Abstract
Pantothenate kinase is the master regulator of CoA biosynthesis and is feedback-inhibited by acetyl-CoA. Comparison of the human PANK3·acetyl-CoA complex to the structures of PANK3 in four catalytically relevant complexes, 5′-adenylyl-β,γ-imidodiphosphate (AMPPNP)·Mg2+, AMPPNP·Mg2+·pantothenate, ADP·Mg2+·phosphopantothenate, and AMP phosphoramidate (AMPPN)·Mg2+, revealed a large conformational change in the dimeric enzyme. The amino-terminal nucleotide binding domain rotates to close the active site, and this allows the P-loop to engage ATP and facilitates required substrate/product interactions at the active site. Biochemical analyses showed that the transition between the inactive and active conformations, as assessed by the binding of either ATP·Mg2+ or acyl-CoA to PANK3, is highly cooperative indicating that both protomers move in concert. PANK3(G19V) cannot bind ATP, and biochemical analyses of an engineered PANK3/PANK3(G19V) heterodimer confirmed that the two active sites are functionally coupled. The communication between the two protomers is mediated by an α-helix that interacts with the ATP-binding site at its amino terminus and with the substrate/inhibitor-binding site of the opposite protomer at its carboxyl terminus. The two α-helices within the dimer together with the bound ligands create a ring that stabilizes the assembly in either the active closed conformation or the inactive open conformation. Thus, both active sites of the dimeric mammalian pantothenate kinases coordinately switch between the on and off states in response to intracellular concentrations of ATP and its key negative regulators, acetyl(acyl)-CoA.
Keywords: acetyl coenzyme A (acetyl-CoA), allosteric regulation, coenzyme A (CoA), cooperativity, enzyme kinetics, vitamin, X-ray crystallography
Introduction
Pantothenate kinase catalyzes the key regulatory step in CoA biosynthesis in bacteria and mammals (1–3). In mammals, there are four distinct kinases that exhibit tissue-specific expression as follows: PANK1α and PANK1β are splice variants of the PANK1 gene, and PANK2 and PANK3 are encoded by the PANK2 and PANK3 genes, respectively (4, 5). The kinase isoforms possess almost identical catalytic cores but differ in their amino termini that direct the enzymes to different subcellular compartments (6). Isoform 1α is targeted to the nucleus, whereas isoform 1β is associated with endosomes in humans and mice. Isoform 2 of mice is cytosolic, but the PANK2 gene in humans encodes a protein with both nuclear localization and mitochondrial targeting sequences. Isoform 3 is cytosolic in both species. All pantothenate kinases operate by a compulsory ordered mechanism with ATP·Mg2+ as the leading substrate followed by pantothenate (Fig. 1) (7). The principal mechanism for controlling mammalian pantothenate kinase activity is through feedback inhibition by acetyl-CoA, and the isoforms differ in their sensitivity to this regulator (Fig. 1) (4, 8, 9). The 1α and 1β isoforms are least sensitive to inhibition, whereas isoforms 2 and 3 are more potently inhibited by acetyl-CoA. Acetyl-CoA inhibition is competitive with ATP·Mg2+, but acetyl-CoA binds far more tightly than ATP (10). Acyl-carnitines antagonize the inhibition of pantothenate kinases by acetyl-CoA (8).
FIGURE 1.
Catalytic cycle and acyl-CoA inhibition of PANK3. PANK3 operates by a compulsory ordered mechanism with ATP as the leading substrate (7). The PANK3 dimer exists in two distinct conformations. The inactive conformation has an “open” carboxyl-terminal nucleotide binding domain that is stabilized by acetyl-CoA binding (red). The active conformation has a “closed” nucleotide binding domain that is stabilized by ATP binding (green). Acetyl-CoA and ATP binding to PANK3 is competitive (10). Pantothenate binds to the PANK3·ATP·Mg2+ complex, and following catalysis, 4-phosphopantothenate is released followed by ADP. 4-Phosphopantothenate is rapidly converted to CoA and its thioesters, which function as feedback regulators of the enzyme. This work describes the structural basis for the highly cooperative transition between the active (ATP-bound) and inactive (acyl-CoA-bound) conformations, and the structural alterations that occur at each intermediate step in the catalytic cycle. The PDB codes for the determined structures are shown next to the model representations. Although ligand-free PANK3 must exist, the structure(s) has not been determined (red/green state).
The importance of pantothenate kinase to mammalian physiology is highlighted by the phenotypes of knock-out mice and their connection to human disease. Isoform 1 is most highly expressed in the liver, and Pank1−/− mice have lower total hepatic CoA and exhibit fatty acid β-oxidation and glucose homeostasis defects in the fasted state (11). In addition, Pank1−/− Lep−/− mice have dramatically lower blood glucose and insulin levels compared with their diabetic Lep−/− counterparts, which highlights the connection between isoform 1 and glucose homeostasis (12). Consistent with this, an association between polymorphisms in the PANK1 gene and insulin levels was uncovered in a cohort of individuals in Finland (13). It has also been shown that the severe neurodegenerative disease pantothenate kinase-associated neurodegeneration arises from mutations in the human PANK2 gene (14). Unfortunately, Pank2−/− mice do not recapitulate the pantothenate kinase-associated neurodegeneration disease phenotype (15, 16), and this may be due either to differences in the levels of isoform expression in mouse and human brains or to the fact that human PANK2 accumulates in the intra-membrane space in mitochondria, whereas mouse isoform 2 is cytosolic (9). Finally, Pank1−/− Pank2−/− double knock-out mice are unable to metabolize fats and ketones resulting in early postnatal death (16), and Pank1−/− Pank3−/− and Pank2−/− Pank3−/− double knock-out mice are both embryonic lethal. A chemical knock-out of all pantothenate kinases in adult mice resulted in an 80% reduction in hepatic CoA levels and death within days (17). It is clear from these various studies that CoA homeostasis is critical to support mammalian oxidative metabolism and neuronal function.
The crystal structures of PANK1β and PANK3 bound to acetyl-CoA show that acetyl-CoA occupies the ATP binding pocket and threads through the putative pantothenate-binding site to contact the dimer interface (10). The adenine moiety binds atypically and maintains each protomer in an “open” catalytically inactive conformation, whereas the pantothenate moiety of acetyl-CoA blocks access to the substrate binding pocket (10, 18, 19). Thus, acetyl-CoA exploits the common feature of kinases with active sites located between two domains that open to allow access to ATP and substrates and then close to exclude water molecules during catalysis. The goals of this study were to analyze the crystal structures of active PANK3 catalytic complexes, define the movements in the active site that occur during the enzyme's catalytic cycle, and characterize the conformational changes that occur during the transition from the inactive state bound to acetyl-CoA to the catalytically active state. A key finding is that PANK3 is a highly cooperative allosteric enzyme that exploits structural changes at the dimer interface to coordinately switch between active and inactive conformations.
Results
Active Conformation of PANK3
The overall structure of PANK3 has previously been described from crystallographic studies of the inhibited PANK3·acetyl-CoA binary complex (10). These studies revealed that the enzyme contains two αβ domains and forms a parallel dimeric assembly in which the carboxyl-terminal domains create the dimer interface placing the amino-terminal domains at the periphery of the assembly. The adenine and pantothenate-like moieties of acetyl-CoA occupy the ATP-binding site and the putative binding pocket of the pantothenate substrate, respectively (Fig. 2A). Comparison of the inhibited PANK3 complex with the structurally related Staphylococcus aureus PanK·ATP·Mg2+ complex (PDB3 code 2EWS) (19) revealed that the presence of the ribose 3′-phosphate in PANK3·acetyl-CoA complex enforces a distinct binding mode that rotates the adenine ring to form a wedge between the PANK3 nucleotide binding domains that prevents their closure. A predictive model of the closed active conformation of substrate-bound PANK3 suggested how the ATP and the pantothenate might be aligned for transfer of the γ-phosphate with Glu-138 acting as the general base (7).
FIGURE 2.

Comparison of the overall structures of the inactive and active PANK3 conformations. A, inactive open PANK3 conformation stabilized by its interactions with acetyl-CoA that binds in the nucleotide pocket and extends through the substrate-binding site to the dimer interface (PDB code 3MK6). The P-loop is shown in magenta, and acetyl-CoA is shown in cyan. B, active closed PANK3 conformation stabilized by AMPPNP·Mg2+ and pantothenate binding. The nucleotide binding pocket is closed with AMPPNP (gray) engaging the P-loop (magenta) and pantothenate (green) in the substrate pocket. The domain closure in the inactive to active transition is mediated by a rotation around the axis of helix α4 (purple). The ∼40° rotation of helix α1 to the closed conformation breaks α1 into two helical segments (α1a and α1b), and the disordered loop at the carboxyl terminus of helix α1 becomes a short β-ribbon that associates with the outside edge of the subdomain's β-sheet.
Incubation of PANK3 in high concentrations of AMPPNP (a non-hydrolysable ATP analog) and pantothenate prior to crystallization was necessary to remove tightly bound acetyl-CoA present in the purified PANK3 samples. The resulting structure of the active PANK3·AMPPNP·Mg2+·pantothenate quaternary complex is similar to the proposed model based on the S. aureus pantothenate kinase structure (Fig. 2B). Most significantly, the adenine ring of AMPPNP rotates by ∼180° compared with the acetyl-CoA mode of binding to allow closure of the two domains (Fig. 2B). This domain closure in the inactive to active transition is mediated by a 13° rotation around the axis of helix α4. At the active site, the P-loop repositions to interact with AMPPNP, the required Mg2+ cation is clearly visible adjacent to the β- and γ-phosphates of the ATP analog, and Glu-138 is repositioned to act as the general base. The active conformation is further organized by a structural change between residues Ile-33 and Arg-64. Helix α1 within this region rotates by ∼40°, and the disordered loop at its carboxyl terminus (Fig. 2A) creates a short β-ribbon that associates with the outside edge of the domain's β-sheet (Fig. 2B). The latter interaction requires the rotation of the subdomain to create the necessary space and is not possible in the inactive state. The domain closure also breaks helix α1 into two smaller helices, α1a and α1b (Fig. 2B).
PANK3 Catalytic Intermediates
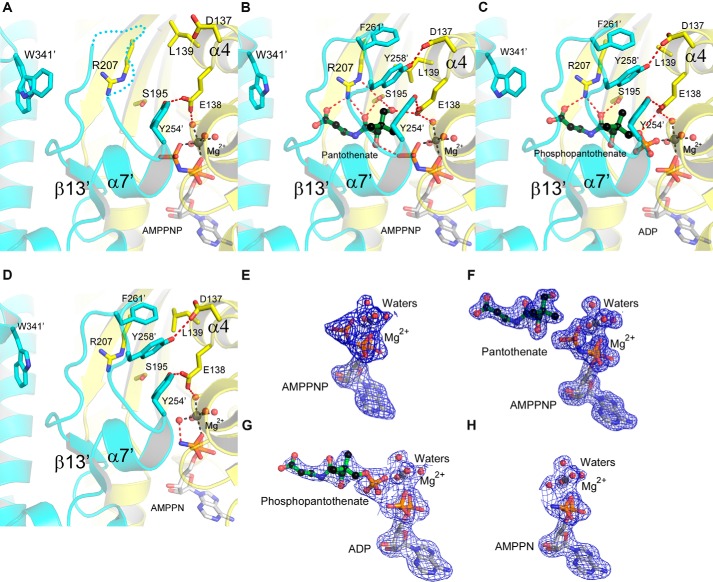
Having grown stable crystals of the active PANK3·AMPPNP·Mg2+ ternary complex (Fig. 3A), we subsequently trapped the remaining three catalytic states of the enzyme: AMPPNP·Mg2+·pantothenate (Fig. 3B), ADP·Mg2+·phosphopantothenate (Fig. 3C), and AMPPN·Mg2+ (Fig. 3D), by manipulating these crystals (see under “Experimental Procedures”). The overall conformation of the four “active” complexes superimpose very well showing that, once in the active conformation, PANK3 can perform the phosphoryl transfer with only minimal structural changes within the active site. Thus, in the ADP·Mg2+·phosphopantothenate product complex, the phosphate on the 4-hydroxyl of the substrate remains close to its original position on AMPPNP and makes the same hydrogen bond interactions (Fig. 3C). The loop between α7′-β13′ from the opposite protomer was disordered in the PANK3·AMPPNP·Mg2+ complex (Fig. 3A). This loop becomes ordered and forms a “lid” that covers and stabilizes the bound pantothenate substrate via residues Tyr-258′ and Phe-261′ in the PANK3·AMPPNP·Mg2+·pantothenate complex (Fig. 3B). The ordering of this loop is apparently synergistic with the movement of helix α4 that repositions residues Asp-137, Glu-138, and Leu-139 to make additional stabilizing interactions adjacent to the pantothenate-binding site. It should be noted that this loop is visible in the PANK3·AMPPN·Mg2+ ternary complex structure without bound pantothenate (Fig. 3D) but is still very mobile. The temperature factors are very high, and the electron density is correspondingly weak. The electron densities contoured at 1σ of the ligands, and structured water molecules in the four active site structures are shown in Fig. 3, E–H.
FIGURE 3.
Substrate/product interactions within the active site during the PANK3 catalytic cycle. A, active site structure of the PANK3·AMPPNP·Mg2+ complex (PDB code 5KPT). B, active site structure of the PANK3·AMPPNP·Mg2+·pantothenate quaternary complex (PDB code 5KPR). C, active site structure of the PANK3·ADP·Mg2+·phosphopantothenate complex (PDB code 5KPZ). D, active site structure of the PANK3·AMPPN·Mg2+ complex (PDB code 5KQ8). The active site is composed of residues derived from ATP-binding protomer shown in yellow, and residues contributed by the opposite protomer are shown in cyan. E–H, 2Fo − Fc electron density maps contoured at 1σ for the ligands and the key structured water molecules in the active site complexes. E, AMPPNP·Mg2+. F, AMPPNP·Mg2+·pantothenate. G, ADP·Mg2+·phosphopantothenate. H, AMPPN·Mg2+.
Role of Glu-138 in PANK3 Catalysis
PANK3 belongs to the ASKHA kinase superfamily, which typically uses either an Asp or Glu residue as the catalytic base to activate the substrate hydroxyl for attack on the γ-phosphate of ATP (20, 21). Glu-138 appears to be the logical candidate for the catalytic base; it is brought into the active site locale by the rotation around helix α4 in the inactive to active conformational change, and Glu-138 moves an additional 1.2 Å closer to pantothenate in the PANK3·AMPPNP·Mg2+·pantothenate complex to more effectively deprotonate the 4-hydroxyl of pantothenate. However, in the PANK3·AMPPNP·Mg2+·pantothenate ternary complex, the 4-hydroxyl is rotated away from the γ-phosphate of AMPPNP and is not positioned appropriately to receive it (Fig. 4A). This repositioning may be due to the presence of AMPPNP rather than ATP in the complex. There is an atypical interaction of the imido group of AMPPNP with the side chain hydroxyl oxygen and the main chain amide nitrogen of Ser-192. The 4-hydroxyl of pantothenate is appropriately positioned in the PANK3·ADP·N7-pantothenamide structure (PDB code 3SMS). The role of Glu-138 was confirmed by biochemical analyses of the PANK3(E138A) mutant. PANK3(E138A) was correctly folded based on its elution position in gel filtration chromatography compared with that of native dimeric PANK3 (Fig. 4B). Kinetic analysis showed that the mutant enzyme was catalytically inactive (Fig. 4C) but was stabilized to thermal denaturation by ATP and thus still bound ATP (Fig. 4D). The circular dichroism spectrum of PANK3(E138A) was the same as PANK3 (Fig. 4E), corroborating the gel filtration and thermal stabilization experiments. In summary, these data confirm that Glu-138 is the catalytic base and lead to the proposed chemical mechanism for the activation of the 4-hydroxyl of pantothenate to facilitate its attack on the γ-phosphate of ATP (Fig. 4F).
FIGURE 4.
Role of Glu-138 in PANK3 catalysis. A, view of the PANK3 active site illustrating the environment of Glu-138 in the PANK3·AMPPNP·Mg2+·pantothenate complex. B, purification of PANK3(E138A) by gel filtration chromatography on a calibrated Sepharose S-200 column (black). The elution profile of PANK3 is shown in red. The purity of the final PANK3(E138A) protein was assessed by gel electrophoresis (inset). C, PANK3(E138A) was catalytically inactive. D, PANK3(E138A) was correctly folded based on the concentration-dependent thermal stabilization of the protein by ATP. These data were compiled from two technical replicates from two independent biological experiments, and the line is the data points fitted to the non-linear Michaelis-Menten equation. A representative experiment comparing the thermal stability of PANK3(E138A) in the presence and absence of 8 mm ATP (inset) is shown. The peaks of the first derivative plots of the thermal denaturation curves identify the temperature at which 50% of the protein is unfolded. The average thermal stabilization was calculated from triplicate measurements, rounded to the nearest degree, and shown in the figure panel inset. E, circular dichroism spectra of PANK3, PANK3(E138A), and PANK3(G19V) show that the mutant proteins were structured. F, “road-kill” diagram of the key interactions in the PANK3 active site and the chemical mechanism for the phosphorylation of pantothenate.
Pantothenate Binding to the Active and Inactive PANK3 Conformations
Comparison of the PANK3·acetyl-CoA and PANK3·AMPPNP·Mg2+·pantothenate structures shows that the pantothenate moiety of CoA lies in the same location and orientation as the pantothenate substrate (Fig. 5A). In both cases, Ser-195, Arg-207, Val-268′, and Ala-269′ form hydrogen bonds with the same atoms of pantothenate. However, the longer acetyl-cysteine portion of acetyl-CoA reaches across the dimer interface to interact with Val-268′, Asn-299′, Tyr-336′, Tyr-340′, and Trp-341′ on the opposite protomer (Fig. 5A). Pantothenate did not increase the thermal stabilization of PANK3 beyond what was achieved in the presence of AMPPNP·Mg2+ alone indicating the weakness of its interaction with the dimer interface (Fig. 5B). Thus, the substrate/protein interactions were not significant enough to further stabilize the protein to thermal denaturation. Pantetheine is another PANK3 substrate that also had little effect on PANK3 stability, increasing the denaturation temperature by only 1 °C (Fig. 5B). However, the N7-pantothenamide substrate significantly stabilized PANK3 by 11 °C (Fig. 5B) in a concentration-dependent manner (Fig. 5C). A comparison of the PANK3·ADP·N7-pantothenamide (PDB code 3SMS) and PANK3·AMPPNP·Mg2+·pantothenate structures provides an understanding of this phenomenon. Unlike pantothenate, the hydrocarbon tail of N7-pantothenamide exits the pantothenate-binding site to interact with Trp-341′ across the dimer interface thereby stabilizing the dimeric assembly (Fig. 5D). The additional structural interactions of N7-pantothenamide with PANK3 compared with pantothenate also explain the potency of N7-pantothenamide as a competitive inhibitor of both mammalian and bacterial pantothenate kinases (22) and its effectiveness as an antibacterial agent against S. aureus (23).
FIGURE 5.
Substrate interactions in PANK3. A, x-ray structure of the PANK3·AMPPNP·Mg2+·pantothenate complex (Pan) (green) is overlaid with the PANK3·acetyl-CoA complex (gray). Pantothenate and the pantothenate portion of acetyl-CoA superimpose and make the same key interactions with Arg-207 and Ser-195. Hydrogen bonds in the PANK3·AMPNP·Mg2+·pantothenate and PANK3·acetyl-CoA structures are shown as red or black dashed lines, respectively. B, representative thermal stabilization profiles displayed as the first derivative of the denaturation curve of PANK3 in the presence of 2 mm AMPPNP·Mg2+ (green), 10 mm pantothenate (Pan) (red), pantetheine (PanSH) (blue), and N-heptylpantothenamide (N7-Pan) (black). The peaks of the first derivative plots of the thermal denaturation curves identify the temperature at which 50% of the protein is unfolded. Thermal stabilization was calculated from triplicate measurements, and the average changes in temperature were rounded to the nearest degree, which were 1 °C for pantetheine and 11 °C for N-heptylpantothenamide. C, concentration-dependent thermal stabilization of PANK3 by pantothenate and N-heptylpantothenamide. These data were compiled from two technical replicates from two independent biological experiments, and the line is the data points fitted to the non-linear Michaelis-Menten equation. D, overlay of the PANK3·AMPPNP·Mg2+·pantothenate complex (green) and the ADP·N-heptylpantothenamide (PDB code 3SMS) structures (orange). The hydrocarbon tail of the N-heptylpantothenamide substrate reaches across the dimer interface to interact with Trp-341′ on helix α10 on the opposite protomer. Pantothenate does not contact helix α10 and Trp-341′ assumes a different conformation.
PANK3 Is an Allosteric Enzyme
A series of experiments was performed to biochemically dissect the cooperative nature of ATP·Mg2+ binding to PANK3 (Fig. 6). First, conventional kinetic analysis of PANK3 using the Hill equation revealed highly cooperative ATP binding with a Hill number of 1.8 and a half-maximal activity (K0.5) occurring at 70 μm ATP (Fig. 6A). This large Hill number approaches the theoretical limit of two for a dimer, which corresponds to the case where the active sites of both protomers are altered in unison. The kinetic analysis also showed that the pantothenate dependence of the reaction was less cooperative than ATP (Fig. 6B). The binding of ATP to PANK3 stabilized the protein to thermal denaturation by a maximum of 9 °C (Fig. 6C), and ATP stabilized PANK3 in a concentration-dependent manner (Fig. 6D). However, the amount of protein required in these experiments (2.5 μm) meant that the K0.5 increased significantly under these conditions, and the cooperativity could not be detected due to the inability to accurately measure thermal stabilization at low ATP concentrations.
FIGURE 6.

Cooperative binding of ATP to PANK3. A, kinetic analysis of PANK3 with respect to the ATP concentration. B, kinetic analysis of PANK3 with respect to the pantothenate concentration. These data were compiled from duplicate technical replicates in two independent biological experiments. The lines are the fit to the Hill equation giving rise to the K0.5 and Hill number (H) parameters shown in the figure panels. C, representative experiment showing that the addition of 8 mm ATP (black) compared with buffer alone (red) stabilizes PANK3 to thermal denaturation. The peaks of the first derivative plots of the thermal denaturation curve identify the temperature at which 50% of the protein is unfolded. Thermal stabilization was calculated from triplicate technical replicates from two independent experiments, and the average changes in temperatures were rounded to the nearest degree and shown in the figure panels. D, concentration dependence of PANK3 thermal stabilization by ATP·Mg2+. These data were compiled from two technical replicates from two independent biological experiments, and the line is the data points fitted to the non-linear Michaelis-Menten equation.
The cooperative nature of PANK3 was further tested by the biochemical analysis of PANK3(G19V) homodimers and PANK3/PANK3(G19V) heterodimers. Gly-19 is located within the P-loop and provides space for the binding of the ATP phosphates (Fig. 7A). Introduction of a bulky valine for Gly-19 should interfere with the binding of ATP and prevent catalysis, but it should not affect the binding of acetyl-CoA that does not involve the P-loop (Fig. 7B). PANK3(G19V) was robustly expressed (Fig. 7C) and was correctly folded based on its identical elution position in gel filtration chromatography as native PANK3 (data not shown). PANK3(G19V) was catalytically inactive as predicted (Fig. 7D). Thermal stabilization confirmed that ATP did not bind the PANK3(G19V) mutant enzyme (Fig. 8A) shifting the curve by 1 °C, whereas PANK3 was stabilized by 9 °C under the same conditions (Fig. 6C). In contrast, acetyl-CoA did bind to PANK3(G19V) and increased the melting temperature by 8 °C (Fig. 8B) showing that mutant protein interacted correctly with acetyl-CoA. The circular dichroism spectrum of PANK3(G19V) was the same as PANK3 (Fig. 4E), corroborating the gel filtration and thermal stabilization experiments.
FIGURE 7.

Characterization of PANK3/PANK3(G19V) heterodimers. A, structure of the PANK3·AMPPNP·Mg2+ complex showing the location of Gly-19 on the P loop. The mutation of Gly-19 to Val-19 introduces a bulky side chain into the P-loop that interferes with ATP binding. B, structure of the PANK3·acetyl-CoA complex illustrating the location of Gly-19 in this conformation. The P-loop has moved away from the adenine pocket, and the introduction of a bulky valine at this position would not be predicted to interfere with acetyl-CoA binding. C, immunoblotting of the PANK3, PANK3/PANK3(G19V) heterodimers, and PANK3(G19V) with anti-His tag and anti-FLAG tag antibodies. In the heterodimer, the PANK3 subunit was His-tagged and the PANK3(G19V) subunit was FLAG-tagged. PANK3 (WW), PANK3/PANK3(G19V) heterodimers (VW), and PANK3(G19V) (VV) immunoblots and CBB is the Coomassie-stained gel with molecular mass markers are shown. D, enzymatic activity of PANK3 compared with PANK3/PANK3(G19V) heterodimers and PANK3(G19V). E, kinetic analysis of PANK3/PANK3(G19V) heterodimers with respect to ATP illustrating the loss of cooperativity and ATP affinity. F, kinetic analysis of PANK3/PANK3(G19V) heterodimers with respect to pantothenate. The kinetic data were derived from two technical replicates from two independent experiments, and the lines are the data fit to the Hill equation as described under “Experimental Procedures.”
FIGURE 8.

Biochemical analysis of ATP and acetyl-CoA binding to PANK3 and PANK3/PANK3(G19V) heterodimers. The peaks of the first derivative plots of the thermal denaturation curves identify the temperatures at which 50% of the protein is unfolded. Representative thermal denaturation curves are shown. The shifts in the thermal denaturation temperatures were calculated from experiments performed in triplicate, and the averages were rounded to the nearest degree and shown in the figure panels. Samples contained buffer alone (red) or buffer plus 10 mm ATP·Mg2+ (black) or 10 μm acetyl-CoA (blue). A, PANK3(G19V) was not stabilized to thermal denaturation by 10 mm ATP·Mg2+. B, acetyl-CoA (10 μm) stabilized PANK3(G19V) to thermal denaturation by 8 °C. C, PANK3/PANK3(G19V) heterodimers exhibited two thermal transitions in the presence of 10 mm ATP·Mg2+. D, acetyl-CoA (10 μm) stabilizes PANK3/PANK3(G19V) heterodimers to thermal denaturation.
We next prepared the PANK3/PANK3(G19V) heterodimer by introducing a His tag on the wild-type subunit and a FLAG tag on the mutant subunit. The two plasmids were co-expressed, and the heterodimers were purified by two sequential affinity chromatography steps. The heterodimer was obtained in high purity, and the presence of both subunits was confirmed by immunoblotting with antibodies against the two different tags (Fig. 7C). Intact mass spectrometry also clearly showed molecular masses corresponding to a His- and FLAG-tagged PANK3 protein lacking the amino-terminal methionine (data not shown). The heterodimer retained catalytic activity (Fig. 7D) but displayed a biphasic thermal denaturation profile in the presence of ATP (Fig. 8C). The two thermal transitions correspond to the denaturation of the PANK(G19V) protomer that cannot interact with ATP and stabilization of the native PANK3 protomer by ATP. The heterodimer also bound acetyl-CoA in a similar fashion to both the wild-type and PANK3(G19V) homodimers (Fig. 8D). Kinetic analysis of the heterodimer showed no cooperativity in ATP binding, a significantly higher ATP K0.5 (3.9 mm; Fig. 7E) compared with the 0.07 mm for wild-type PANK3 (Fig. 6A), and a pantothenate K0.5 similar to that of the wild-type enzyme (Figs. 7F and 6B). These data showed that the cooperative property of PANK3 was lost when one of the subunits cannot bind ATP leading to an enzyme that is catalytically impaired due to the presence of only a single low affinity ATP-binding site.
Structural Basis for PANK3 Cooperativity
A closer comparison of the inactive PANK3 complex bound to acetyl-CoA and the active PANK3 complexes revealed a movement of helix α10 in the carboxyl-terminal domain at the dimer interface that is associated with rearrangements of the loops at the helix termini (Fig. 9). Helix α10 occupies a unique location in the dimer; its amino terminus is adjacent to the ATP-binding site in one protomer, although its carboxyl terminus is adjacent to the binding pocket of the acetyl group of acetyl-CoA in the opposite protomer. Asn-322 and Arg-325 at the amino terminus and Tyr-336, Tyr-340, and Trp-341 at the carboxyl terminus are in very different conformations in the “off” conformation (Fig. 9A; PANK3·acetyl-CoA complex) compared with the “on” conformation (Fig. 9B; PANK3·AMPPNP·Mg2+·pantothenate complex). The paired acetyl-CoA molecules and α10 helices within the inactive PANK3·acetyl-CoA dimer and their counterparts in the active PANK3·AMPPNP·Mg2+·pantothenate complex form a ring that encircles the dimer (Fig. 9, A and B). Helix α10 is the structural element that communicates the acetyl-CoA/ATP binding status between the two protomers. An overlay of the two complexes illustrates the movements and side-chain interactions of helix α10 with both active sites in the two PANK3 conformations (Fig. 9C). The ribose 3′-phosphate of acetyl-CoA in the inactive state and the α-phosphate of AMPPNP in the active state both bind to the amino terminus of helix α10 to stabilize these two alternative conformations. At the carboxyl terminus of helix α10, the acyl group of acyl-CoA interacts with Tyr-340′ and Trp-341′ from the opposite protomer to tilt the helix toward the regulatory ligand, which in turn positions Arg-325 in the active site of the opposite protomer to interact with the 3′-phosphate of CoA instead of the adenine of ATP. This coordinate movement of helix α10 in response to ligands directly connects the active sites of the two PANK3 protomers and explains the high degree of positive cooperativity in PANK3. Tyr-254 also has a key role in allosterically activating the enzyme. It functions across the dimer interface in the active state and positions Glu-138′ from the opposite protomer to act as the general base (Fig. 3).
FIGURE 9.

Realignment of the dimer interface simultaneously alters the conformation of both protomers. View of the communication between the active and regulatory sites of the two PANK3 protomers through ligand interactions with helix α10. Helix α10 of one protomer is colored orange and the other is colored teal. A, in the PANK3·acetyl-CoA complex, the ribose 3′-phosphate of acetyl-CoA interacts with the amino terminus of helix α10 in the ATP-binding site, and the CoA thioester interacts with the carboxyl terminus of helix α10 on the adjacent protomer. B, in the PANK3·AMPPNP·Mg2+·pantothenate complex, ATP makes strong interactions with the amino terminus of helix α10 in the nucleotide-binding site, but the substrate pantothenate does not interact with the carboxyl terminus of helix α10 on the opposite protomer resulting in a different organization of the residues in this region. C, overlay of the PANK3·AMPPNP·Mg2+·pantothenate (green ligands, yellow and blue structure) with the PANK3·acetyl-CoA complex (gray ligands and structure) showing the movement and side-chain rearrangements in helix α10 associated with the inactive (acetyl-CoA bound) and active (ATP bound) conformations that link the active sites of the two protomers.
Interaction of PANK3 with Palmitoyl-CoA
We next examined the effect of chain length on the stringency of PANK3 inhibition and found that the potency of acyl-CoA inhibition increased with increasing chain length (Fig. 10A). Long-chain acyl-CoA bound to PANK3 with very high affinity resulting in the isolation of the PANK3·oleoyl-CoA complex by gel filtration chromatography (Fig. 10B). Octanoyl-(1,N6)-etheno-CoA was prepared as a fluorescent probe to directly assess the binding of acyl-CoA to PANK3 (Fig. 10C). Like ATP, acyl-CoA binding to PANK3 was highly cooperative with a Hill number close to the theoretical maximum of two illustrating that the transition to the inhibited conformation was also highly cooperative. We purified the PANK3·palmitoyl-CoA complex using the gel filtration chromatography, obtained crystals, and determined the structure (Fig. 10D). Overall, the PANK3·palmitoyl-CoA structure is very similar to that of the PANK3·acetyl-CoA complex (Fig. 10E), but there are two significant differences. First, the protomer domains are more closed in the palmitoyl-CoA complex, and the 3′-phosphate of CoA moiety adopts a different conformation that allows this more closed state. In the PANK3·acetyl-CoA complex, the 3′-phosphate forms a salt bridge with Arg-325, but in the PANK3·palmitoyl-CoA complex, it forms a salt bridge with Lys-24. This new conformation is not compatible with the P21 space group observed in the PANK3·acetyl-CoA complex crystals and generates a more loosely packed P23 cubic space group with a solvent content of 65%. This explains why the temperature factors are abnormally high for the PANK3·palmitoyl-CoA complex compared with the other PANK3 structures presented here. Second, the longer palmitoyl group at the distal end of the ligand extends across the dimer interface deeper into the other protomer. The interfacial residues are in similar conformations with the exceptions of Tyr-336′ that rotates by ∼120° to accommodate the invading acyl chain and the adjacent loop spanning residues 165′–172′ that becomes disordered (Fig. 10D). The terminal four carbons of the hydrocarbon tail of palmitoyl-CoA were not visible in the electron density (Fig. 10F). The more extensive interactions across the dimer interface and condensed structure of the PANK3·palmitoyl-CoA complex are consistent with the higher potency of longer chain acyl-CoAs as feedback inhibitors of PANK3 (Fig. 10A).
FIGURE 10.
Cooperative binding of acyl-CoA to PANK3. A, inhibition of PANK3 activity by acetyl-CoA, octanoyl-CoA, and palmitoyl-CoA in the presence of 2 mm ATP. The IC50 values in these experiments were as follows: acetyl-CoA, 4.6 ± 0.6 μm; octanoyl-CoA, 1.7 ± 0.08 μm, and palmitoyl-CoA, 0.5 ± 0.02 μm. B, tight binding of [3H]oleoyl-CoA to PANK3 illustrated by the co-elution of the protein and the labeled ligand on a Sepharose S-200 gel filtration column. C, cooperative binding of the fluorescent probe octanoyl-(1,N6)-etheno-CoA to PANK3. These data were combined from duplicate technical replicates in two independent experiments, and the average values were analyzed using the Hill equation. The lines in the graphs show the fit to the equation using the K0.5 and Hill number parameters. D, structure of the PANK3·palmitoyl-CoA complex illustrating the intercalation of the acyl chain into the dimer interface and the altered interactions in the nucleotide binding pocket (PDB code 5KQD). E, same view of the PANK3·acetyl-CoA complex illustrate the differences with the PANK3·palmitoyl-CoA complex. The two PANK3 protomers are colored blue and yellow. F, 2Fo − Fc electron density map for the palmitoyl-CoA ligand contoured at 1σ.
Discussion
The mammalian pantothenate kinases exist in two distinct, structurally connected conformational states stabilized by ligand binding. The catalytically incompetent (or open) conformation is stabilized by the binding of acetyl(acyl)-CoA, and the catalytically competent (or closed) conformation is stabilized by ATP·Mg2+ binding (Fig. 1). The transition between these two conformational states involves a rotation around helix α4 that closes the amino-terminal nucleotide binding domain. This hinge motion of helix α4 is similar to the rotation of the “αC” helix in protein kinases (24). In the inactive conformation, the catalytic Glu-138 at the end of helix α4 is rotated out of the pocket, and in the active conformation, it rotates into the active site. In addition, helix α10 directly connects the nucleotide- and pantothenate-binding sites providing the structural basis for the coordinated movement of the active sites of both protomers. The high degree of positive cooperativity exhibited by PANK3 means that PANK3 acts like a switch that is either on or off. When PANK3 is switched on, both active sites proceed through the catalytic cycle returning the protein to a state where it may either bind acyl-CoA or ATP (Fig. 1). When bound to an acyl-CoA regulatory ligand, both protomers are inactive (Fig. 1). Although the connection between the active sites via helix α10 is clearly a major element connecting the two active sites, the solvation states of the conformers and structural rearrangements distant from the active site, like the marked differences in the structure of helix α1 in the two PANK3 states, all contribute to the degree of cooperativity in the transition between the on and off states. Although our biochemical and structural analyses provide an accurate description of the key conformations pertinent to PANK3 regulation in vivo, it is not a complete biochemical description of all possible PANK3 states. Whereas the allosteric behavior of PANK3 is most easily understood in light of the Monod, Wyman, and Changeux model for cooperativity that posits that the ligand-free protein exists in two structurally coupled conformational states (25, 26), we have no specific structural data that directly support the existence of two distinct ligand-free states. Instead, the ligand-free state may be more dynamic with one or more intermediate state(s) where localized unfolding may exist (27–29).
The similarities between the mammalian pantothenate kinases suggest that they all operate by a similar mechanism and undergo nearly identical conformational changes during catalysis and inhibition. All four mammalian pantothenate kinases have >85% sequence identity in their catalytic cores, and the x-ray structures of the PANK3·acetyl-CoA and PANK1β·acetyl-CoA complexes overlay with an overall root mean square deviation of only 1.5 Å (10). Likewise, the PANK3·ADP·N7-pantothenamide (PDB code 3SMS) and the PANK2·ADP·pantothenate (PDB code 5E26) complexes are in the closed active conformation. The mammalian pantothenate kinases have different sensitivities to acetyl-CoA inhibition and exhibit tissue-specific expression profiles (6). Minor sequence differences in the flexible loop that covers the pantothenate-binding site are candidate residues that may account for the affinity differences between the isoforms, but additional comparative research is needed to test this idea. Nonetheless, it is clear that the combination of the expression levels and regulatory properties are the major determinants of the tissue CoA levels. The biochemical perspective provided by this work leads to the conclusion that, when the steady state tissue CoA level is reached, most of the cellular pantothenate kinases are bound to regulatory acyl-CoAs and are thus in the off conformation. Although the acyl-CoA feedback regulatory loop is the major determinant of tissue CoA levels, it does not completely explain the regulation of CoA content due to the contributions of allosteric effectors that antagonize the inhibition of the kinases by acetyl-CoA. Acyl-carnitines are an established activating ligand (8), and other signaling molecules like the N-acylethanolamines (7) may also play a role. We have previously demonstrated that small molecule effectors can modulate the activity of PANK3 in a manner that appears to be allosteric in nature (7, 30). The studies reported here have confirmed the allosteric nature of PANK3 and now provide the framework for a structure-based optimization of these small molecule modulators of pantothenate kinase with potential therapeutic applications.
Experimental Procedures
Materials
Sources of supplies were as follows: oligonucleotides were ordered from Thermo Fisher Scientific; d-[1-14C]pantothenate (specific activity, 55 mCi/mol) was from American Radiolabeled Chemicals; Ni2+-NTA resin was from Qiagen; anti-FLAG M2 affinity gel was from Sigma; FLAG peptide was from Hartwell Center at St. Jude Children's Research Hospital; Sypro Orange dye was from Thermo Fisher Scientific; acetyl-CoA, octanoyl-CoA, and palmitoyl-CoA were from Avanti Polar lipids.
Protein Purification
PANK3 was expressed and purified as described earlier (7) and was used for thermal shift and enzyme assays. For generating the PANK3/PANK3(G19V) heterodimer, PANK3 was PCR-cloned into pET21a using NheI-FLAG for 5′-GCTAGCGACTACAAAGACGATGACGACAAGCCATGGTTTGGCATG and HindIII reverse 5′-AAGCTTTCATCAATTTGGCAGCC to generate an amino-terminal FLAG-tagged PANK3. A six-glycine linker between FLAG and the coding sequence of PANK3 was added by site-directed mutagenesis using complementary primers 5′-ATGACGACAAGGGTGGAGGCGGTGGAGGCCCATGGTTTGGCAT. The G19V mutant was generated by site-directed mutagenesis on pET28a-PANK3 by changing the codon for glycine to a valine. Both the FLAG-PANK3 and His-PANK3(G19V) plasmids were co-transformed into BL21(DE3)-pGroE cells and selected on chloramphenicol, kanamycin, and carbenicillin triple selection plates. Cells were grown in Luria Broth and induced at 16 °C overnight with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and the cells were then harvested, resuspended in 20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 10 mm imidazole, 10% glycerol along with a protease mixture, and lysed with a microfluidizer. The cleared lysates were applied onto Ni-NTA columns and purified using standard non-denaturing Ni2+-NTA column chromatography and eluted with 20 mm Tris-HCl, pH 7.9, 500 mm NaCl, 300 mm imidazole, and 10% glycerol. The eluate was diluted with 50 mm Tris-HCl, pH 7.5, to decrease the NaCl concentration to 150 mm and then loaded onto an anti-FLAG affinity matrix. The column was washed with 10 column volumes of Tris-buffered saline and eluted with 5 column volumes of 0.4 mg/ml FLAG-peptide and then with 7 column volumes of 0.1 m glycine, pH 3.5. To neutralize the glycine, fractions were eluted into 300 μl of 1 m Tris-HCl, pH 7.5. The fractions containing the protein were pooled and dialyzed overnight in 20 mm Tris-HCl, pH 7.5, 300 mm NaCl. The proteins were stored in 50% glycerol at −20 °C until further use.
Western Blotting
The purified heterodimer was resolved on a 10% bis-Tris Bio-Rad Criterion gel and transferred to a polyvinylidene difluoride membrane by electroblotting. The His-tagged protein was probed using an anti-His epitope antibody (Santa Cruz Biotechnology Inc.), and the FLAG-tagged protein was probed using an anti-FLAG-tag antibody (Sigma) at a 1:1000 dilution. This was followed by a goat anti-rabbit secondary antibody IgG conjugated with alkaline phosphatase at a 1:5000 dilution and detected using the ECF substrate (GE Healthcare). The fluorescent signal was visualized using a Typhoon 9200 PhosphorImager.
Protein Thermal Shifts
Protein thermal shift experiments were conducted in a mixture containing 100 mm HEPES, pH 7.0, Sypro Orange dye, 10 mm MgCl2, 2.5 μm PANK3 and ATP, or acetyl-CoA at the indicated concentrations. The reaction mixtures (100 μl) were aliquoted into optically clear 96-well PCR plates. The plates were centrifuged and placed in an ABI 7500 real time PCR system for thermal shift analysis. The temperature was ramped from 25 to 95 °C at a rate of 1 °C/min, and fluorescence was measured using a TAMRA filter set (excitation = 560 nm and emission = 582 nm). The data were plotted as fluorescence intensity as a function of temperature and were fit to the first derivative of the Boltzmann sigmoidal equation to determine the temperature corresponding to the denaturation of 50% of the protein in the presence or absence of the indicated ligands. Each experiment was performed in triplicate, and the data represented are an average of three data sets. For determining the K0.5, the changes in melting temperatures from three data sets were plotted against concentration of the ligand, and those data were fit to the Michaelis-Menten equation using GraphPad software. The thermal shift differences were calculated from the averaged data sets and rounded to the nearest degree.
PANK3 Activity Assay
A typical PANK3 activity assay reaction mixture contained 100 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 2.5 mm ATP, 45 μm d-[1-14C]pantothenate (specific activity 22.5 mCi/mmol), and 7.8 ng of PANK3. The assay was done for 10 min at 37 °C unless specified. The K0.5 of ATP was determined by varying the concentration of ATP from 3.9 to 500 μm. The K0.5 of pantothenate was determined by varying the concentration of pantothenate from 5.6 to 180 μm. The PANK3/PANK3(G19V) heterodimer assays were performed with 250 ng of enzyme, and the K0.5 of ATP was determined by varying the concentration of ATP from 0.078 to 10 mm. The K0.5 of pantothenate was determined similar to the PANK3 homodimer. The inhibition assays to determine the IC50 for acetyl-CoA, octanoyl-CoA, and palmitoyl-CoA were performed with 2.5 mm ATP and increasing concentrations of the ligands from 0.078 to 10 μm. All experiments were repeated twice in duplicate, and the data were fitted to the Hill equation using GraphPad software, and the kinetic parameters were an average ± S.E.
Fluorescent Polarization
Octanoyl-CoA was incubated with chloroacetaldehyde solution (Sigma) for 24 h at 37 °C (31–33) and purified using a 2-(2-pyridyl)ethyl column (Supelco) (34). The reaction was monitored, and the purity of the product was assessed using a Waters e2695 Separations Module equipped with a fluorescence detector and by MS/MS using a Finnigan TSQ Quantum mass spectrometer (Thermo Electron). The binding of octanoyl-(1,N6-etheno)-CoA to the PANK3 protein was measured via fluorescence polarization on a HORIBA Jobin Yvon Fluoromax-4 instrument using a 200-μl cuvette. Increasing concentrations of PANK3 were titrated into a 200-μl solution of 1 μm octanoyl-(1,N6-etheno)-CoA in 200 mm Tris-HCl, pH 7.5, 300 mm NaCl. The excitation wavelength was 230 nm, and the emission wavelength was 420 nm with 5-s integration times for the anisotropy measurements. The change in fluorescence polarization versus concentration of PANK3 was fitted using a one site-specific binding with Hill slope model in GraphPad Prism 5. K0.5 is defined as the concentration of PANK3 protein where half the probe is bound, whereas the H is the Hill coefficient that describes the cooperativity of binding.
Crystallization and Structure Determination
DNA encoding PANK3 with four amino acids (FSDD) added to the carboxyl terminus was subcloned into pET-28a, and the purified protein (7) dialyzed against Tris-HCl, pH 7.5, 300 mm NaCl. The PANK3(FSDD) construct was used for x-ray crystallography because higher quality crystals can be obtained (35). To remove bound acetyl-CoA from the PANK3(FSDD) protein sample and create the PANK3·AMPPNP·Mg2+·pantothenate quaternary complex, the protein solution (6.7 mg/ml in 20 mm Tris-HCl, pH 7.5, 0.15 m NaCl, 0.5 mm DTT) was incubated for 2 h with 25 mm AMPPNP, 25 mm d-pantothenate, and 50 mm MgCl2. The ternary complex was crystallized using the sitting-drop vapor-diffusion method at 18 °C with 0.2 m ammonium acetate, 0.1 m citrate, pH 5.6, and 30% PEG 4K, in a 1:1 ratio with the protein complex solution. To obtain the PANK3·AMPPNP·Mg2+ ternary complex structure, the ternary complex crystals were soaked in mother liquor (0.2 m ammonium acetate, 0.1 m citrate, pH 5.6, 50 mm MgCl2, 32% PEG 4K) and 10 mm AMPPNP for 24 h. The PANK3·ADP·Mg2+·phosphopantothenate product quaternary complex structure was obtained by soaking the crystals in 20 mm 4′-phosphopantothenate and 20 mm ADP in mother liquor for 4 days. After six months, the initial PANK3·AMPPNP·Mg2+·pantothenate crystals were found to contain AMPPN, which provided the PANK3·AMPPN·Mg2+ complex structure. All of these crystals were cryoprotected in 25–27% ethylene glycol prior to data collection. The PANK3·palmitoyl-CoA complex was prepared by incubating PANK3 with palmitoyl-CoA and re-purifying the complex by gel filtration chromatography. Crystals of the PANK3·palmitoyl-CoA complex were grown from the purified complex (6.5 mg/ml in 20 mm Tris-HCl, pH 7.5, and 0.2 m NaCl) using 0.5 m ammonium sulfate, 0.1 m Tris-HCl, pH 7.0, 23% PEG 600, and 10% glycerol. These crystals were cryoprotected with 35% glycerol. Diffraction data were collected at the SER-CAT beam line 22-ID at the Advanced Photon Source and processed using HKL2000 (36). Structures were solved by molecular replacement using the program PHASER (37). The protein molecules of PANK3·ADP·N7-pantothenamide complex (PDB code 3SMS) and PANK3·acetyl-CoA complex (PDB code 3MK6) were used as the search models to solve the PANK3·AMPPNP·Mg2+·pantothenate and PANK3·palmitoyl-CoA structures, respectively. Structures were refined and optimized using PHENIX (38) and COOT (39, 40), respectively. Data collection and refinement statistics and the PDB accession codes are presented in Table 1. All structures were rendered with PyMOL (version 1.8, Schrödinger, LLC).
TABLE 1.
Data collection and refinement statistics
| Parameter | AMPPNP·Pan | AMPPNP | ADP·P-Pan | AMPPN | Palmitoyl-CoA |
|---|---|---|---|---|---|
| Data collectiona | |||||
| Wavelength (Å) | 0.9712 | 1.0 | 1.0 | 1.0 | 0.9795 |
| Space group | P3121 | P3121 | P3121 | P3121 | P23 |
| Cell dimensions | |||||
| a (Å) | 99.0 | 97.5 | 99.5 | 98.5 | 121.7 |
| b (Å) | 99.0 | 97.5 | 99.5 | 98.5 | 121.7 |
| c (Å) | 68.8 | 68.2 | 68.5 | 68.5 | 121.7 |
| α (°) | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| γ (°) | 120.0 | 120.0 | 120.0 | 120.0 | 90.0 |
| Resolution range (Å) | 50.0–1.83 | 50.0–2.3 | 50.0–2.4 | 50.0–2.0 | 50.0–2.6 |
| (1.88–1.83) | (2.36–2.30) | (2.46–2.40) | (2.05–2.00) | (2.66–2.60) | |
| Rmergeb | 0.087 (0.466) | 0.097 (0.578) | 0.086 (0.447) | 0.105 (0.992) | 0.091 (0.968) |
| CC1/2 | 0.997 (0.844) | 0.998 (0.800) | 0.999 (0.898) | 0.997 (0.749) | 0.997 (0.423) |
| Completeness (%) | 99.1 (91.2) | 98.8 (89.9) | 98.7 (89.8) | 99.9 (100.0) | 99.9 (99.7) |
| Redundancy | 6.7 (4.9) | 10.4 (5.8) | 8.3 (5.5) | 8.8 (7.0) | 5.4 (4.4) |
| I/σ(I) | 23.5 (2.2) | 22.4 (1.9) | 29.6 (2.9) | 24.6 (2.0) | 21.2 (1.4) |
| Unique reflections | 34,465 | 16,737 | 15,535 | 26,151 | 18,791 |
| Refinement | |||||
| Resolution range (Å) | 36.4–1.83 | 40.0–2.3 | 36.5–2.4 | 42.6–2.0 | 38.5–2.6 |
| No. of reflections | 34,445 | 16,723 | 15,452 | 26,133 | 18.791 |
| No. of atoms | |||||
| Protein | 2,737 | 2,639 | 2,716 | 2,717 | 2,365 |
| Ligand | 51 | 36 | 47 | 54 | 66 |
| Water | 161 | 68 | 46 | 32 | 0 |
| Rwork | 0.171 | 0.176 | 0.173 | 0.181 | 0.188 |
| Rfreec | 0.193 | 0.236 | 0.216 | 0.219 | 0.209 |
| Average B factors (Å2) | |||||
| Protein | 27.9 | 42.6 | 61.5 | 54.9 | 81.7 |
| Ligand | 22.4 | 29.8 | 58.1 | 39.8 | 71.6 |
| Water | 35.8 | 43.9 | 56.8 | 53.6 | |
| Root mean square deviations from ideal values | |||||
| Bond lengths (Å) | 0.007 | 0.010 | 0.010 | 0.008 | 0.010 |
| Bond angles (°) | 0.962 | 1.091 | 0.858 | 1.034 | 1.150 |
| Ramachandran plot | |||||
| Favored (%) | 98.02 | 97.31 | 96.87 | 96.58 | 95.30 |
| Allowed (%) | 1.98 | 2.69 | 3.13 | 3.13 | 4.70 |
| Outliers (%) | 0.00 | 0.00 | 0.00 | 0.28 | 0.00 |
| PDB code | 5KPR | 5KPT | 5KPZ | 5KQ8 | 5KQD |
a Values in parentheses refer to the highest resolution shell.
b Rmerge = Σ|(I−〈I〉)|/Σ(I), where I is the observed intensity.
c Rfree is the R value obtained for a test set of reflections consisting of randomly selected 5% subset of the data set excluded from refinement.
Author Contributions
C. S., M. Y., L. K. S., R. E. L., S. W. W., S. J., and C. O. R. designed the study. M. Y. and S. W. W. determined and interpreted the crystal structures. C. S., S. J., and C. O. R. performed and analyzed the biochemical experiments. J. Y. performed the fluorescent CoA binding experiments. All authors contributed to and approved the manuscript.
Acknowledgments
We thank the Protein Production and Hartwell Center DNA Sequencing Shared Resources for protein expression/purification and DNA sequencing, respectively. We thank Matthew Frank for the preparation of fluorescent octanoyl-CoA, Katie Creed for biochemical assays, and Karen Miller for protein purification.
This work was supported in whole or in part by National Institutes of Health Grants GM034496 (to C. O. R.), GM062896 (to S. J.), and Cancer Center Support Grant CA21765 and the American Lebanese Syrian Associated Charities. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PDB
- Protein Data Bank
- AMPPNP
- adenosine 5′-(β,γ-imido)triphosphate tetralithium salt
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Jackowski S., and Rock C. O. (1981) Regulation of coenzyme A biosynthesis. J. Bacteriol. 148, 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rock C. O., Calder R. B., Karim M. A., and Jackowski S. (2000) Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J. Biol. Chem. 275, 1377–1383 [DOI] [PubMed] [Google Scholar]
- 3. Leonardi R., Zhang Y.-M., Rock C. O., and Jackowski S. (2005) Coenzyme A: back in action. Prog. Lipid Res. 44, 125–153 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y.-M., Rock C. O., and Jackowski S. (2005) Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J. Biol. Chem. 280, 32594–32601 [DOI] [PubMed] [Google Scholar]
- 5. Dansie L. E., Reeves S., Miller K., Zano S. P., Frank M., Pate C., Wang J., and Jackowski S. (2014) Physiological roles of the pantothenate kinases. Biochem. Soc. Trans. 42, 1033–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alfonso-Pecchio A., Garcia M., Leonardi R., and Jackowski S. (2012) Compartmentalization of mammalian pantothenate kinases. PLoS ONE 7, e49509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leonardi R., Zhang Y.-M., Yun M.-K., Zhou R., Zeng F.-Y., Lin W., Cui J., Chen T., Rock C. O., White S. W., and Jackowski S. (2010) Modulation of pantothenate kinase 3 activity by small molecules that interact with the substrate/allosteric regulatory domain. Chem. Biol. 17, 892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leonardi R., Rock C. O., Jackowski S., and Zhang Y.-M. (2007) Activation of human mitochondrial pantothenate kinase 2 by palmitoylcarnitine. Proc. Natl. Acad. Sci. U.S.A. 104, 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leonardi R., Zhang Y. M., Lykidis A., Rock C. O., and Jackowski S. (2007) Localization and regulation of mouse pantothenate kinase 2. FEBS Lett. 581, 4639–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hong B. S., Senisterra G., Rabeh W. M., Vedadi M., Leonardi R., Zhang Y. M., Rock C. O., Jackowski S., and Park H. W. (2007) Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J. Biol. Chem. 282, 27984–27993 [DOI] [PubMed] [Google Scholar]
- 11. Leonardi R., Rehg J. E., Rock C. O., and Jackowski S. (2010) Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS ONE 5, e11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leonardi R., Rock C. O., and Jackowski S. (2014) Pank1 deletion in leptin-deficient mice reduces hyperglycaemia and hyperinsulinaemia and modifies global metabolism without affecting insulin resistance. Diabetologia 57, 1466–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., Sovio U., Ruokonen A., Laitinen J., Jakkula E., Coin L., et al. (2009) Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou B., Westaway S. K., Levinson B., Johnson M. A., Gitschier J., and Hayflick S. J. (2001) A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat. Genet. 28, 345–349 [DOI] [PubMed] [Google Scholar]
- 15. Kuo Y. M., Duncan J. L., Westaway S. K., Yang H., Nune G., Xu E. Y., Hayflick S. J., and Gitschier J. (2005) Deficiency of pantothenate kinase 2 (Pank2) in mice leads to retinal degeneration and azoospermia. Hum. Mol. Genet. 14, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia M., Leonardi R., Zhang Y. M., Rehg J. E., and Jackowski S. (2012) Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS ONE 7, e40871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y. M., Chohnan S., Virga K. G., Stevens R. D., Ilkayeva O. R., Wenner B. R., Bain J. R., Newgard C. B., Lee R. E., Rock C. O., and Jackowski S. (2007) Chemical knockout of pantothenate kinase reveals the metabolic and genetic program responsible for hepatic coenzyme A homeostasis. Chem. Biol. 14, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leonardi R., Chohnan S., Zhang Y.-M., Virga K. G., Lee R. E., Rock C. O., and Jackowski S. (2005) A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J. Biol. Chem. 280, 3314–3322 [DOI] [PubMed] [Google Scholar]
- 19. Hong B. S., Yun M. K., Zhang Y.-M., Chohnan S., Rock C. O., White S. W., Jackowski S., Park H. W., and Leonardi R. (2006) Prokaryotic type II and type III pantothenate kinases: the same monomer fold creates dimers with distinct catalytic properties. Structure 14, 1251–1261 [DOI] [PubMed] [Google Scholar]
- 20. Bork P., Sander C., and Valencia A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheek S., Zhang H., and Grishin N. V. (2002) Sequence and structure classification of kinases. J. Mol. Biol. 320, 855–881 [DOI] [PubMed] [Google Scholar]
- 22. Virga K. G., Zhang Y.-M., Leonardi R., Ivey R. A., Hevener K., Park H.-W., Jackowski S., Rock C. O., and Lee R. E. (2006) Structure-activity relationships and enzyme inhibition of pantothenamide-type pantothenate kinase inhibitors. Bioorg. Med. Chem. 14, 1007–1020 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y.-M., Frank M. W., Virga K. G., Lee R. E., Rock C. O., and Jackowski S. (2004) Acyl carrier protein is a cellular target for the antibacterial action of the pantothenamide class of pantothenate antimetabolites. J. Biol. Chem. 279, 50969–50975 [DOI] [PubMed] [Google Scholar]
- 24. Papaleo E., Saladino G., Lambrughi M., Lindorff-Larsen K., Gervasio F. L., and Nussinov R. (2016) The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 116, 6391–6423 [DOI] [PubMed] [Google Scholar]
- 25. Monod J., Wyman J., and Changeux J. P. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 26. Changeux J. P. (2012) Allostery and the Monod-Wyman-Changeux model after 50 years. Annu. Rev. Biophys. 41, 103–133 [DOI] [PubMed] [Google Scholar]
- 27. Motlagh H. N., Wrabl J. O., Li J., and Hilser V. J. (2014) The ensemble nature of allostery. Nature 508, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo J., and Zhou H. X. (2016) Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., and Nussinov R. (2016) Allostery: An overview of its history, concepts, methods, and applications. PLoS Comput. Biol. 12, e1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma L. K., Leonardi R., Lin W., Boyd V. A., Goktug A., Shelat A. A., Chen T., Jackowski S., and Rock C. O. (2015) A high-throughput screen reveals new small-molecule activators and inhibitors of pantothenate kinases. J. Med. Chem. 58, 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsu K. H., and Powell G. L. (1975) Inhibition of citrate synthase by oleoyl-CoA: a regulatory phenomenon. Proc. Natl. Acad. Sci. U.S.A. 72, 4729–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrio J. R., Secrist J. A. 3rd, and Leonard N. J. (1972) A fluorescent analog of nicotinamide adenine dinucleotide. Proc. Natl. Acad. Sci. U.S.A. 69, 2039–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Secrist J. A. 3rd, Barrio J. R., Leonard N. J., Villar-Palasi C., and Gilman A. G. (1972) Fluorescent modification of adenosine 3′,5′-monophosphate: spectroscopic properties and activity in enzyme systems. Science 177, 279–280 [DOI] [PubMed] [Google Scholar]
- 34. Zano S. P., Pate C., Frank M., Rock C. O., and Jackowski S. (2015) Correction of a genetic deficiency in pantothenate kinase 1 using phosphopantothenate replacement therapy. Mol. Genet. Metab. 116, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mottaghi K. (2013) Structure-based Development of Vitamin B5 Analogs and Evaluation of Their Antimicrobial Efficiency against S. aureus and E. coli. M.Sc. thesis, University of Toronto [Google Scholar]
- 36. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., et al. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]