or presence (C) of 0.5 μm 5-IP7. The obtained data were normalized to Vtc4 levels determined as in Fig. 3; n = 3. D, levels of full-length Vtc4 and Vtc3 were determined during the time course of the polyP synthesis reaction by Western blotting. Isolated vacuoles from BY4741 and the vtc5 mutants were incubated as in A in the absence of 5-IP7. At the indicated time points, aliquots were withdrawn, and proteins were TCA-precipitated before SDS-PAGE analysis. In addition to Vtc3 and Vtc4, stability of one of the most protease-sensitive proteins, Vam3, is shown. The migration of relevant molecular mass markers (size in kDa) is indicated on the right-hand side of the panels.
Vtc5-mediated Stimulation of PolyP Synthesis Activates the PHO Pathway
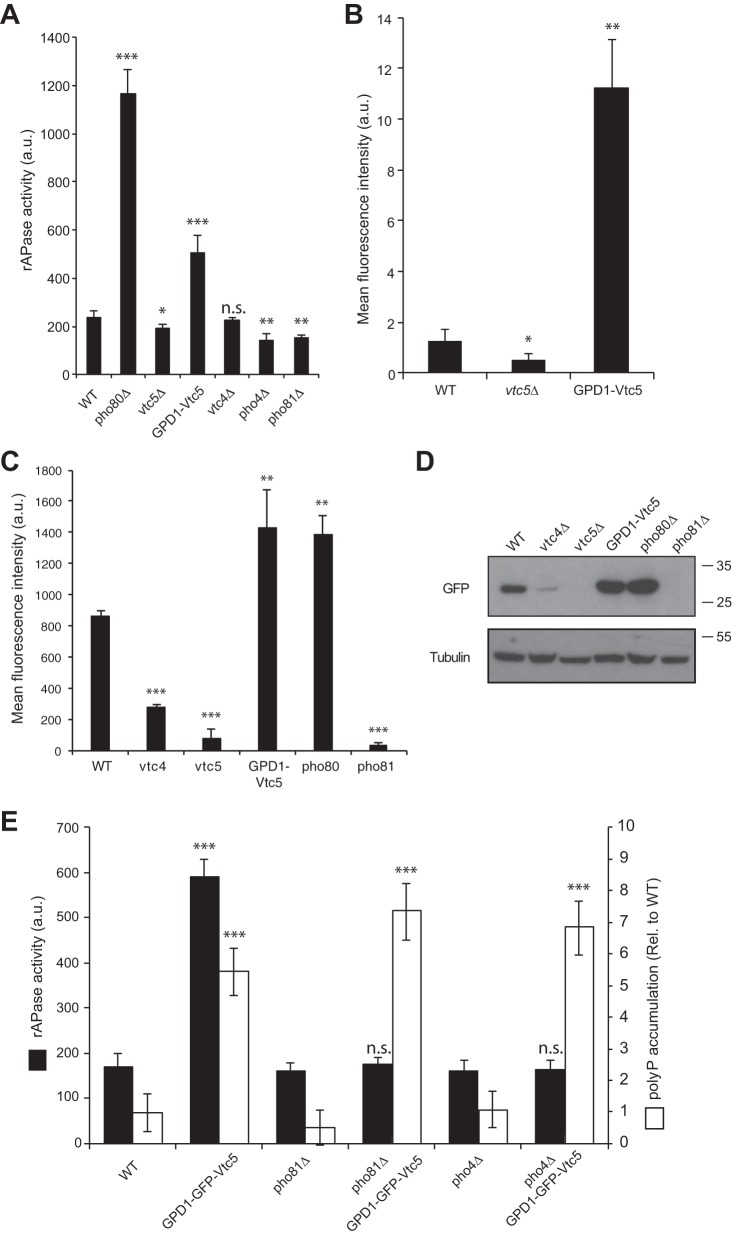
Because overexpression of VTC5 allowed us to uncouple VTC activity from its inositol pyrophosphate-mediated control by intracellular Pi levels, we exploited this feature to study the impact of polyP synthesis on cytosolic Pi homeostasis. The transcriptional program of the PHO pathway is induced by a decline in intracellular Pi levels (39). This induction can be read out via the production of Pho5, which is the major PHO pathway-controlled acid phosphatase that is secreted into the medium (40, 41). Deletion of VTC5 led to a low but reproducible reduction of secreted phosphatase activity by 20% (Fig. 6A). By contrast, overexpression of VTC5 increased it 2-fold, indicating activation of the PHO pathway. As controls, we used a pho80Δ strain, in which the PHO pathway is constantly active, and the pho4Δ and pho81Δ strains, in which the PHO pathway is constitutively inactive. We also measured the expression level of Pho84, a high affinity Pi importer that is strongly induced under low Pi conditions (1, 42, 43). vtc5Δ mutants carrying Pho84-GFP were grown to exponential phase in YPD, i.e. under Pi-rich conditions, and Pho84-GFP fluorescence intensity was measured by fluorescence-activated cell sorting (FACS). Deletion of VTC5 reduced Pho84-GFP by 70%, whereas its overexpression increased fluorescence intensity 10-fold (Fig. 6B). To verify that these observations resulted from increased transcription of the PHO genes and not from defective degradation of Pho84, we measured activation of the Pho84 promoter via a GFP reporter (PHO84pr-GFP) (1), using FACS (Fig. 6C) and Western blotting (Fig. 6D). In wild-type cells grown in Pi-rich medium, GFP expression was low. Deletion of VTC4 or VTC5 reduced expression of the reporter to 30 and 9% of the wild type, respectively. This is equivalent to the levels observed in a pho81Δ strain, in which the PHO pathway is constitutively repressed. Overexpression of VTC5 increased GFP levels similarly as deletion of PHO80, which renders the PHO pathway constitutively active. Thus, up- or down-regulation of Vtc5 levels activates or inactivates the PHO pathway, respectively.
FIGURE 6.
Vtc5 influences the expression of the PHO genes. A, Pho5 activity was measured in the indicated mutants. Cells were grown in YPD to logarithmic phase, and acid phosphatase secreted into the medium was measured using a colorimetric assay; n = 4. B, Pho84 expression levels were measured by FACS, using a strain expressing Pho84-GFP. The mean fluorescence intensity of the distribution is given, n = 3. C, mean fluorescence intensity of cells expressing GFP under the control of the PHO84 promoter was quantified by FACS, n = 3. ***, p < 0.0001; **, p < 0.002. D, GFP levels in extracts of cells used in C were quantified by Western blotting, n = 3. Tubulin was used as a loading control. E, Pho5 activity (black bars) and polyP levels (white bars) were measured in a wild-type strain (BY4741) and in the indicated deletion or overexpression strains; n = 3. Statistical differences between the indicated strain and the same strain overexpressing Vtc5 (GPD1-GFP-Vtc5) are shown. ***, p < 0.0001; **, p < 0.005; *, p < 0.05, n.s. not significant.
Given that the PHO genes, including the Pi importer Pho84, are up-regulated in cells overexpressing Vtc5, the increased polyP levels in this mutant might result from higher abundance of PHO proteins and higher import of Pi into the cytosol. We tested this hypothesis in pho81Δ and pho4Δ cells, which cannot up-regulate the PHO genes. Overexpression of Vtc5 in these mutants did not induce Pho5, but it did induce polyP overaccumulation (Fig. 6E). This suggests that Vtc5 regulates polyP synthesis independently of the PHO pathway.
Vtc5-induced Activation of the PHO Pathway Depends on PolyP Accumulation
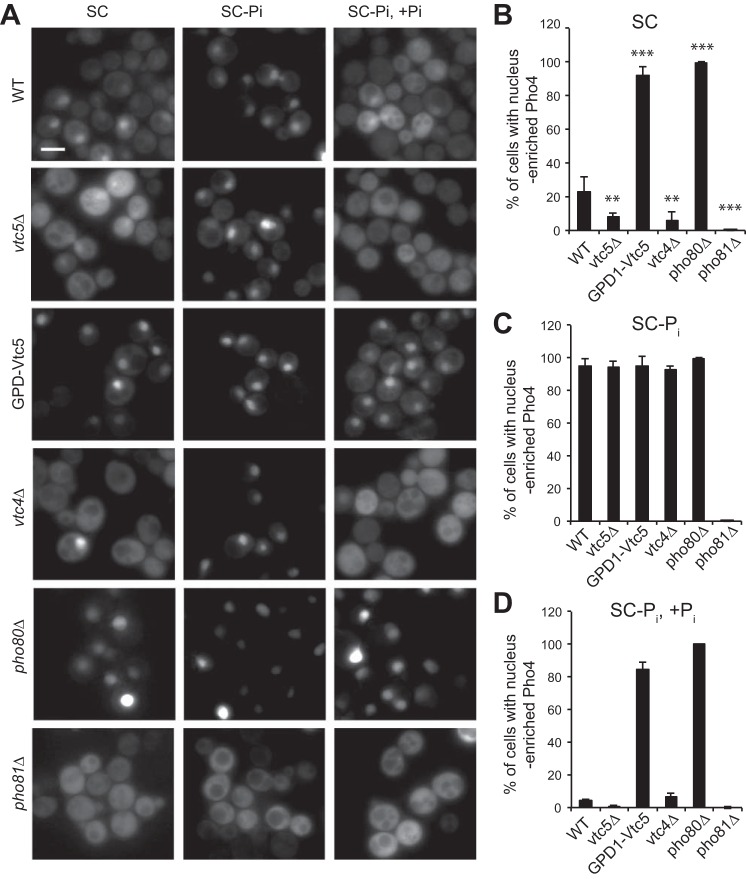
A GFP-tagged version of the Pi-sensitive transcription factor Pho4 (Pho4-GFP) shifts to the cytosol in Pi-rich conditions and accumulates in the nucleus under Pi limitation, providing a rapidly responding bona fide readout for cytosolic Pi concentration (1). In line with this, pho81Δ cells, in which the phosphate starvation program is constitutively inactive, showed a strictly cytosolic staining in Pi-rich as well as Pi-limiting SC medium (Fig. 7). Conversely, pho80Δ cells, in which the phosphate starvation program is constitutively active, carried Pho4-GFP in the nucleus even on Pi-rich media. Wild-type cells showed an increase in nuclear Pho4-GFP localization from 20 to 100% when shifted from Pi-rich to Pi-limiting medium for 15 min, indicating that cytosolic Pi concentration had dropped, and the starvation response had been initiated. vtc5Δ and vtc4Δ cells behaved similarly but showed nuclear Pho4-GFP in less than 10% of the cells on Pi-rich medium. By contrast, among cells overexpressing Vtc5, more than 90% showed nuclear Pho4-GFP under Pi-rich as well as under Pi-limited conditions. This nuclear accumulation could not be reverted by addition of 10 mm Pi, although the same treatment led to rapid redistribution of Pho4-GFP to the cytosol in wild-type, vtc4Δ, and vtc5Δ cells. Thus, similar to a pho80Δ mutation, VTC5-overexpression constitutively activates the Pi starvation response, suggesting that it leads to a constitutive depletion of cytosolic Pi.
FIGURE 7.
Vtc5 expression changes the localization of the transcription factor Pho4. A, indicated mutants had been transformed with a centromeric plasmid encoding Pho4 under the control of its own promoter. The localization of Pho4-GFP was assessed in living cells cultured in SC medium (SC), after 15 min of Pi starvation (SC-Pi), and 15 min after re-addition of 10 mm Pi to Pi-starved cells (SC-Pi, + Pi). Scale bar, 5 μm. B–D, percentage of cells showing enrichment in nuclear Pho4 was determined for these conditions. At least 200 cells were counted per condition. n = 3. ***, p < 0.0001; **, p < 0.05.
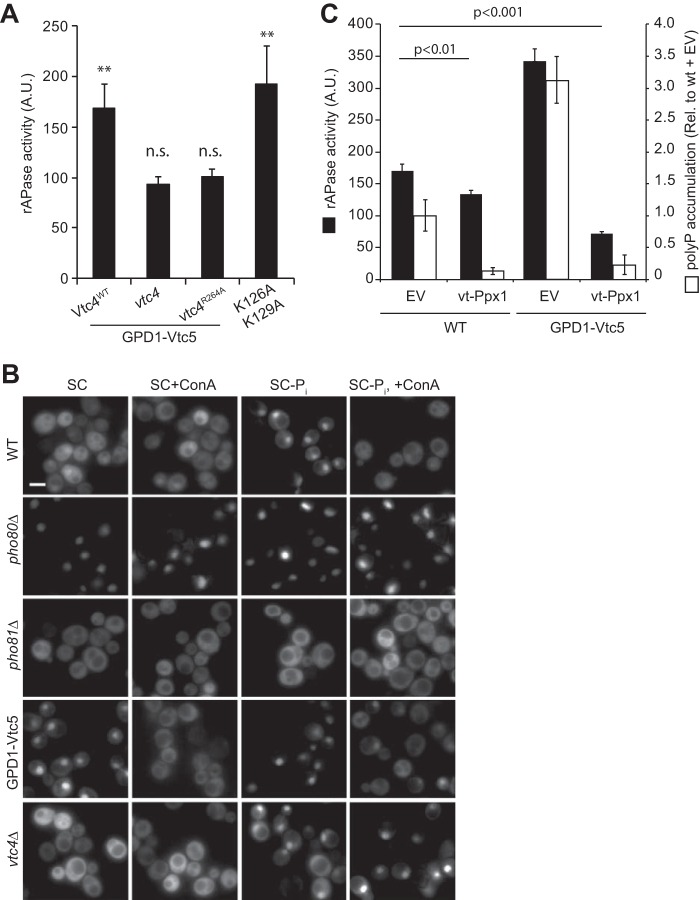
Next, we asked whether this change in Pho4-GFP localization could be ascribed to altered polyP synthesis or whether it might be due to other effects of Vtc5, such as a direct interaction with cytosolic Pi signaling components. To this end, we manipulated polyP synthesis pharmacologically and also through mutations that leave the VTC complex intact but increase or decrease its catalytic activity (8, 19). VTC5 was overexpressed in vtc4Δ strains that were reconstituted with plasmids expressing the wild-type allele Vtc4WT or a catalytically inactive vtc4R264A allele. In addition, we used a hyperactive VTC variant that carries a point mutation in the SPX domains of Vtc3 and Vtc4 (vtc3K126A/vtc4K129A) (8). As observed before, VTC5 overexpression increased Pho5 activity (Fig. 8A). This effect was not observed upon VTC5 overexpression in vtc4Δ or vtc4R264A cells. Conversely, if polyP synthesis was raised independently of Vtc5 by using the vtc3 K126A/vtc4K129A alleles, Pho5 activity rose to a similar degree as in VTC5-overexpressing wild-type cells.
FIGURE 8.
Regulation of the PHO pathway depends on polyP synthesis. A, Pho5 activity was measured in the indicated vtc4 mutants overexpressing GFP-Vtc5 from the GPD1 promoter and from a vtc3K126A/vtc4K129A double mutant that shows constitutively active polyP synthesis. Samples were prepared as in Fig. 6A. n = 3. Values are relative to the wild-type BY4741. Statistical differences between the indicated mutant and the wild type are as follows: **, p < 0.005, n.s. not significant. B, Pho4 localization was determined in the indicated mutants grown in SC medium, after 15 min in 2 μm ConA, after 15 min in Pi starvation medium (SC-Pi), and 15 min after addition of 2 μm of ConA to Pi-starved cells (SC-Pi, +ConA). Scale bar, 5 μm. Pictures are representative of at least 200 cells observed in three independent experiments. C, Pho5 activity (black bars) and polyP levels (white bars) were measured in strains expressing vt-Ppx1 and carrying an empty vector or a GPD1-based overexpression plasmid for VTC5; n = 3.
The rapidly responding Pho4-GFP allowed us to test the effect of an acute pharmacological shutdown of polyP synthesis. Because polyP synthesis and translocation are driven by the V-ATPase-dependent electrochemical gradient across the vacuolar membrane (21), disruption of this gradient immediately arrests polyP synthesis. Within less than 15 min, addition of the V-ATPase pump inhibitor concanamycin A (ConA) to wild-type cells growing on SC-Pi led to a re-localization of Pho4-GFP from the nucleus to the cytosol, suggesting that a sudden arrest in polyP synthesis rapidly increases cytosolic Pi concentration. Pho4-GFP did not re-localize in Pi-limited vtc4Δ cells. Furthermore, ConA did not change the predominantly cytosolic localization of Pho4-GFP in wild-type cells growing in Pi-rich SC (Fig. 8B), and it had no effect on the localization of Pho4-GFP in pho80Δ and pho81Δ mutants, indicating that the drug itself does not promote changes in Pho4-GFP localization. However, ConA abolished the nuclear accumulation of Pho4-GFP in VTC5-overexpressing cells.
To better understand the link between polyP accumulation and activation of the PHO pathway, we targeted an exopolyphosphatase (vt-Ppx1) to the vacuole lumen of a strain overexpressing VTC5. This is expected to result in the degradation of polyP (21) and export of released Pi to the cytosol. Expression of vt-Ppx1 in a wild-type strain reduced polyP content by 80% but decreased rAPase activity by less than 20% (Fig. 8C). In VTC5 overexpressing cells, vt-PPX1 reduced polyP by >90%. In these cells, rAPase activity was also reduced by >75%, bringing the absolute rAPase activity to a level only half as high as in wild-type cells. This suggests that polyP production, when followed by uncontrolled degradation in the vacuole, increases cytosolic Pi above the normal level and suppresses the PHO pathway.
Discussion
Although the response to phosphate starvation has been well characterized in yeast, the mechanisms of sensing Pi and managing Pi stores remain poorly understood. PolyP is the main Pi reservoir in yeast. The VTC complex, which is mainly localized on the vacuole membrane (15–17, 44), synthesizes polyP from ATP and translocates the nascent polyP chain into the lumen (19, 21). Here, we show that the ORF YDR089W encodes a previously unknown subunit of the VTC complex, Vtc5. Vtc5 is not essential for catalytic activity of VTC because a vtc5Δ strain retains ∼20% of polyP, and vacuoles isolated from this strain still accumulate polyP, although at a lower rate. Conversely, overexpression of VTC5 increases polyP accumulation in vivo but is not sufficient for it. VTC complexes containing Vtc3 are mainly located on vacuoles, but Vtc2-containing complexes are abundant in the cell periphery, probably in the peripheral ER (20). Even when overexpressed, we found Vtc5 only in the vacuole membrane, and its sorting to the vacuole was independent of the presence of other VTC subunits. This suggests that Vtc5 stimulates polyP synthesis specifically on vacuoles, which is required to accumulate the major polyP stores of the cell. In vitro, VTC5 overexpression accelerates polyP accumulation drastically and partially substitutes for the stimulation of VTC by 5-IP7. The abundance of Vtc5 is low compared with the other subunits (40). In line with this, we found that the amount of Vtc5 interacting with the other subunits increases with the increased expression of VTC5 on the vacuole membrane. This suggests that VTC complexes are not saturated with Vtc5 and that Vtc5 may regulate their activity by interacting with them transiently.
This invites the consideration that inositol pyrophosphates might regulate polyP synthesis in the context of cytosolic Pi homeostasis, whereas Vtc5 might regulate polyP synthesis with respect to other cellular parameters. PolyP synthesis is activated in correlation with Pi availability by the binding of inositol pyrophosphates to SPX domains of the VTC complex (8). BecauseVtc2, Vtc3, Vtc4, and Vtc5 carry SPX domains, we tested whether their interactions might be affected by 5-IP7 (data not shown). This was not the case, although polyP synthesis on the isolated vacuoles used for this assay strongly responded to overexpression of VTC5 and to 5-IP7. Neither overexpression nor deletion of VTC5 abolished the stimulation of VTC by 5-IP7. Therefore, we presume that association with Vtc5 might represent an independent mechanism for regulating polyP synthesis. We speculate that this might constitute a means to integrate multiple regulatory inputs on the VTC complex. This is relevant because polyP synthesis is not the only determinant of cytosolic Pi homeostasis, where the SPX domains and inositol pyrophosphates play a role. In amino acid and metal homeostasis, polyP also constitutes an important polymeric counter-ion to basic amino acids and divalent metal ions (45, 46). Basic amino acids can be stored in vacuoles and acidocalcisomes in concentrations of several hundred millimolars. This accumulation is only possible if sufficient polyP is synthesized, probably because polyP is needed to adsorb and osmotically inactivate the amino acids, which otherwise could build up strong osmotic pressure inside the organelle (47). Regulated association of Vtc5 with the other VTC subunits might modify polyP synthesis in response to such requirements that are independent of Pi levels. In line with this, VTC5 has not been found among genes undergoing strong transcriptional activation upon phosphate starvation, whereas the other four VTC genes are all strongly induced (15). We confirmed the lack of induction of Vtc5 by quantifying the levels of Vtc5-GFP expressed from the native VTC5 promoter and found no change upon transferring the cells to low-Pi medium (data not shown). Furthermore, Vtc2, Vtc3, and Vtc4 share a similar central domain, whereas the central domain of Vtc5 is of similar size, and yet its primary sequence and the secondary structure prediction reveal no conservation between this domain and the central domains of Vtc2, Vtc3, and Vtc4.
PolyP also serves as a Pi source to facilitate rapid DNA synthesis in S-phase, which transiently drains more Pi than the cells can take up via their transporters (48). During S-phase, cytosolic Pi concentrations are held constant, although the polyP reserves are consumed, which suggests that net polyP synthesis should be down-regulated. This might implicate Vtc5 because the hydrophilic domain that is found between its SPX and transmembrane domains is extensively phosphorylated and, for at least one of these phospho-sites, the cell cycle regulator Cdc28 has been identified as the kinase (49). Furthermore, both polyP synthesis and the cell cycle-regulating proteins Cdc28 and Cdc42 influence vacuolar structure through the vacuolar fusion machinery (50–52). This, together with the cyclic consumption of polyP (48), can explain the transient decay of vacuoles into multiple smaller fragments during S-phase (53), because vacuole structure is determined by an equilibrium of fusion and fission activities (54–56). Our working hypothesis, which we will explore in future studies, is that Cdc28 influences vacuolar morphology by regulating VTC and the polyP store produced by it.
Several studies have implicated enzymes of polyP metabolism in the onset and stabilization of the yeast phosphate starvation program, the PHO pathway (1, 7, 57). Consistent with this, we found that de-regulation of VTC5 expression alters activation of the PHO pathway. vtc5Δ cells show the transcription factor Pho4 in the cytosol, like cells on Pi-rich medium, whereas cells overexpressing VTC5 show Pho4 in the nucleus and hence mimic cells starving for Pi, even in Pi-rich conditions. The fact that this effect depends on PHO81 provides a genetic argument suggesting that VTC5 acts upstream of PHO81. Straightforward hypotheses to be derived from this observation would be that either cytosolic Pi is strongly affected in vtc5 mutants or that the vacuolar polyP that is produced from it serves as an indirect readout of cytosolic Pi. A signal about vacuolar polyP levels would then have to be communicated to the PHO pathway. An argument against the latter possibility is that a vtc4Δ strain, which does not make any polyP, does not show Pho4-GFP constitutively in the nucleus and hence does not induce the PHO pathway. Also the vacuole-targeted exopolyphosphatase vt-Ppx1, which degrades most of the vacuolar polyP store, does not induce the PHO pathway. To the contrary, vt-Ppx1 even represses it when used in a strain overexpressing VTC5. This suggests that the increased production of polyP in VTC5-overexpressing cells, in conjunction with non-controlled degradation of this pool by vt-Ppx1, increases cytosolic Pi as compared with a wild-type strain. This argues for the notion that cytosolic Pi determines the regulation of the PHO pathway.
Several mechanisms to detect changes in Pi have been identified in yeast. The high affinity Pi transporter Pho84 was proposed to be a Pi transceptor meaning that it would be able to sense ambient Pi and transport it to the cytosol. Pho84 can indeed transduce information about extracellular Pi availability to the protein kinase A pathway (58). Subsequent studies attributed regulation of the PHO pathway to the levels of cytosolic Pi, independently of Pho84, and postulated the existence of an unidentified cytosolic Pi receptor (39, 59). How cells measure their cytosolic Pi concentration and ensure its homeostasis remains poorly understood. This work reinforces the idea that cytosolic Pi is sensed by the cells. We believe that our finding that Vtc5 may modify the PHO pathway by altering the intracellular Pi concentration provides a good tool to tackle this problem.
Author Contributions
Y. D. and R. G. designed and performed the experiments. H. J. J. synthesized 5-IP7. Y. D., R. G., and A. M. analyzed the data and wrote the manuscript.
Acknowledgments
We thank Toshikazu Shiba (Regenetiss Inc., Japan) for synthetic polyP-60, Erin O'Shea for the kind gift of the EY1109 yeast strain, and Adolfo Saiardi and Cristina Azevedo for critical reading of the manuscript.
This work was supported by European Research Council Grant 233458 and Schweizerischer Nationalfonds zur Förderung der Wissenschaften Grants 144258 and 163477/1 (to A. M.) and PP00P2-157607 (to H. J. J.). The authors declare that they have no conflicts of interest with the contents of this article.
- polyP
- polyphosphate
- VTC
- vacuole transporter chaperone
- ER
- endoplasmic reticulum
- FM4-64
- N-(3-triethylammoniumpropyl)- 4-(p-diethylaminophenylhexatrienyl)-pyridinium dibromide
- ConA
- concanamycin A
- 5-IP7
- 5-diphospho-myo-inositol pentakisphosphate
- 1-IP7
- 1-diphospho-myo-inositol pentakisphosphate.
References
- 1. Thomas M. R., and O'Shea E. K. (2005) An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 9565–9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ljungdahl P. O., and Daignan-Fornier B. (2012) Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190, 885–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu C., Yang Z., Yang J., Xia Z., and Ao S. (2000) Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J. Biol. Chem. 275, 31972–31978 [DOI] [PubMed] [Google Scholar]
- 4. Kaffman A., Herskowitz I., Tjian R., and O'Shea E. K. (1994) Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263, 1153–1156 [DOI] [PubMed] [Google Scholar]
- 5. O'Neill E. M., Kaffman A., Jolly E. R., and O'Shea E. K. (1996) Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271, 209–212 [DOI] [PubMed] [Google Scholar]
- 6. Lee Y. S., Mulugu S., York J. D., and O'Shea E. K. (2007) Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vardi N., Levy S., Gurvich Y., Polacheck T., Carmi M., Jaitin D., Amit I., and Barkai N. (2014) Sequential feedback induction stabilizes the phosphate starvation response in budding yeast. Cell Rep. 9, 1122–1134 [DOI] [PubMed] [Google Scholar]
- 8. Wild R., Gerasimaite R., Jung J.-Y., Truffault V., Pavlovic I., Schmidt A., Saiardi A., Jessen H. J., Poirier Y., Hothorn M., and Mayer A. (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990 [DOI] [PubMed] [Google Scholar]
- 9. Secco D., Wang C., Shou H., and Whelan J. (2012) Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586, 289–295 [DOI] [PubMed] [Google Scholar]
- 10. Bun-ya M., Shikata K., Nakade S., Yompakdee C., Harashima S., and Oshima Y. (1996) Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr. Genet. 29, 344–351 [PubMed] [Google Scholar]
- 11. Wykoff D. D., and O'Shea E. K. (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hürlimann H. C., Stadler-Waibel M., Werner T. P., and Freimoser F. M. (2007) Pho91 is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4438–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schneider K. R., Smith R. L., and O'Shea E. K. (1994) Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266, 122–126 [DOI] [PubMed] [Google Scholar]
- 14. Fisher E., Almaguer C., Holic R., Griac P., and Patton-Vogt J. (2005) Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280, 36110–36117 [DOI] [PubMed] [Google Scholar]
- 15. Ogawa N., DeRisi J., and Brown P. O. (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11, 4309–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen A., Perzov N., Nelson H., and Nelson N. (1999) A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 274, 26885–26893 [DOI] [PubMed] [Google Scholar]
- 17. Müller O., Neumann H., Bayer M. J., and Mayer A. (2003) Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 116, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 18. Müller O., Bayer M. J., Peters C., Andersen J. S., Mann M., and Mayer A. (2002) The Vtc proteins in vacuole fusion: coupling NSF activity to V(0) trans-complex formation. EMBO J. 21, 259–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hothorn M., Neumann H., Lenherr E. D., Wehner M., Rybin V., Hassa P. O., Uttenweiler A., Reinhardt M., Schmidt A., Seiler J., Ladurner A. G., Herrmann C., Scheffzek K., and Mayer A. (2009) Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516 [DOI] [PubMed] [Google Scholar]
- 20. Uttenweiler A., Schwarz H., Neumann H., and Mayer A. (2007) The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell 18, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerasimaite R., Sharma S., Desfougères Y., Schmidt A., and Mayer A. (2014) Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 127, 5093–5104 [DOI] [PubMed] [Google Scholar]
- 22. Shirahama K., Yazaki Y., Sakano K., Wada Y., and Ohsumi Y. (1996) Vacuolar function in the phosphate homeostasis of the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 37, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 23. Spain B. H., Koo D., Ramakrishnan M., Dzudzor B., and Colicelli J. (1995) Truncated forms of a novel yeast protein suppress the lethality of a G protein α subunit deficiency by interacting with the β subunit. J. Biol. Chem. 270, 25435–25444 [DOI] [PubMed] [Google Scholar]
- 24. Vaughan A. E., Mendoza R., Aranda R., Battini J.-L., and Miller A. D. (2012) Xpr1 is an atypical G-protein-coupled receptor that mediates xenotropic and polytropic murine retrovirus neurotoxicity. J. Virol. 86, 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giovannini D., Touhami J., Charnet P., Sitbon M., and Battini J.-L. (2013) Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep. 3, 1866–1873 [DOI] [PubMed] [Google Scholar]
- 26. Pieren M., Schmidt A., and Mayer A. (2010) The SM protein Vps33 and the t-SNARE H(abc) domain promote fusion pore opening. Nat. Struct. Mol. Biol. 17, 710–717 [DOI] [PubMed] [Google Scholar]
- 27. Reese C., Heise F., and Mayer A. (2005) Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature 436, 410–414 [DOI] [PubMed] [Google Scholar]
- 28. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 29. Sheff M. A., and Thorn K. S. (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 [DOI] [PubMed] [Google Scholar]
- 30. Longtine M. S., McKenzie A. 3rd., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 31. Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., and Saiardi A. (2011) Identification of an evolutionary conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith S. A., and Morrissey J. H. (2007) Sensitive fluorescence detection of polyphosphate in polyacrylamide gels using 4′,6-diamidino-2-phenylindol. Electrophoresis 28, 3461–3465 [DOI] [PubMed] [Google Scholar]
- 33. Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 34. Werner T. P., Amrhein N., and Freimoser F. M. (2005) Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch. Microbiol. 184, 129–136 [DOI] [PubMed] [Google Scholar]
- 35. Wurst H., and Kornberg A. (1994) A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 269, 10996–11001 [PubMed] [Google Scholar]
- 36. Kumble K. D., and Kornberg A. (1996) Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 271, 27146–27151 [DOI] [PubMed] [Google Scholar]
- 37. Sethuraman A., Rao N. N., and Kornberg A. (2001) The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 8542–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Auesukaree C., Tochio H., Shirakawa M., Kaneko Y., and Harashima S. (2005) Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280, 25127–25133 [DOI] [PubMed] [Google Scholar]
- 39. Auesukaree C., Homma T., Tochio H., Shirakawa M., Kaneko Y., and Harashima S. (2004) Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 279, 17289–17294 [DOI] [PubMed] [Google Scholar]
- 40. To-E A., Ueda Y., Kakimoto S. I., and Oshima Y. (1973) Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J. Bacteriol. 113, 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ueda Y., To-E A., and Oshima Y. (1975) Isolation and characterization of recessive, constitutive mutations for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J. Bacteriol. 122, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Springer M., Wykoff D. D., Miller N., and O'Shea E. K. (2003) Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1, E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lau W. T., Howson R. W., Malkus P., Schekman R., and O'Shea E. K. (2000) Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc. Natl. Acad. Sci. U.S.A. 97, 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kornberg A., Rao N. N., and Ault-Riché D. (1999) Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125 [DOI] [PubMed] [Google Scholar]
- 45. Li Z.-H., Alvarez V. E., De Gaudenzi J. G., Sant'Anna C., Frasch A. C., Cazzulo J. J., and Docampo R. (2011) Hyperosmotic stress induces aquaporin-dependent cell shrinkage, polyphosphate synthesis, amino acid accumulation, and global gene expression changes in Trypanosoma cruzi. J. Biol. Chem. 286, 43959–43971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fang J., Ruiz F. A., Docampo M., Luo S., Rodrigues J. C., Motta L. S., Rohloff P., and Docampo R. (2007) Overexpression of a Zn2+-sensitive soluble exopolyphosphatase from Trypanosoma cruzi depletes polyphosphate and affects osmoregulation. J. Biol. Chem. 282, 32501–32510 [DOI] [PubMed] [Google Scholar]
- 47. Dürr M., Urech K., Boller T., Wiemken A., Schwencke J., and Nagy M. (1979) Sequestration of arginine by polyphosphate in vacuoles of yeast (Saccharomyces cerevisiae). Arch. Microbiol. 121, 169–175 [DOI] [PubMed] [Google Scholar]
- 48. Bru S., Martínez-Laínez J. M., Hernández-Ortega S., Quandt E., Torres-Torronteras J., Martí R., Canadell D., Ariño J., Sharma S., Jiménez J., and Clotet J. (2016) Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol. Microbiol. 101, 367–380 [DOI] [PubMed] [Google Scholar]
- 49. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Müller O., Johnson D. I., and Mayer A. (2001) Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 20, 5657–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han B.-K., Bogomolnaya L. M., Totten J. M., Blank H. M., Dangott L. J., and Polymenis M. (2005) Bem1p, a scaffold signaling protein, mediates cyclin-dependent control of vacuolar homeostasis in Saccharomyces cerevisiae. Genes Dev. 19, 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Desfougères Y., Neumann H., and Mayer A. (2016) Organelle size control: Accumulating vacuole content activates SNAREs to augment organelle volume by homotypic fusion. J. Cell Sci. 129, 2817–2828 [DOI] [PubMed] [Google Scholar]
- 53. Wiemken A., Matile P., and Moor H. (1970) Vacuolar dynamics in synchronously budding yeast. Arch. Mikrobiol. 70, 89–103 [DOI] [PubMed] [Google Scholar]
- 54. Peters C., Baars T. L., Bühler S., and Mayer A. (2004) Mutual control of membrane fission and fusion proteins. Cell 119, 667–678 [DOI] [PubMed] [Google Scholar]
- 55. Baars T. L., Petri S., Peters C., and Mayer A. (2007) Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol. Biol. Cell 18, 3873–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Michaillat L., Baars T. L., and Mayer A. (2012) Cell-free reconstitution of vacuole membrane fragmentation reveals regulation of vacuole size and number by TORC1. Mol. Biol. Cell 23, 881–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vardi N., Levy S., Assaf M., Carmi M., and Barkai N. (2013) Budding yeast escape commitment to the phosphate starvation program using gene expression noise. Curr. Biol. 23, 2051–2057 [DOI] [PubMed] [Google Scholar]
- 58. Giots F., Donaton M. C., and Thevelein J. M. (2003) Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181 [DOI] [PubMed] [Google Scholar]
- 59. Pinson B., Merle M., Franconi J.-M., and Daignan-Fornier B. (2004) Low affinity orthophosphate carriers regulate PHO gene expression independently of internal orthophosphate concentration in Saccharomyces cerevisiae. J. Biol. Chem. 279, 35273–35280 [DOI] [PubMed] [Google Scholar]
- 60. Jones E. W., Zubenko G. S., and Parker R. R. (1982) PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 102, 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haas A., Conradt B., and Wickner W. (1994) G-protein ligands inhibit in vitro reactions of vacuole inheritance. J. Cell Biol. 126, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]