Abstract
Accumulation of amyloid-β peptide (Aβ) in the brain is regarded as central to Alzheimer's disease (AD) pathogenesis. Aβ is generated by a sequential cleavage of amyloid precursor protein (APP) by β-secretase 1 (BACE-1) followed by γ-secretase. BACE-1 cleavage of APP is the committed step in Aβ synthesis. Understanding the mechanism by which BACE-1 is activated leading to Aβ synthesis in the brain can provide better understanding of AD pathology and help to develop novel therapies. In this study, we found that the levels of Aβ and BACE-1 are significantly reduced in the brains of mice lacking transcription factor early growth response 1 (Egr-1) when compared with the WT. We demonstrate that in COS-7 cells, Egr-1 binds to the BACE-1 promoter and activates BACE-1 transcription. In rat hippocampal primary neurons, overexpression of Egr-1 induces BACE-1 expression, activates BACE-1, promotes amyloidogenic APP processing, and enhances Aβ synthesis. In mouse hippocampal primary neurons, knockdown of BACE-1 almost completely blocks Egr-1-induced amyloidogenic APP processing and Aβ synthesis. Our data indicate that Egr-1 promotes Aβ synthesis via transcriptional activation of BACE-1 and suggest that Egr-1 plays role in activation of BACE-1 and acceleration of Aβ synthesis in AD brain. Egr-1 is a potential therapeutic target for AD.
Keywords: amyloid-β (Aβ), early growth response protein 1 (EGR1), gene regulation, neuroscience, promoter, transcription factor
Introduction
Progressive accumulation of Aβ2 in the brain is strongly implicated in the pathogenesis of Alzheimer's disease (AD). Aβ is derived from amyloid precursor protein (APP) (1). In the amyloidogenic pathway, APP is cleaved by β-secretase (BACE-1) to generate a 99-amino acid membrane-bound protein C99 and soluble APPβ. C99 is further cleaved by γ-secretase to produce the Aβ peptide. APP is also cleaved by α-secretase within the Aβ domain generating an 83-kDa protein C83. Subsequent cleavage of C83 by γ-secretase generates a nontoxic short peptide P3 containing the C-terminal region of Aβ. BACE-1 cleavage of APP is the rate-limiting step in Aβ production. BACE-1 is regarded as the essential enzyme in the Aβ synthesis in the brain (1).
A number of studies have shown that the level of BACE-1 protein is significantly up-regulated in AD brain (2–6). It has been suggested that the up-regulation of BACE-1 initiates and/or accelerates Aβ synthesis and promotes AD pathogenesis (1). Determining the mechanism by which BACE-1 is activated in AD brain can provide better understanding of AD pathology and help develop therapies against AD. However, what activates BACE-1 in AD brain is not known. Interestingly, in rat brain, BACE-1 level is elevated following experimental traumatic brain injury (7), transient cerebral ischemia (8), and following occlusion of the middle cerebral artery (9). Oxidative stress induces BACE-1 expression in mouse brains (10) and vascular smooth and HEK-293 cells (11, 12). Thus, several cellular events triggered by vascular insults elevate BACE-1 levels in the brain. It has been suggested that the biochemical pathway that is activated by vascular injuries may be involved in up-regulating BACE-1 and initiating AD pathology in the brain (13).
Early growth response 1 (Egr-1) is a zinc finger transcription factor and is involved in regulation of a number of cell functions, including cell proliferation, apoptosis, cell growth, and signal transduction. The cellular level of Egr-1 is relatively low, but its biosynthesis is induced by many environmental signals, including growth factors, hormones, and neurotransmitters (14, 15). Once induced, it binds to targets genes and regulates their expression. More importantly, in the CNS, Egr-1 is induced during ischemic stroke, hypoxia, brain injury, and inflammation and plays an important role in the induction and maintenance of various vascular pathologies (14–17). A number of recent studies have found that Egr-1 is elevated in AD brain and AD mouse models (18–23). As discussed above, BACE-1 expression is up-regulated during vascular injuries (13). This observation promoted us to examine whether Egr-1 regulates BACE-1 expression in the brain. Herein, we show that Egr-1 is a transcriptional activator of the BACE-1 gene. We demonstrate that Egr-1 promotes Aβ synthesis by activating BACE-1 in neurons. Our data suggest that Egr-1 may be involved in the activation of BACE-1 leading to acceleration of Aβ synthesis in AD brain.
Results
Egr-1 KO Mice Have Reduced Aβ Levels in the Brain
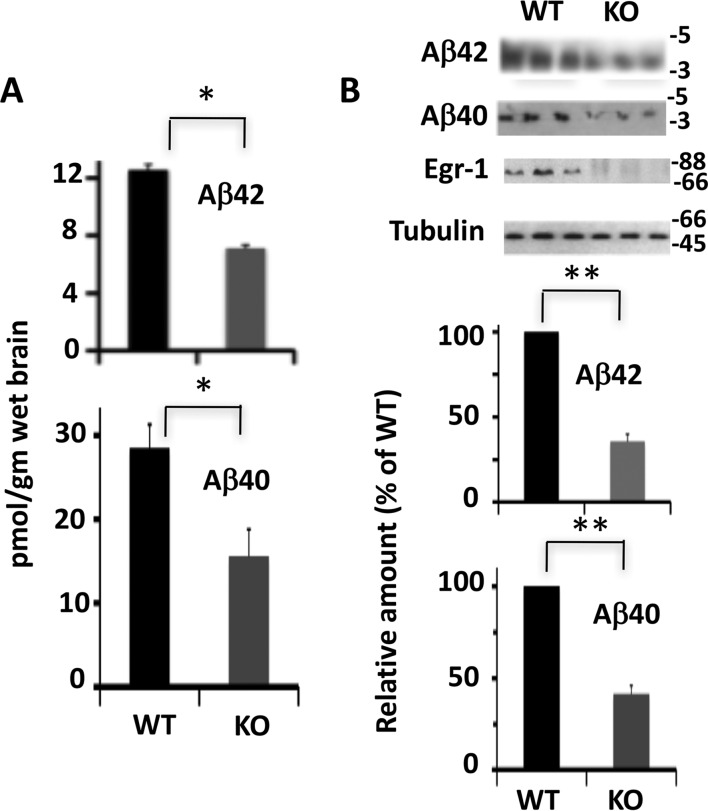
By sandwich ELISA, we determined that the amount of Aβ42 in the brain extract of WT mice was 12.1 pmol/g wet brain (Fig. 1A). In contrast, the amount of Aβ42 in the age-matched KO mice was 6.9 pmol/g wet brain. Likewise, the amount of Aβ40 was 28.5 pmol/g wet brain in the WT mice and 15.6 pmol/g wet brain in the KO mice. Thus, compared with the WT, Egr-1 KO mice had 43% less Aβ42 and 45.3% less Aβ40 in the brain. Western blotting analysis confirmed the ELISA data and showed that compared with the WT, Egr-1 KO mice had 68.5 and 58.8% reduced Aβ42 and Aβ40, respectively, in the brain (Fig. 1B). This result determined that Egr-1 KO mice have significantly reduced levels of Aβ40 and Aβ42 in the brain.
FIGURE 1.
Levels of Aβ40 and Aβ42 are reduced in Egr-1 KO mice. Levels of Aβ40 and Aβ42 in the brain extracts of Egr-1 KO and WT mice were analyzed by sandwich ELISA and Western blotting. A, sandwich ELISA. B, Western blots. Western blotting was performed using a 4–20% Tricine gel. Data are mean ± S.E. from six age-matched animals in each group. Compared with the WT, significantly lower levels of Aβ40 and Aβ42 were observed in KO mice brain by sandwich ELISA (*, p < 0.05) and Western blotting analysis (**, p < 0.01) (t test).
BACE-1 Level Is Reduced in the Brain of Egr-1 KO Mice
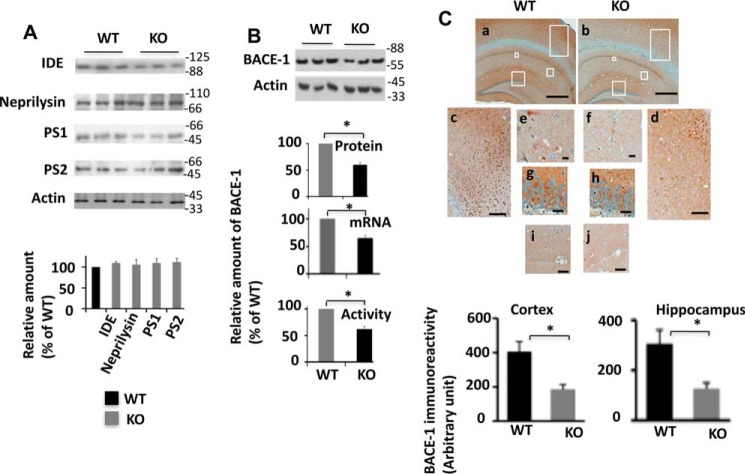
Aβ is generated and eliminated continuously, and its level in the brain is mainly controlled by its synthesis and clearance. The proteolytic degradation is a major route of Aβ clearance (24). Neprilysin and insulin-degrading enzyme (IDE) are among the main Aβ-degrading enzymes that have been identified to date (24–27). Western blotting analysis determined that levels of both neprilysin and IDE were similar in Egr-1 KO and WT mouse brains (Fig. 2A). These data indicated that genetic deletion of Egr-1 does not affect levels of neprilysin and IDE in the brain.
FIGURE 2.
BACE-1 level is reduced in Egr-1 KO mice. A, levels of PS1, PS2, neprilysin, and IDE in the brains of Egr-1 KO and WT mice. Upper panel shows representative Western blots. Lower panel is quantification. No significant difference was found among the groups (mean ± S.E., n = 6/group). B, BACE-1 level is reduced in Egr-1 KO mouse brain. BACE-1 protein and mRNA and activity levels in Egr-1 KO and WT mice were determined and compared by Western blotting, qPCR, and fluorogenic BACE-1 activity assay. Data are mean ± S.E. from five different animals in each group. Upper panel is a representative Western blot. Lower panels are quantifications of BACE-1 protein from Western blottings, qPCR analysis for mRNA, and BACE-1 activity assay. Compared with the WT, significantly reduced BACE-1 protein (*, p < 0.05), mRNA (*, p < 0.04), and activity (*, p < 0.05) is observed in Egr-1 KO mouse brain (t test). C, BACE-1 immunostaining is reduced in Egr-1 KO mouse brain. Brain sections of Egr-1 KO and WT mice were immunostained against BACE-1. Upper panel is a representative BACE-1 immunostained micrographs of brain sections. Boxed areas of panel a (WT) and panel b (KO) are shown in panels c–j in higher magnifications. WT: panel c, cortex; panels e, g, and i, hippocampus. KO: panel d, cortex; panels f, h, and j, hippocampus. BACE-1 is widely expressed all over the brain. Significant staining is seen in the CA1, CA2, and dentate gyrus subfields of hippocampus. Egr-1 KO mice also display similar distribution patterns but significantly reduced levels. Lower panel is quantification of BACE-1 immunoreactivity from micrographs of five mice in each group using Spectrum Analysis algorithm package and ImageScope analysis software (version 11.2, Aperio Technologies). Compared with the WT, Egr-1 KO mice have a significantly lower level of BACE-1 in the cortex (*, < 0.05) and the hippocampus (*, < 0.05) (t test). Scale bars,500 μm (panels a and b), 200 μm (panels c and d), 50 μm (panels i and j), 20 μm (panels g and h), and 10 μm (panels e and f).
Synthesis of Aβ occurs by a sequential cleavage of APP by BACE-1 and γ-secretase. Although BACE-1 is a monomeric enzyme, γ-secretase is a multimeric enzyme with presenilin 1 (PS1) or presenilin 2 (PS2) being the catalytic subunit (1, 28). Therefore, to examine whether Egr-1 regulates any of the enzymes involved in Aβ synthesis, we analyzed levels of PS1 and PS2 in mice brains (Fig. 2A). Western blotting analysis determined that the levels of both PS1 and PS2 were similar in KO and WT mice. However, in Egr-1 KO mice, the BACE-1 protein level was 40% less than in the WT. Likewise, qPCR analysis showed that compared with the WT, the level of BACE-1 mRNA was 35% less in KO mice (Fig. 2B). Fluorogenic activity assay confirmed the Western blotting and qPCR data and showed that BACE-1 activity was 37% less in Egr-1 KO than in the WT mice. Immunohistochemistry of the brain sections showed BACE-1 immunoreactivity was 1.8- and 2.1-fold less in the hippocampus and cortex, respectively, of the KO than the WT mice (Fig. 2C). Based on these data, we concluded that compared with the WT, Egr-1 KO mice have significantly reduced BACE-1 in the brain.
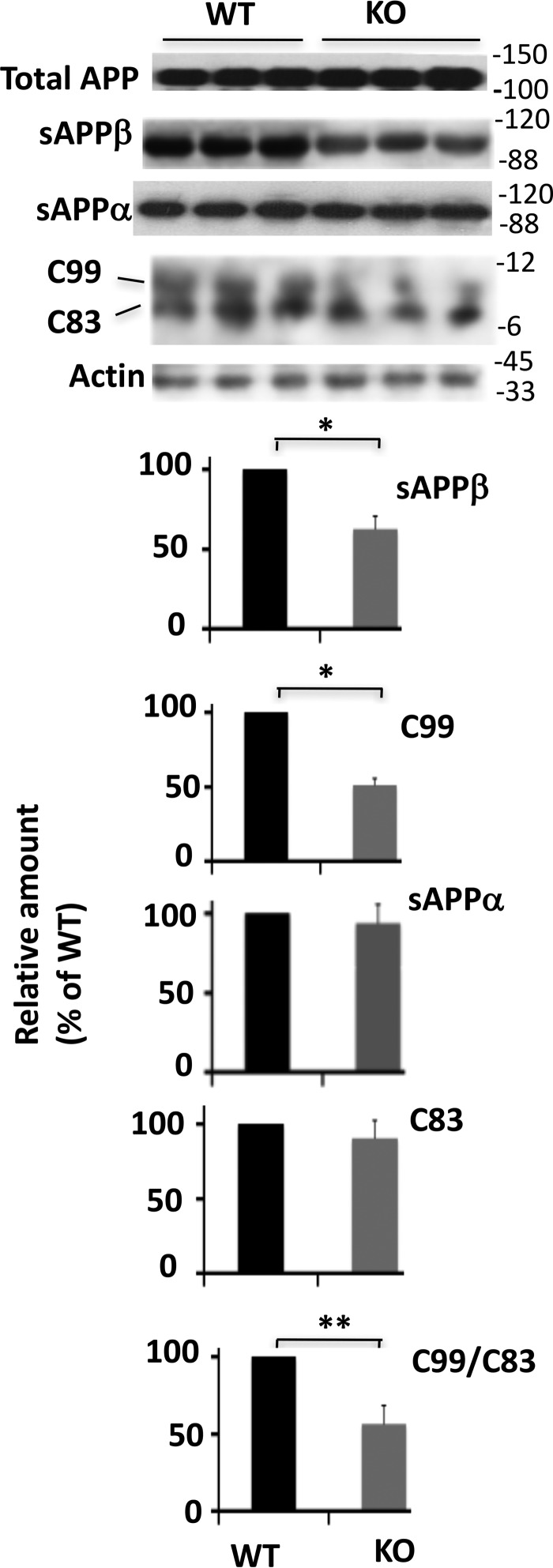
APP is cleaved by either α-secretase or BACE-1 followed by γ-secretase. While BACE-1 cleavage generates C-terminal C99 and N-terminal sAPPβ, α-secretase cleavage generates C83 and sAPPα (1). To substantiate the above data, we analyzed levels of APP metabolites in the brain extracts of mice. As shown in Fig. 3, the levels of both C99 and sAPPβ were 48.8 and 37.6%, respectively, less in KO mice when compared with WT. In contrast, α-secretase-generated sAPPα and C83 were similar in the two experimental groups. The amount of C99/C83 ratio, which is an indicator of amyloidogenic APP metabolism, was 47.8% less in KO mice than the WT.
FIGURE 3.
Levels of sAPPβ and C99 are reduced in Egr-1 KO mice. Western blotting was performed using antibodies against sAPPβ, sAPPα, and APP-CTF. C99 and C83 were separated on a 4–12% gradient BisTris gel. Data are mean ± S.E., n = 6 each group; *, p < 0.05 (sAPPβ); *, p < 0.04 (C99); **, p < 0.01(C99/C83) (t test).
To further substantiate the brain data, we analyzed rat hippocampal primary neurons infected with Ln-Egr-1. Neurons expressing Egr-1 displayed significantly elevated levels of BACE-1, C99, sAPPβ, and Aβ42 when compared with Ln vector-infected neurons (see below). Based on these results, we concluded that Egr-1 KO mice have significantly reduced BACE-1 level in the brain, and overexpression of Egr-1 promotes BACE-1 expression in neurons.
Egr-1 Is a Transcriptional Activator of BACE-1 Gene
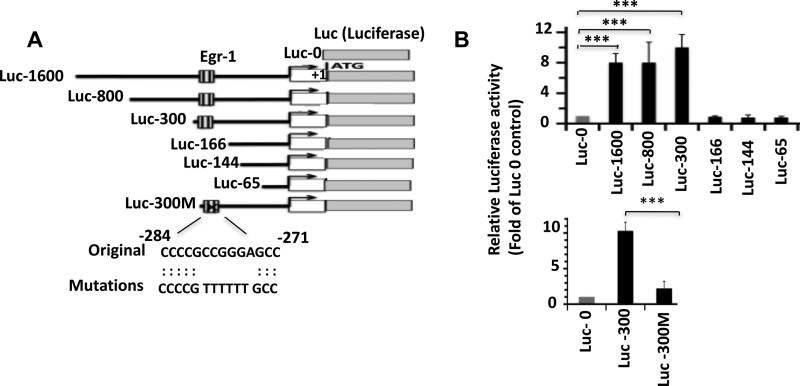
Recently, a genome-wide investigation of in vivo Egr-1-binding sites determined that Egr-1 binds to a GC-rich sequence of its target genes (29). Based on the criteria deduced by this study, we identified five putative Egr-1-binding sites within the BACE-1 promoter located upstream from the transcription initiation site. To determine whether Egr-1 binds to any of these sites and induces BACE-1 transcription, we cloned the following five BACE-1 promoter fragments upstream from the luciferase reporter gene PGL3: Luc-1600 containing the promoter; most of the UTR sequence and all five putative Egr-1-binding sites; and deletion mutants Luc-800, Luc-300, Luc-166, Luc-144, and Luc-65 (Fig. 4A). We co-transfected each of these fragments and control Luc-0 (basic PGL3 empty vector) with Egr-1 in COS-7 cells and monitored luciferase activity (Fig. 4B). Compared with Luc-0, Luc-1600, Luc-800, and Luc-300 displayed 8.3-, 8.1-, and 10.3-fold more luciferase activity, respectively (Fig. 4B, upper panel). However, luciferase activity of Luc-166 was similar to that of Luc-0. Likewise, Luc-144 and Luc-65 also displayed luciferase activity similar to Luc-0 control. These data determined that the Egr-1-binding site is located in-between 166 and 300 bp upstream of the transcription initiation site. A GC-rich sequence is located within the −275–283 segment of the BACE-1 promoter (Fig. 4A). To determine whether this segment contributes to Egr-1 binding, we mutated six bases within this segment of Luc-300 (Luc-300M) (Fig. 4A) and performed luciferase assay. Although Luc-300 showed 10.2-fold higher activity, Luc-300M displayed luciferase activity similar to that of Luc-0 (Fig. 4B, lower panel). Thus, either deletion or mutation of the above segment abolished Egr-1-induced luciferase activity of the BACE-1 promoter. This result determined that Egr-1 binds to the putative segments of BACE-1 promoter and induces BACE-1 expression.
FIGURE 4.
Egr-1 binds to BACE-1 promoter and activates BACE-1 transcription. Luciferase assay was performed to evaluate the binding of Egr-1 onto the BACE-1 promoter. A, schematic diagram of the constructs used to identify the Egr-1-binding site on BACE-1 promoter by luciferase assay. Putative Egr-1-binding site and mutations used to disrupt the binding sequence are shown. These constructs in pGL3 plasmid were co-transfected with Egr-1 and Renilla luciferase reporter plasmid in COS-7 cells. After 24 h, cells were harvested and subjected to luciferase activity assay as described under “Materials and Methods.” Values are expressed as the fold of Luc-0 control. B, bar graphs represent data average ±S.E. from four independent experiments. ***, p < 0.001 against Luc-0- and Egr-1-transfected cells (one-way ANOVA).
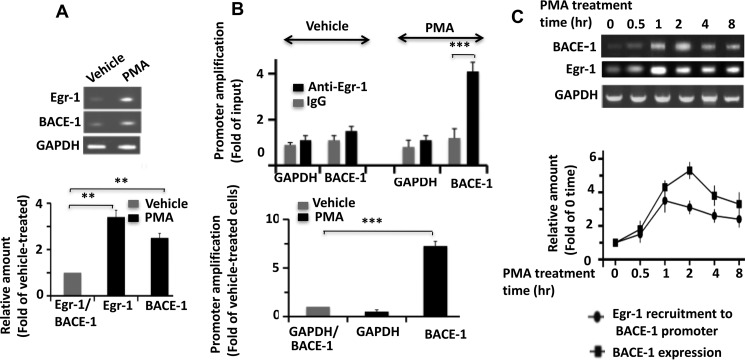
Egr-1 Is Recruited to BACE-1 Promoter in Response to PMA Exposure in COS-7 Cells
Egr-1 regulates the expression of various genes by binding to their promoters (30). Egr-1 expression is induced when COS-7 cells are exposed to PMA (31). PMA also promotes BACE-1 expression in U937 cells and primary CD14+ human monocytes (32). By semiquantitative PCR, we found that PMA induces BACE-1 expression in COS-7 cells also (Fig. 5A). Therefore, to determine whether Egr-1 binds to the BACE-1 promoter in response to PMA exposure, we performed a ChIP assay using anti-Egr-1 antibody and primers flanking the Egr-1-binding site on the BACE-1 promoter, identified by luciferase assay (Table 1 for list of primers used). As the control, we amplified GAPDH (Fig. 5B). In vehicle-treated cells, we observed a basal level of promoter amplification from both IgG and anti-Egr-1 immunoprecipitates (Fig. 5B, upper panel). However, from PMA-treated cells, the BACE-1 promoter amplification was 4.1-fold higher from anti-Egr-1 than from IgG control. No amplification was observed for GAPDH control. More importantly, compared with vehicle-treated cells, PMA-treated cells displayed 7.3-fold more BACE-1 amplification (Fig. 5B, lower panel). These data indicated that 7.3-fold more Egr-1 is complex with the BACE-1 promoter in cells treated with PMA than those treated with vehicle. This in turn indicated that PMA exposure causes recruitment of Egr-1 onto the BACE-1 promoter.
FIGURE 5.
Egr-1 is recruited to BACE-1 promoter in response to PMA exposure in COS-7 cells. COS-7 cells were treated with PMA to induce Egr-1 expression for various time points and then analyzed either for expressions of Egr-1 and BACE-1 by semiquantitative PCR or subjected to ChIP assay using anti-Egr-1 antibody or IgG control. For semiquantitative PCR, RNA purified from cells treated with PMA or vehicle was purified and subjected to RT-PCR for cDNA synthesis. Each synthesized cDNA was amplified using primers against Egr-1, BACE-1, or GAPDH control by PCR. Products were analyzed on agarose gels and quantified by normalizing against GAPDH. A, semiquantitative PCR analysis of cells treated with PMA for 1 h. Upper panel is a representative agarose gel. Lower panel is quantification. Values are the average ± S.E. of four independent determinations. **, p < 0.01 (t test). B, ChIP assay. Cells treated with PMA for 1 h were immunoprecipitated using either anti-Egr-1 antibody or IgG control. BACE-1 promoter in the immunoprecipitate was amplified by qPCR using primers specific to Egr-1-binding site of BACE-1 promoter identified by luciferase assay (Table 1). GAPDH was amplified as the internal control. Bar graphs were generated from qPCR data from four independent cultures. ***, p < 0.0005 (t test). Lower panel compares BACE-1 promoter amplification between vehicle and PMA-treated cells. To make the comparison, qPCR value obtained for the IgG from the ChIP data of a sample was subtracted from the corresponding value for the anti-Egr-1. Resulting values were used to generate the graph. Values are the average of three determinations. ***, p < 0.001 (one-way ANOVA). C, time course of Egr-1 recruitment to BACE-1 promoter and BACE-1 expression during PMA exposure. Cells treated with PMA for each indicated time point were either analyzed by semiquantitative PCR or subjected to ChIP assay for Egr-1 binding to the BACE-1 promoter as in B. Upper panel is a representative agarose DNA gel showing expressions of BACE-1 and Egr-1. Lower panel is a graph showing recruitment of Egr-1 onto the BACE-1 promoter (by ChIP assay) and BACE-1 expression (by semiquantitative PCR). Data are average of three determinations and are expressed as the fold of zero time point.
TABLE 1.
Primer list

If Egr-1 induces BACE-1 transcription, BACE-1 expression will be expected to follow binding of Egr-1 to the BACE-1 promoter. To demonstrate this, we monitored the time course of Egr-1 recruitment to the BACE-1 promoter and BACE-1 expression (Fig. 5C). PMA exposures caused a rapid recruitment of Egr-1 to the BACE-1 promoter that peaked at 1 h and slowly declined over the next 8 h (Fig. 5C, lower). BACE-1 expression, however, was relatively slow, peaked at 2 h, and then declined. Thus, recruitment of Egr-1 to BACE-1 promoter preceded BACE-1 expression in PMA-treated COS-7 cells. Based on these data, we concluded that Egr-1 binds to the BACE-1 promoter and induces BACE-1 expression in PMA-exposed COS-7 cells.
Egr-1 Activates BACE-1, Accelerates APP Metabolism, and Promotes Aβ Synthesis in Primary Neurons and COS-7 Cells
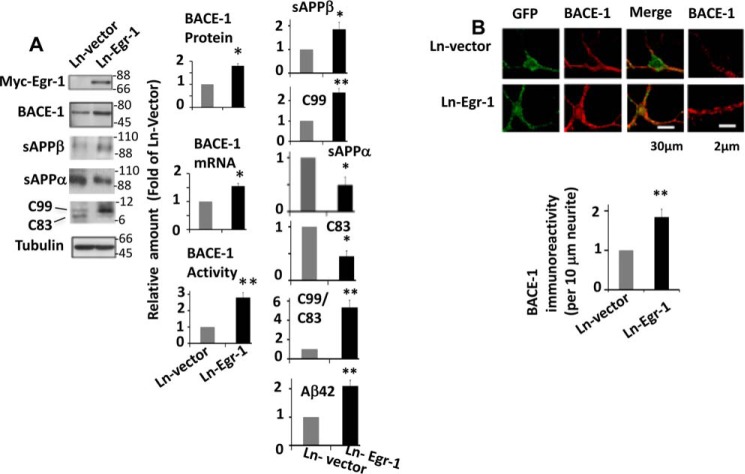
We found that Egr-1 KO mice have reduced levels of both Aβ and BACE-1 (Figs. 1 and 2). In addition, our data showed that Egr-1 is a transcriptional activator of the BACE-1 gene (Fig. 4). BACE-1 catalyzes the committed step in Aβ synthesis (1). However, the level of Aβ in the brain is regulated by its synthesis and clearance (24). Therefore, to determine whether Egr-1 promotes Aβ synthesis via activating BACE-1, we infected rat hippocampal primary neurons with Ln-Egr-1 and analyzed neuronal lysate and conditional medium. Western blotting, BACE-1 activity assay, and qPCR analysis determined that the protein, mRNA, and activity levels of BACE-1 were 1.7-, 1.6-, and 2.8-fold, respectively, higher in Egr-1-overexpressing neurons when compared with those expressing vector (Fig. 6A). Immunocytochemistry confirmed the Western data and showed that the BACE-1 level was 1.85-fold higher in neurons expressing Egr-1 than vector (Fig. 6B). Thus, Egr-1 overexpression enhanced the levels of BACE-1 mRNA, protein, and activity in primary neurons.
FIGURE 6.
Overexpression of Egr-1 activates BACE-1 in the rat hippocampal primary neurons. Rat hippocampal primary neurons in culture for 14 days were infected with Ln-Egr-1 or Ln vector. Cell were analyzed for level of BACE-1 by Western blotting, qPCR, immunocytochemistry, and fluorogenic BACE-1 activity assay. Western blotting was performed for C99 and C83 as in Fig. 3 using anti-APP-CTF antibody. Culture medium was analyzed for sAPPβ, sAPPα (Western blotting), and Aβ42 (ELISA). A, representative Western blots and bar graphs of quantification of BACE-1 protein (Western blots), mRNA (qPCR), and activity (fluorogenic BACE-1 activity assay), C99 and C83 from lysates and levels of sAPPβ (Western blots) and Aβ42 (mouse Aβ42 ELISA kit) in the medium from four different cultures. To determine C99/C83 ratio, C99 value of a sample was divided by C83 value of that sample. Data are mean ± S.E. BACE-1 protein (*, p < 0.05), BACE-1-mRNA (*, p < 0.04), BACE-1 activity (**, p < 0.005), C99 (**, p < 0.01), C99/C83 (**, p < 0.001), sAPPβ (*, p < 0.05), and Aβ42 (**, p < 0.01), C83 (*, p < 0.05), and sAPPα (*p < 0.05) (t test). B, confocal micrograph of immunocytochemistry of infected (GFP-positive) neurons showing BACE-1 (red) and Egr-1 (green). Quantification of BACE-1 level is from micrographs of 15 randomly chosen infected neurons from four different cultures in each group along at least 30 μm of each neurite. Compared with Ln vector-infected neurons, Ln-Egr-1-infected neurons show significantly increased BACE-1 immunoreactivity (**, p < 0.01) (t test).
We then analyzed levels of APP metabolites in the neuronal lysates and medium. In the lysates of Ln-Egr-1-infected neurons, although the level of C99 was 2.4-fold higher, that of C83 was 2.2-fold reduced when compared with vector-infected counterparts (Fig. 6A). As a result, the C99/C83 ratio in neurons infected with Ln-Egr-1 was 5.2-fold higher than in neurons infected with Ln vector. Western blots showed that in the medium of neurons infected with Ln-Egr-1, the level of sAPPβ was 1.9-fold higher than in the medium of those infected with Ln vector. In contrast, in the medium of neurons infected with Ln-Egr-1, the level of sAPPα was 2.1-fold lower than in the medium of those infected with Ln vector. Likewise, ELISA of medium determined that amount of secreted Aβ42 was 2.1-fold more in Egr-1-expressing neurons when compared with those expressing vector. Thus, overexpression of Egr-1 activated BACE-1, accelerated formation of BACE-1-generated C99 and sAPPβ, reduced α-secretase-cleaved C83 and sAPPα, and increased Aβ42 in neurons.
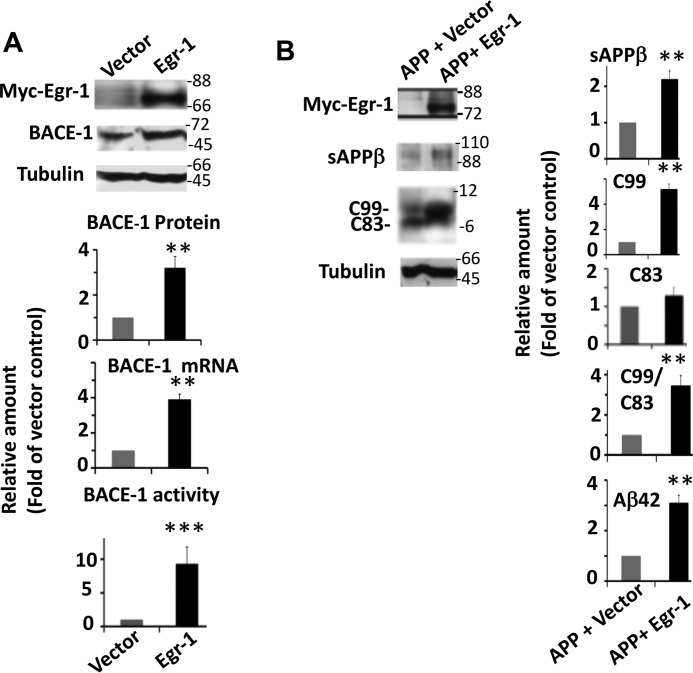
To substantiate the neuron data, we co-transfected COS-7 cells with Egr-1 and APPSwe in different combinations and analyzed them. In cells expressing Egr-1, BACE-1 protein, mRNA, and activity levels were 3.2-, 3.9-, and 9.3-fold higher, respectively, than in vector-expressing cells (Fig. 7A). Thus, as in neurons Egr-1 overexpression activated BACE-1. In cells expressing APPSwe and vector, the major APP-cleaved product was C83 generated by α-secretase (Fig. 7B). In contrast, in cells expressing APPSwe and Egr-1, the major APP-cleaved product was C99 generated by BACE-1. Furthermore, compared with APPSwe-expressing cells, those expressing APPSwe and Egr-1 had a C99/C83 ratio 3.46-fold higher and secreted 2.2- and 3.1-fold more sAPPβ and Aβ42, respectively, in the medium (Fig. 7B). These data indicated that Egr-1 overexpression activates BACE-1 and accelerates amyloidogenic APP cleavage and Aβ42 synthesis in COS-7 cells.
FIGURE 7.
Egr-1 activates BACE-1 and promotes amyloidogenic APP processing in COS-7 cells. A, Egr-1 activates BACE-1. COS-7 cells transfected with Egr-1 or vector were analyzed for BACE-1 protein (Western blotting), mRNA (qPCR), and activity (fluorogenic BACE-1 activity assay). Upper panel is a representative Western blot. Data are mean ± S.E. from four different cultures. Compared with vector-transfected cells, Egr-1-transfected cells show significantly enhanced levels of BACE-1 protein (**, p < 0.01), BACE-mRNA (**, p < 0.005), and BACE-1 activity (***, p < 0.001) (t test). B, Egr-1 accelerates amyloidogenic APP processing. APPSwe was co-transfected in COS-7 cells with either vector or Egr-1. Transfected cell lysates were analyzed for levels of C83 and C99 by Western blotting. Culture media were used to analyze sAPPβ by Western blotting and Aβ42 by human Aβ42 ELISA kit. Data are mean ± S.E. from four different cultures. Compared with cells expressing APPSwe, those expressing APP and Egr-1 had significantly increased levels of sAPPβ (**, p < 0.005), C99 (**, p < 0.001), and Aβ42 (**, p < 0.001) (t test).
Silencing BACE-1 Blocks Egr-1-induced Amyloidogenic APP Processing in Mouse Hippocampal Primary Neurons
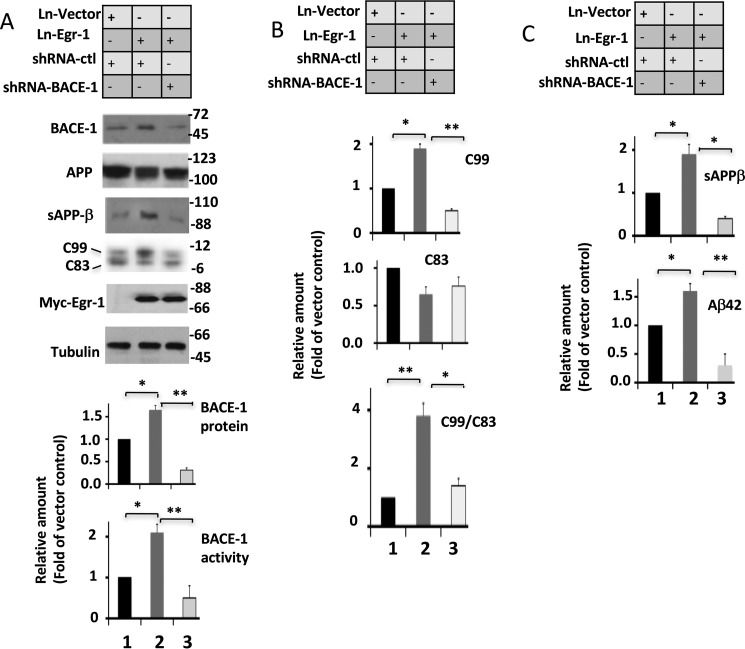
If Egr-1 promotes Aβ synthesis by activating BACE-1, knockdown of BACE-1 will be expected to block Egr-1-induced amyloidogenic APP processing. To demonstrate this, we silenced BACE-1 in mouse hippocampal primary neurons using shRNA-BACE-1 and analyzed them (Fig. 8).
FIGURE 8.
Knockdown of BACE-1 blocks Egr-1-induced amyloidogenic APP processing in mouse hippocampal primary neurons. Neurons co-infected with shRNA-BACE-1, shRNA-ctl, Ln-Egr-1, and Ln-vector in various combinations were lysed and analyzed as in Fig. 6. A, representative Western blots and quantification of BACE-1 protein and activity. Values are the average of four independent cultures. *, p < 0.05; **, p < 0.005, one-way ANOVA. B, quantification of C99, C83, and C99/C83. Quantifications were performed from Western blots from A. Values are the average of four independent cultures. *, p < 0.05; **, p < 0.005, one-way ANOVA. C, quantification of sAPPβ and Aβ42. Culture medium was analyzed by Western blotting for sAPPβ and sandwich ELISA for Aβ42. Values are from four different cultures. *, p < 0.04; **, p < 0.001, one-way ANOVA.
Compared with neurons infected with shRNA-ctl, those infected with shRNA-BACE-1 displayed 58 and 65% reduction in levels of BACE-1 protein and BACE-1 activity, respectively, but had similar levels of tubulin (compare lane 3 with lane 1 in Fig. 8A). Likewise, compared with control neurons co-infected with Ln vector and shRNA-ctl, those co-infected with Ln-Egr-1 and shRNA-ctl displayed 1.6- and 2.1-fold higher BACE-1 protein and activity, respectively, a 1.8-fold higher level of C99 but a reduced level of C83, a 2.6-fold higher C99/C83, a 1.8-fold more sAPPβ, and 1.5-fold more Aβ42 (compare lane 2 with lane 1 in Fig. 8B). These data are as expected and showed that Egr-1 activates BACE-1 and accelerates amyloidogenic APP processing, and shRNA-BACE-1 had specifically silenced BACE-1 in these neurons. Importantly, compared with neurons co-infected with Ln-Egr-1 and shRNA-ctl, those co-infected with Ln-Egr-1 and shRNA-BACE-1 had C99 levels 75% less (compare lane 3 with lane 2 in Fig. 8B). Likewise, levels of sAPPβ and Aβ42 were 72 and 69%, respectively, less in neurons co-infected with Ln-Egr-1 and shRNA-BACE-1 when compared with those co-infected with Ln-Egr-1 and shRNA-ctl (compare lane 3 with lane 2 in Fig. 8C). Thus, silencing BACE-1 action almost completely blocked Egr-1-induced amyloidogenic APP processing in neurons. Based on these data, we concluded that Egr-1-promoted amyloidogenic APP processing leading to Aβ synthesis requires BACE-1.
Aβ42 Exposure Enhances Levels of Egr-1 and BACE-1 in Rat Hippocampal Primary Neurons
Previous studies have shown that Aβ42 exposure activates BACE-1 in human neuroblastoma SH-SY5Y cells (33, 34). Because BACE-1 catalyzes a committed step in Aβ synthesis and Aβ causes neurodegeneration in various in vitro and in vivo models (35), it was suggested that Aβ may lead to a vicious cycle of neurodegeneration by promoting its own synthesis (36). Aβ also induces Egr-1 expression in rat cortical neurons (21). Therefore, to test whether Egr-1 plays any role in this process, we exposed rat hippocampal primary neurons to oligomeric Aβ42 and analyzed them. Aβ42 exposure caused a 2.1- and 1.8-fold increase in the levels of Egr-1 and in BACE-1, respectively. However, the levels of sAPPβ, C83, C99, and C99/C83 remained similar in neurons treated with Aβ42 and β42 scramble control (data not included). Thus, under our experimental conditions, Aβ42 exposure did not promote amyloidogenic APP processing in neurons.
Discussion
BACE-1 is the essential enzyme in Aβ production, and both mRNA and protein levels of BACE-1 are elevated in sporadic AD (2–4). It was suggested that up-regulation of BACE-1 is one of the main causes of Aβ accumulation in AD brain (1). The mechanism by which BACE-1 expression is increased in AD brain is not known. In this study, we showed that Egr-1 binds to the BACE-1 promoter and induces BACE-1 expression in neurons and COS-7 cells (Figs. 4–7). Overexpression of Egr-1 activates BACE-1 and accelerates Aβ synthesis in primary neuron and COS-7 cells (Figs. 6 and 7). Previous studies have shown that the Egr-1 level is up-regulated in AD brains (18–21, 37). Furthermore, levels of both BACE-1 and Egr-1 are elevated in the brain during aging (22, 38, 39), hypoxia, brain injury, ischemia, and inflammation (13, 14) and in AD mouse models (13, 21, 23). These observations together indicate that Egr-1 is an activator of BACE-1 in the CNS and suggest that Egr-1 plays role in the up-regulation of levels of BACE-1 and Aβ in AD brain.
Aβ induces BACE-1 expression in neurons (33, 34) and hence by promoting its own synthesis may lead to a vicious cycle of neurodegeneration (36). Recently, prion protein (PrPc), ephrin type B receptor (EphB2), IgG Fcγ receptor II-b (FcγIIRb), and paired immunoglobulin-like receptor B (PirB) (human homologue LilrB2) were found to act as the cell surface receptors for Aβ (40–44). Interestingly, among these receptors, PrPc and EphB2 act through the N-methyl-d-aspartate receptor (NMDAR) (42, 43), whereas PirB2 (and LilrB2) acts via the early gene protein Arc, whose expression is NMDAR-dependent (45). Thus, these receptors act via NMDAR, which is the main inducer of Egr-1 in the CNS (14). NMDAR activation recruits Egr-1 to the promoter of its target gene in neurons (31). BACE-1 is one target gene of Egr-1 (this study). Aβ may activate Egr-1 via NMDAR, which subsequently may bind to the BACE-1 promoter and may induce BACE-1 expression. BACE-1 then may accelerate Aβ synthesis. Consistent with this idea, Aβ exposure induces expressions of Egr-1 and BACE-1 in neurons (21, 34). In contrast to this hypothesis, we found that Aβ42 exposure does not significantly promote amyloidogenic APP processing in rat hippocampal primary neurons (data not shown). BACE-1 is synthesized in the endoplasmic reticulum and is then subjected to several post-translational modifications such as glycosylation, phosphorylation, S-palmitoylation, and acetylation (1). Aβ causes many cellular dysfunctions (35). It is possible that Aβ induces BACE-1 synthesis but disrupts any of the BACE-1 post-translational modifications, which affect BACE-1 activity and/or amyloidogenic APP processing.
Growing evidence suggests that synthesis and proteolytic degradation are the main determinants of cerebral Aβ level. A number of proteases, including matrix metallopeptidase-9, IDE, neprilysin, endothelin-converting enzyme, angiotensin-converting enzyme, plasmin, α2-macroglobulin, and matrix metallopeptidase-9 proteolyze Aβ in vitro (24). In addition, Aβ was reported to be a substrate of the proteasome (46). In this study, we find that the levels of neprilysin and IDE are comparable in the brain of Egr-1 KO and WT mouse brains (Fig. 2A). In addition, a previous study showed that Egr-1 down-regulates proteasome activity by transcriptional suppression of proteasome subunits psmb9 (Lmp2) and psme2 (PA28β) along with the proteasome-regulatory kinase serum/glucocorticoid-regulated kinase and the proteasome-associated antigen peptide transporter subunit 1 (Tap1) (47). Although we did not analyze all proteolytic enzymes implicated to degrade Aβ in the brain, in primary neurons, and COS-7 cells, overexpression of Egr-1 activates BACE-1 and enhances Aβ levels (Figs. 6 and 7). When BACE-1 function is blocked, Egr-1-induced enhancement in the Aβ level is almost completely blocked (Fig. 8). These results suggest that Egr-1 regulates brain Aβ level via controlling the Aβ synthesis.
Aβ is generated by a sequential cleavage of APP by BACE-1 followed by γ-secretase. APP is also cleaved by α-secretase, which generates sAPPα and C83 and precludes Aβ formation (1). Thus, in the brain, levels of Aβ could increase either by activation of BACE-1, inhibition of α-secretase, or both. As shown in Fig. 6, Egr-1 overexpression causes a 1.9-fold increase in BACE-1-cleaved sAPPβ and a 2.4-fold increase in C99. In addition, the levels of α-secretase-cleaved sAPPα and C83 are also reduced by 2.1- and 2.2-fold, respectively, in neurons overexpressing Egr-1 (Fig. 6). The data appear to suggest that Egr-1 promotes Aβ synthesis by both activating BACE-1 and inhibiting α-secretase-cleavage of APP. However, in neurons, when BACE-1 is silenced, Egr-1-induced Aβ synthesis is almost completely blocked (Fig. 8). These data indicate that Egr-1 promotes Aβ synthesis by activating BACE-1 and suggest that reduction in sAPPα and C83 in neurons overexpressing Egr-1 is likely caused by competition between BACE-1 and α-secretase for APP.
The level of PS2 is similar in Egr-1 KO and WT mouse brains (Fig. 2A). These data suggest that Egr-1 does not regulate PS2 expression. Consistent with our data, a previous study showed that Egr-1 does not bind to the murine PS2 promoter and does not regulate PS2 gene expression (48). However, another study reported that Egr-1 is a transcriptional activator of human PS2 genes in neural cells (49). It appears that the PS2 gene expression is regulated by different mechanisms in mice and humans.
This study emphasizes Egr-1-mediated Aβ synthesis via activation of BACE-1 and suggests that lower level of Aβ in Egr-1 KO mice is due to the reduced BACE-1 activity. However, apoE, a risk for sporadic AD associates with extracellular amyloid plaques, affects Aβ aggregation and clearance in the brain (50). Interestingly, in a murine model of atherosclerosis, aorta retrieved from apoE−/− mice demonstrated increased Egr-1 transcripts in an age-dependent manner (51). Human subjects carrying the T2238C ANP gene variant have a higher risk to suffer a stroke or myocardial infarction. C2238/αANP modulates apoE through Egr-1/miR199a in vascular smooth muscle cells in vitro (52). It will be interesting to investigate whether Egr-1 regulates Aβ clearance via apoE in the brain.
A number of transcription factors have been identified that act as activators or repressors of BACE-1 transcription. These include SP1, NF-κB, YY1, HIF-1, PPARγ, GADD153 (53), and Egr-1 (this study). SP1 binds to the BACE-1 promoter, induces BACE-1 expression in PC12 and HEK-293 cells (54), and mediates BACE-1 transcription induced by 12/15-lipoxygenase in the CNS (55). Likewise, NF-κB acts as a repressor for BACE-1 transcription in differentiated neuronal cells and non-activated glia cells but as an activator in activated astrocytes and the Aβ-exposed neuronal cultures (56). In neuroblastoma SH-SY5 cells, NF-κB interacts with GADD153 to regulate BACE-1 expression induced by 27-hydroxycholesterol (57). Similarly, YY1 binds to the BACE-1 promoter in PC12 cells and neurons in culture, but in the brain, BACE-1 and YY1 are co-expressed in only a minor population of neurons (58). Finally, HIF-1 binds to the BACE-1 promoter only under hypoxic conditions (10), and PPARγ was suggested to repress BACE-1 expression by antagonizing activities of other transcription factors (54, 59). These observations suggest that BACE-1 expression is regulated by complex mechanisms. A detailed study will be required to determine how these various transcription factors regulate BACE-1 expression in different cells under different physiological conditions.
Materials and Methods
Animals
All experiments involving animals were performed according to Canadian Council of Animal Care and Lady Davis Institute for Medical Research guidelines. Egr-1 KO and WT mice (all 6 months old and in C57BL6 background) were described previously (20, 60). All mice were genotyped by PCR.
Antibodies and Chemicals
Rabbit polyclonal antibodies against GFP, Egr-1, and Myc were described previously (20). Polyclonal antibodies against neprilysin (AB5458), IDE (AB9210), and PS1 (AB5757) were obtained from Millipore. Anti-PS2 antibody was from Sigma (P0070). Monoclonal antibody against sAPPβ was from BioLegend (9138005). Monoclonal antibody against sAPPα was from BioSource (MBS533522). Anti-APP C-terminal antibody was from Invitrogen (51-2700). Polyclonal antibody raised against the C-terminal six amino acids of human Aβ42 was from Millipore (AB5078P). Polyclonal antibody against human Aβ40 was also from Millipore (ABN240). PMA was obtained from Sigma.
Histochemistry
Mice were anesthetized and transcardially perfused with cold PBS. Coronal sections (50 μm thick) corresponding to the hippocampus and cortex were used for histology. The procedure for immunohistochemistry using immunostainer “Discovery XT” from Ventana and quantification of images was described previously (31). Briefly, fixed sections were immunolabeled with rabbit anti-BACE-1 antibody (1:50 mouse Millipore, MAB5308) followed by an HRP-conjugated secondary antibody. Sections were developed using a diaminobenzidine histochemistry kit (Molecular Probes), counterstained with hematoxylin-eosin, and viewed under a microscope. For quantification, slides were scanned at ×400 magnification (resolution of 0.25 μm/pixel (100,000 pixels/inch)) using an Aperio ScanScope AT Turbo (Leica Biosystems). The background illumination levels were calibrated using a prescan procedure. Acquired digital images representing whole tissue sections were analyzed applying the Spectrum Analysis algorithm package and the ImageScope analysis software (version 11.2, Aperio Technologies). A rectangular region of interest (ROI) 25 μm2 in size was defined. Immunoreactivity value for each ROI was obtained. The immunoreactivity within each ROI in each section was averaged to generate a mean immunoreactive value for each animal. Data were averaged from six mice in each group with four regions (n = 24) for each genotype and is expressed as the fold of WT.
Lentiviral Production, Cell Culture, and Viral Infection
Construction and production of lentivirus expressing human Egr-1 with a Myc tag at the C terminus was described previously (31). Hippocampal neurons were prepared from rat or mouse pups P0 (Charles River) (31). Neurons 2 weeks in culture were infected with Ln vector or Ln-Egr-1 (multiplicity of infection of 20 each). After 72 h, neurons were fixed for immunocytochemistry or homogenized in extraction buffer for Western blotting analysis. COS-7 cells were transfected with human APPSwe (APPsw KM670/671NL) and Egr-1 using Lipofectamine. Human APPSwe clone in pcDNA vector was a gift from Dr. Andrea Leblanc of the Lady Davis Institute.
Cell Culture, Mouse Short Hairpin RNA (shRNA) Construction, and Viral Infection
shRNA against mouse BACE-1 was constructed as described previously (31). Briefly, 19-mer of either BACE-1 targeting sequence siBACE-1 (5′-GTT CGC TGT CTC ACA GTC A-3′) (61) or non-effective scrambled control siCtl (5′-caa agc caa gca aac caa t-3′) were subcloned into lentiviral vector pLVX-IRWS-ZsGreen1 (Clontech) at the EcoRI/XbaI site. Lentivirus was produced as described previously (31). Briefly, the DNA was transfected along with a Lenti-X HTX packaging mix into the Lenti-X 293T cells, and the virus was harvested after 48 and 72 h from the supernatant and purified. The titer of purified virus was determined by flow cytometry, and the virus was stored at −80 °C. To validate the shRNA construct, lentivirus was infected into mouse hippocampal primary neurons. After 48 h of infection, neurons were analyzed by Western blotting analysis. Compared with shRNA-ctl-infected neurons, those infected with shRNA-BACE-1 had BACE-1 level reduced by ∼60% (data not shown but see Fig. 8).
Immunocytochemistry, Image Acquisition, and Quantification
Immunocytochemistry was carried out as described previously (31). Briefly, Ln-Egr-1 or Ln vector-infected neurons were fixed and labeled with rabbit anti-BACE-1 or GFP (mouse 1:1000 Cell Signaling) followed by Alexa Fluor 488- or 594- conjugated goat anti-rabbit or goat anti-mouse secondary antibody (Invitrogen). Labeled neurons were viewed by using LSM Pascal (Zeiss, Germany) confocal microscopy system. At least 15 labeled neurons were randomly chosen from each group for quantification from 3 to 4 coverslips. Immunointensity was analyzed along 30–40 μm of dendritic length of each neuron by Volocity image analysis software (Improvision). Data were averaged from four independent cultures.
Quantitative Real Time PCR (qPCR) and Semiquantitative PCR
One microgram of total RNA purified from brain extract, neurons, or COS-7 cells was used for cDNA synthesis using oligo(dT) or random hexamer primer and reverse transcriptase. SYBR Green-based (Qiagen) real time PCR was carried out with gene-specific primer for BACE-1, Egr-1, and GAPDH (see list of primers in Table 1). Data were analyzed by real time PCR software 7500 version 2.0.4 (Applied Biosystems). The relative RNA expression of the gene of interest was determined using comparative ΔΔCt method with GAPDH as the internal control as described previously (31). For semiquantitative PCR, specific primers for Egr-1, BACE-1, and GAPDH control (Table 1) were used for PCR amplification. PCR products were resolved on ethidium bromide-stained agarose gels, and bands were quantified by normalizing against the corresponding GAPDH band.
Constructions of Plasmids for Luciferase Activity and Chromatin Immunoprecipitation (ChIP) Assay
Luciferase assay was performed as described previously (31). For the plasmid constructions, a 1600-bp BACE-1 promoter construct (Luc-1600) containing most of the 5′UTR and all of the putative Egr-1-binding sites and deletion constructs, Luc-800, Luc-300, Luc-166, Luc-144, Luc-65, and a Luc-300M with mutated putative Egr-1-binding site (see Fig. 4A), were cloned by PCR using human genomic DNA as the template (see list of primers used in Table 1). Mutations were introduced into the plasmid by overlap extension PCR. Genetic constructs were assembled through site-specific promoter swapping approaches involving successive PCRs, as described previously (31). All PCR products were subcloned into pGL3-Promoter Vector (Promega) and were confirmed by DNA sequencing. For luciferase activity, COS-7 cells were co-transfected with pcDNA3-Egr-1 or pcDNA3 vector and one of the constructs and pRL-CMV Renilla luciferase reporter plasmid (Promega) using Lipofectamine. After 24 h, cells were lysed and assayed for luciferase activity (Promega kit). Firefly luciferase activity was normalized to Renilla luciferase activity. For ChIP assay, COS-7 cells were treated with PMA (30 ng/ml) to induce Egr-1 (31). After 1 h of treatment, ChIP assay was performed using Magna ChIP T/MA/G Millipore kit (Millipore) following the manufacturer's instructions. Cells were cross-linked, and the DNA was fragmented to 200–1000 bp in length. Chromatin solution (100 μl each) was immunoprecipitated using 5 μg of either monoclonal anti-Egr-1 (Millipore, MABC 1023) or IgG control. Input and immunoprecipitated samples were digested with proteinase K to reverse the cross-linking. The DNA was purified and analyzed by qPCR using primers against the BACE-1 promoter (Table 1).
BACE-1 Activity Assay and Sandwich ELISA for Aβ40 and Aβ42
BACE-1 activity was measured by using the β-secretase fluorogenic activity assay kit (catalog no. 565785 from Calbiochem) following the manufacturer's instruction manual. Briefly, cells or brain samples were homogenized in the extraction buffer and centrifuged, and the supernatant (50 μl each) was transferred to a fluorescence plate reader. β-Secretase substrate was added to each well, and the plate was incubated for 1 h at 37 °C in the dark. After incubation, fluorescence was measured with excitation at 335–355 and emission at 495–570 nm. Background reading from buffer and the substrate was subtracted.
Aβ42 in the cell culture medium and brain extract was measured using either the mouse (KMB3441) or the human (KHB3441) Aβ42 ELISA kit from Invitrogen following the manufacturer's instruction manual. For estimation of Aβ40 in the mouse brain, the ELISA kit (KMB3481) from Invitrogen was used. Briefly, mouse brain (100 mg) was homogenized in 1 ml of casein buffer (0.25% casein, 0.05% sodium azide, 20 μg/ml of aprotinin, 5 mm EDTA, and 10 μg/ml leupeptin in PBS). After centrifugation, supernatant (50 μl each) was pipetted to the ELISA plate well coated with mouse antibody specific to the N terminus of Aβ42. After incubation and washing, rabbit polyclonal antibody specific to the Aβ42 C terminus was added. Bound antibody was detected by using HRP-labeled second antibody and an ELISA plate reader at A450 nm. Aβ42 measurement from conditional medium was also performed as described above using 50 μl of each sample. Measurement of Aβ40 from the brain extract was performed as described above except N- and C-terminal antibodies against Aβ40 were used. For Western blotting, samples were separated on 10–20% Tricine gel (Invitrogen) and probed against anti-Aβ40 or anti-Aβ42 antibody.
Preparation of Aβ42 Oligomers
Aβ42 oligomers were prepared as described previously (21). Briefly, synthetic Aβ42 peptide (American Peptides) was dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol and dried by evaporation in a SpeedVac. Dried peptide was suspended in DMEM (without glutamine) to a 100-μm final concentration and incubated overnight at 4 °C. Incubated peptide was warmed using a water bath set at 37 °C. An aliquot of warm peptide was added to the medium of rat hippocampal primary neurons (14 days in culture) to a final concentration of 5 μm, and neurons were incubated for 4 h. Incubated neurons were analyzed by Western blotting. Synthetic scrambled Aβ42 peptide (American Peptides) was also prepared as above and used as control.
Statistics
All data were analyzed by one-way ANOVA followed by Newman Keuls post hoc test for multiple group comparisons and Student's t test for comparisons between two groups. All data are presented as mean ± S.E., and differences with p values <0.05 were considered significant.
Author Contributions
H. K. P. conceived the project, designed the experiments, supervised the study, analyzed the data, and wrote the manuscript. X. Q. designed and performed most of the experiments and assisted in data analysis. Y. W. designed and validated shRNA-BACE-1 construct and prepared lentivirus.
Acknowledgment
We thank Heather Pekeles for editing the manuscript.
This work was supported by grants from Canadian Institute for Health Research and Alzheimer's Society of Canada. The authors declare that they have no conflicts of interest with the contents of this article.
- Aβ
- amyloid-β
- AD
- Alzheimer's disease
- APP
- amyloid precursor protein
- BACE-1
- β-secretase 1
- PMA
- phorbol 12-myristate 13-acetate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR
- sAPP
- soluble APP
- PS
- presenilin
- NMDAR
- N-methyl-d-aspartate receptor
- IDE
- insulin-degrading enzyme
- ROI
- region of interest.
References
- 1. Vassar R., Kovacs D. M., Yan R., and Wong P. C. (2009) The β-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., and Shen Y. (2003) Elevated β-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 3. Fukumoto H., Cheung B. S., Hyman B. T., and Irizarry M. C. (2002) β-Secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 59, 1381–1389 [DOI] [PubMed] [Google Scholar]
- 4. Holsinger R. M., McLean C. A., Beyreuther K., Masters C. L., and Evin G. (2002) Increased expression of the amyloid precursor β-secretase in Alzheimer's disease. Ann. Neurol. 51, 783–786 [DOI] [PubMed] [Google Scholar]
- 5. Tyler S. J., Dawbarn D., Wilcock G. K., and Allen S. J. (2002) α- and β-secretase: profound changes in Alzheimer's disease. Biochem. Biophys. Res. Commun. 299, 373–376 [DOI] [PubMed] [Google Scholar]
- 6. Zhao J., Fu Y., Yasvoina M., Shao P., Hitt B., O'Connor T., Logan S., Maus E., Citron M., Berry R., Binder L., and Vassar R. (2007) β-Site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer's disease pathogenesis. J. Neurosci. 27, 3639–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blasko I., Beer R., Bigl M., Apelt J., Franz G., Rudzki D., Ransmayr G., Kampfl A., and Schliebs R. (2004) Experimental traumatic brain injury in rats stimulates the expression, production and activity of Alzheimer's disease β-secretase (BACE-1). J. Neural. Transm. 111, 523–536 [DOI] [PubMed] [Google Scholar]
- 8. Wen Y., Onyewuchi O., Yang S., Liu R., and Simpkins J. W. (2004) Increased β-secretase activity and expression in rats following transient cerebral ischemia. Brain Res. 1009, 1–8 [DOI] [PubMed] [Google Scholar]
- 9. Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., and Tanzi R. E. (2007) Depletion of GGA3 stabilizes BACE and enhances β-secretase activity. Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X., Zhou K., Wang R., Cui J., Lipton S. A., Liao F. F., Xu H., and Zhang Y. W. (2007) Hypoxia-inducible factor 1α (HIF-1α)-mediated hypoxia increases BACE1 expression and β-amyloid generation. J. Biol. Chem. 282, 10873–10880 [DOI] [PubMed] [Google Scholar]
- 11. Coma M., Guix F. X., Ill-Raga G., Uribesalgo I., Alameda F., Valverde M. A., and Muñoz F. J. (2008) Oxidative stress triggers the amyloidogenic pathway in human vascular smooth muscle cells. Neurobiol. Aging 29, 969–980 [DOI] [PubMed] [Google Scholar]
- 12. Tong Y., Zhou W., Fung V., Christensen M. A., Qing H., Sun X., and Song W. (2005) Oxidative stress potentiates BACE1 gene expression and Aβ generation. J. Neural. Transm. 112, 455–469 [DOI] [PubMed] [Google Scholar]
- 13. Cole S. L., and Vassar R. (2009) Linking vascular disorders and Alzheimer's disease: potential involvement of BACE1. Neurobiol. Aging 30, 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beckmann A. M., and Wilce P. A. (1997) Egr transcription factors in the nervous system. Neurochem. Int. 31, 477–510 [DOI] [PubMed] [Google Scholar]
- 15. Yan S. F., Fujita T., Lu J., Okada K., Shan Zou Y., Mackman N., Pinsky D. J., and Stern D. M. (2000) Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat. Med. 6, 1355–1361 [DOI] [PubMed] [Google Scholar]
- 16. Khachigian L. M. (2006) Early growth response-1 in cardiovascular pathobiology. Circ. Res. 98, 186–191 [DOI] [PubMed] [Google Scholar]
- 17. Khachigian L. M., Lindner V., Williams A. J., and Collins T. (1996) Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science 271, 1427–1431 [DOI] [PubMed] [Google Scholar]
- 18. Gómez Ravetti M., Rosso O. A., Berretta R., and Moscato P. (2010) Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus' gene expression profiles in Alzheimer's disease. PLoS ONE 5, e10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. MacGibbon G. A., Lawlor P. A., Walton M., Sirimanne E., Faull R. L., Synek B., Mee E., Connor B., and Dragunow M. (1997) Expression of Fos, Jun, and Krox family proteins in Alzheimer's disease. Exp. Neurol. 147, 316–332 [DOI] [PubMed] [Google Scholar]
- 20. Lu Y., Li T., Qureshi H. Y., Han D., and Paudel H. K. (2011) Early growth response 1 (Egr-1) regulates phosphorylation of microtubule-associated protein Tau in mammalian brain. J. Biol. Chem. 286, 20569–20581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Killick R., Ribe E. M., Al-Shawi R., Malik B., Hooper C., Fernandes C., Dobson R., Nolan P. M., Lourdusamy A., Furney S., Lin K., Breen G., Wroe R., To A. W., Leroy K., et al. (2014) Clusterin regulates β-amyloid toxicity via Dickkopf-1-driven induction of the wnt-PCP-JNK pathway. Mol. Psychiatry 19, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bakalash S., Pham M., Koronyo Y., Salumbides B. C., Kramerov A., Seidenberg H., Berel D., Black K. L., and Koronyo-Hamaoui M. (2011) Egr1 expression is induced following glatiramer acetate immunotherapy in rodent models of glaucoma and Alzheimer's disease. Invest. Ophthalmol. Vis. Sci. 52, 9033–9046 [DOI] [PubMed] [Google Scholar]
- 23. Gatta V., D'Aurora M., Granzotto A., Stuppia L., and Sensi S. L. (2014) Early and sustained altered expression of aging-related genes in young 3xTg-AD mice. Cell Death Dis. 5, e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mukherjee A., and Hersh L. B. (2002) Regulation of amyloid β-peptide levels by enzymatic degradation. J. Alzheimers Dis. 4, 341–348 [DOI] [PubMed] [Google Scholar]
- 25. Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N. P., Gerard C., Hama E., Lee H. J., and Saido T. C. (2001) Metabolic regulation of brain Aβ by neprilysin. Science 292, 1550–1552 [DOI] [PubMed] [Google Scholar]
- 26. Miller B. C., Eckman E. A., Sambamurti K., Dobbs N., Chow K. M., Eckman C. B., Hersh L. B., and Thiele D. L. (2003) Amyloid-β peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 6221–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E. A., Frosch M. P., Eckman C. B., Tanzi R. E., Selkoe D. J., and Guenette S. (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid β-protein, and the β-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sisodia S. S., and St George-Hyslop P. H. (2002) γ-Secretase, Notch, Aβ and Alzheimer's disease: where do the presenilins fit in? Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 29. Kubosaki A., Tomaru Y., Tagami M., Arner E., Miura H., Suzuki T., Suzuki M., Suzuki H., and Hayashizaki Y. (2009) Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 10, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knapska E., and Kaczmarek L. (2004) A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog. Neurobiol. 74, 183–211 [DOI] [PubMed] [Google Scholar]
- 31. Qin X., Jiang Y., Tse Y. C., Wang Y., Wong T. P., and Paudel H. K. (2015) Early growth response 1 (Egr-1) regulates N-methyl-d-aspartate receptor (NMDAR)-dependent transcription of PSD-95 and α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor (AMPAR) trafficking in hippocampal primary neurons. J. Biol. Chem. 290, 29603–29616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woodard-Grice A. V., McBrayer A. C., Wakefield J. K., Zhuo Y., and Bellis S. L. (2008) Proteolytic shedding of ST6Gal-I by BACE1 regulates the glycosylation and function of α4β1 integrins. J. Biol. Chem. 283, 26364–26373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faghihi M. A., Modarresi F., Khalil A. M., Wood D. E., Sahagan B. G., Morgan T. E., Finch C. E., St Laurent G. 3rd, Kenny P. J., and Wahlestedt C. (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piccini A., Borghi R., Guglielmotto M., Tamagno E., Cirmena G., Garuti A., Pollero V., Cammarata S., Fornaro M., Messa M., Colombo L., Salmona M., Perry G., and Tabaton M. (2012) β-amyloid 1–42 induces physiological transcriptional regulation of BACE1. J. Neurochem. 122, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 35. Hardy J., Bogdanovic N., Winblad B., Portelius E., Andreasen N., Cedazo-Minguez A., and Zetterberg H. (2014) Pathways to Alzheimer's disease. J. Intern. Med. 275, 296–303 [DOI] [PubMed] [Google Scholar]
- 36. Chami L., and Checler F. (2012) BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and β-amyloid production in Alzheimer's disease. Mol. Neurodegener. 7, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendrickx A., Pierrot N., Tasiaux B., Schakman O., Brion J. P., Kienlen-Campard P., De Smet C., and Octave J. N. (2013) Epigenetic induction of EGR-1 expression by the amyloid precursor protein during exposure to novelty. PLoS ONE 8, e74305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zimmerman S. M., and Kim S. K. (2014) The GATA transcription factor/MTA-1 homolog egr-1 promotes longevity and stress resistance in Caenorhabditis elegans. Aging Cell 13, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukumoto H., Rosene D. L., Moss M. B., Raju S., Hyman B. T., and Irizarry M. C. (2004) β-Secretase activity increases with aging in human, monkey, and mouse brain. Am. J. Pathol. 164, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim T., Vidal G. S., Djurisic M., William C. M., Birnbaum M. E., Garcia K. C., Hyman B. T., and Shatz C. J. (2013) Human LilrB2 is a β-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science 341, 1399–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., and Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Um J. W., Nygaard H. B., Heiss J. K., Kostylev M. A., Stagi M., Vortmeyer A., Wisniewski T., Gunther E. C., and Strittmatter S. M. (2012) Alzheimer amyloid-β oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat. Neurosci. 15, 1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cissé M., Halabisky B., Harris J., Devidze N., Dubal D. B., Sun B., Orr A., Lotz G., Kim D. H., Hamto P., Ho K., Yu G. Q., and Mucke L. (2011) Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature 469, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kam T. I., Song S., Gwon Y., Park H., Yan J. J., Im I., Choi J. W., Choi T. Y., Kim J., Song D. K., Takai T., Kim Y. C., Kim K. S., Choi S. Y., Choi S., Klein W. L., Yuan J., and Jung Y. K. (2013) FcγRIIb mediates amyloid-β neurotoxicity and memory impairment in Alzheimer's disease. J. Clin. Invest. 123, 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mokin M., Lindahl J. S., and Keifer J. (2006) Immediate-early gene-encoded protein Arc is associated with synaptic delivery of GluR4-containing AMPA receptors during in vitro classical conditioning. J. Neurophysiol. 95, 215–224 [DOI] [PubMed] [Google Scholar]
- 46. Lopez Salon M., Pasquini L., Besio Moreno M., Pasquini J. M., and Soto E. (2003) Relationship between β-amyloid degradation and the 26S proteasome in neural cells. Exp. Neurol. 180, 131–143 [DOI] [PubMed] [Google Scholar]
- 47. James A. B., Conway A. M., and Morris B. J. (2006) Regulation of the neuronal proteasome by Zif268 (Egr1). J. Neurosci. 26, 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ounallah-Saad H., Beeri R., Goshen I., Yirmiya R., Renbaum P., and Levy-Lahad E. (2009) Transcriptional regulation of the murine Presenilin-2 gene reveals similarities and differences to its human orthologue. Gene 446, 81–89 [DOI] [PubMed] [Google Scholar]
- 49. Renbaum P., Beeri R., Gabai E., Amiel M., Gal M., Ehrengruber M. U., and Levy-Lahad E. (2003) Egr-1 upregulates the Alzheimer's disease presenilin-2 gene in neuronal cells. Gene 318, 113–124 [DOI] [PubMed] [Google Scholar]
- 50. Liu C. C., Liu C. C., Kanekiyo T., Xu H., and Bu G. (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harja E., Bucciarelli L. G., Lu Y., Stern D. M., Zou Y. S., Schmidt A. M., and Yan S. F. (2004) Early growth response-1 promotes atherogenesis: mice deficient in early growth response-1 and apolipoprotein E display decreased atherosclerosis and vascular inflammation. Circ. Res. 94, 333–339 [DOI] [PubMed] [Google Scholar]
- 52. Stanzione R., Sciarretta S., Marchitti S., Bianchi F., Di Castro S., Scarpino S., Cotugno M., Frati G., Volpe M., and Rubattu S. (2015) C2238/αANP modulates apolipoprotein E through Egr-1/miR199a in vascular smooth muscle cells in vitro. Cell Death Dis. 6, e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rossner S., Sastre M., Bourne K., and Lichtenthaler S. F. (2006) Transcriptional and translational regulation of BACE1 expression–implications for Alzheimer's disease. Prog. Neurobiol. 79, 95–111 [DOI] [PubMed] [Google Scholar]
- 54. Christensen M. A., Zhou W., Qing H., Lehman A., Philipsen S., and Song W. (2004) Transcriptional regulation of BACE1, the β-amyloid precursor protein β-secretase, by Sp1. Mol. Cell. Biol. 24, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chu J., Zhuo J. M., and Praticò D. (2012) Transcriptional regulation of β-secretase-1 by 12/15-lipoxygenase results in enhanced amyloidogenesis and cognitive impairments. Ann. Neurol. 71, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Bourne K. Z., Ferrari D. C., Lange-Dohna C., Rossner S., Wood T. G., and Perez-Polo J. R. (2007) Differential regulation of BACE1 promoter activity by nuclear factor-κB in neurons and glia upon exposure to β-amyloid peptides. J. Neurosci. Res. 85, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 57. Marwarha G., Raza S., Prasanthi J. R., and Ghribi O. (2013) Gadd153 and NF-κB crosstalk regulates 27-hydroxycholesterol-induced increase in BACE1 and β-amyloid production in human neuroblastoma SH-SY5Y cells. PLoS ONE 8, e70773. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Nowak K., Lange-Dohna C., Zeitschel U., Günther A., Lüscher B., Robitzki A., Perez-Polo R., and Rossner S. (2006) The transcription factor Yin Yang 1 is an activator of BACE1 expression. J. Neurochem. 96, 1696–1707 [DOI] [PubMed] [Google Scholar]
- 59. Sastre M., Dewachter I., Rossner S., Bogdanovic N., Rosen E., Borghgraef P., Evert B. O., Dumitrescu-Ozimek L., Thal D. R., Landreth G., Walter J., Klockgether T., van Leuven F., and Heneka M. T. (2006) Nonsteroidal anti-inflammatory drugs repress β-secretase gene promoter activity by the activation of PPARγ. Proc. Natl. Acad. Sci. U.S.A. 103, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee S. L., Sadovsky Y., Swirnoff A. H., Polish J. A., Goda P., Gavrilina G., and Milbrandt J. (1996) Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 273, 1219–1221 [DOI] [PubMed] [Google Scholar]
- 61. Laird F. M., Cai H., Savonenko A. V., Farah M. H., He K., Melnikova T., Wen H., Chiang H. C., Xu G., Koliatsos V. E., Borchelt D. R., Price D. L., Lee H. K., and Wong P. C. (2005) BACE1, a major determinant of selective vulnerability of the brain to amyloid-β amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 25, 11693–11709 [DOI] [PMC free article] [PubMed] [Google Scholar]