Abstract
Chibby1 (Cby1) is a small, conserved coiled-coil protein that localizes to centrioles/basal bodies and plays a crucial role in the formation and function of cilia. During early stages of ciliogenesis, Cby1 is required for the efficient recruitment of small vesicles at the distal end of centrioles to facilitate basal body docking to the plasma membrane. Here, we identified family with sequence similarity 92, member A (FAM92A) and FAM92B, which harbor predicted lipid-binding BAR domains, as novel Cby1-interacting partners using tandem affinity purification and mass spectrometry. We found that in cultured cell lines, FAM92A colocalizes with Cby1 at the centrioles/basal bodies of primary cilia, while FAM92B is undetectable. In airway multiciliated cells, both FAM92A and -92B colocalize with Cby1 at the base of cilia. Notably, the centriolar localization of FAM92A and -92B depends largely on Cby1. Knockdown of FAM92A in RPE1 cells impairs ciliogenesis. Consistent with the membrane-remodeling properties of BAR domains, FAM92A and -92B in cooperation with Cby1 induce deformed membrane-like structures containing the small GTPase Rab8 in cultured cells. Our results therefore suggest that FAM92 proteins interact with Cby1 to promote ciliogenesis via regulation of membrane-remodeling processes.
INTRODUCTION
Cilia are evolutionarily conserved microtubule-based structures that project from the apical surface of many different cell types and function in a wide range of essential biological processes (1–3). Primary cilia, which are found on a variety of cell types, are generally nonmotile and contain a 9+0 axonemal microtubule arrangement. They are involved in chemo- and mechanosensation, photoreception, and intracellular signaling. In contrast, motile multicilia consist of a 9+2 axonemal structure and are present in a restricted set of tissues such as the airways, oviducts, and brain ventricles. The synchronized beating of multicilia generates fluid flow over epithelia, which is important for clearing airway mucus and debris, transporting ova from the ovary to the uterus, and circulating cerebrospinal fluid in the brain. Ciliary dysfunction is associated with a growing class of pleiotropic disorders termed ciliopathies (1–3). Defective primary cilia have been linked to various diseases such as polycystic kidney disease (PKD) and Bardet-Biedl syndrome (BBS). Their clinical features are variable but include reversal of body organs (situs inversus), retinal degeneration, mental retardation, and cystic kidney, liver, and pancreas. On the other hand, defects in multicilia are most prominently associated with primary ciliary dyskinesia (PCD). PCD patients typically display chronic respiratory infections, infertility, and hydrocephalus (4).
As cells exit the cell cycle, cilia are assembled from the basal body, which is derived from the mother centriole. The mother centrioles are distinguished from immature daughter centrioles by the presence of accessory structures, including the subdistal and distal appendages at their distal end. While the subdistal appendages anchor cytoplasmic microtubules, the distal appendages (also called “transition fibers” at the ciliary base) are thought to be critical for the recruitment of small vesicles and subsequent docking of basal bodies to the plasma membrane (5–8). Since no protein synthesis occurs in cilia, ciliary proteins are transported from the cell body via polarized vesicle trafficking (9, 10). The extension and maintenance of cilia require intraflagellar transport (IFT), a bidirectional transport system along the axonemal microtubules (11). The precise functions of ciliary components and the molecular mechanisms of ciliogenesis remain poorly understood.
Chibby1 (Cby1) is a conserved 15-kDa coiled-coil protein that predominantly localizes to mother centrioles/basal bodies and plays a pivotal role in the formation and function of cilia (6, 12–19). Cby1 knockout (Cby1-KO) mice display several hallmarks of ciliary defects, including chronic upper airway infection (12), subfertility, and polycystic kidneys (15) as well as polydactyly and hydrocephalus at low frequencies. In Drosophila melanogaster, Cby1 is expressed in sensory neurons and male germ cells, the only ciliated cell types in this organism, and is required for the proper formation of neuronal cilia and sperm flagella, respectively (16). Similarly, in Xenopus laevis, Cby1 is indispensable for ciliogenesis in multiciliated cells of the embryonic epidermis (17). These studies highlight a critical, evolutionarily conserved function for Cby1 in ciliogenesis. More recently, using superresolution microscopy and immuno-transmission electron microscopy (TEM), we demonstrated that in airway multiciliated cells, the Cby1 protein localizes to the base of mature cilia as a ring with a diameter of 300 nm and a height of 100 nm, which is representative of a distal appendage/transition fiber protein (6). Cby1 rings are slightly smaller and positioned more apically than the rings of the distal appendage protein CEP164 (7, 20, 21). During early differentiation stages of multiciliated cells, Cby1 is recruited to the distal appendages of nascent centrioles through its physical interaction with CEP164. Cby1 then interacts with the membrane trafficking machinery component Rabin8, a guanine nucleotide exchange factor (GEF) for the small GTPase Rab8, to stabilize a CEP164-Rabin8 complex. This promotes the recruitment of Rab8 and small vesicles for the efficient assembly of a large membranous cap called the “ciliary vesicle” (CV). Subsequently, the CV is thought to fuse with apical membranes (22), thereby facilitating efficient basal body docking. However, the precise mechanisms by which Cby1 regulates vesicle fusion and trafficking during ciliogenesis remain largely unknown.
Here, we report the identification and characterization of novel Cby1-interacting proteins, family with sequence similarity 92, member A (FAM92A) and FAM92B. FAM92 proteins contain predicted Bin/amphiphysin/Rvs (BAR) domains that are known to bind to lipid membrane surfaces and generate membrane curvature (23–26). BAR domain proteins have been shown to play diverse roles in endocytosis, vesicle fission and fusion, and regulation of the actin cytoskeleton (23–26). We found that the BAR domains of FAM92A and -92B are responsible for binding to Cby1 and also mediate their homo- and heterodimerization. FAM92A and -92B colocalize with Cby1 at the mother centrioles/basal bodies of cilia. FAM92A is the predominant family member in human retinal pigment epithelial (RPE1) cells, and small interfering RNA (siRNA)-mediated knockdown (KD) of FAM92A abrogates the formation of primary cilia. Notably, the coexpression of FAM92 proteins and Cby1 in U2OS cells induces either large membrane globule-like or extensive membrane tubule-like structures containing Rab8. Our data suggest that FAM92A and -92B collaborate with Cby1 to facilitate ciliogenesis, likely through membrane remodeling.
MATERIALS AND METHODS
Mice and MTEC preparation.
The Cby1-KO mouse line was generated by replacing the entire coding region with a neomycin cassette, as described previously (12), and was maintained on a mixed C57BL/6J-129/SvJ background. All mice were handled according to NIH guidelines, and all experimental procedures were approved by the Institutional Animal Care and Use Committee of Stony Brook University.
Mouse tracheal epithelial cell (MTEC) cultures were established on membranes under air-liquid interface (ALI) conditions as described previously (6, 27). Briefly, tracheas were excised from 2- to 6-month-old wild-type Cby1 (Cby1-WT) and Cby1-KO mice, epithelial cells were harvested by pronase digestion and seeded onto collagen-coated permeable polycarbonate or polyester Transwell membranes (6.5-mm diameter; Corning), and the cells proliferated to confluence in growth medium. Subsequently, an ALI was created by removing the apical chamber medium and switching the medium in the lower compartment to differentiation medium. For viral infection of MTECs, isolated epithelial cells were mixed with lentiviral supernatants (1:1) in the presence of 10 μg/ml protamine sulfate (Sigma) at the time of seeding.
Plasmids.
pCS2+Flag-Cby1 and pCS2+Flag-Cby1Δ1–22 (28), pCS2+Flag-Cby1-4A and pCS2+HA-Cby1 (29), maltose-binding protein (MBP)–Cby1 (30), human Cby1 short hairpin RNA (shRNA) (14), hemagglutinin (HA)-Rab8a and HA–Rab8a-T22N (6), and HA–arfaptin-1 and HA–arfaptin-2 (31) expression plasmids were described previously. Tandem affinity purification (TAP)-tagged Cby1 and green fluorescent protein (GFP) expression constructs were generated by PCR amplification of the individual coding sequences, followed by subcloning into an N-terminally TAP-tagged vector. Subsequently, the TAP-Cby1 and TAP-GFP fusions were amplified by PCR and ligated into the tetracycline-inducible pcDNA5/FRT/TO vector with a hygromycin resistance cassette (Invitrogen). Full-length human FAM92A (catalog number HsCD00333592) and FAM92B (catalog number HsCD00340837) plasmids were purchased from the Harvard PlasmID Repository. For the generation of Flag-, HA-, and GFP-tagged FAM92 expression constructs, the corresponding inserts were amplified by PCR and subcloned into pCS2+Flag or pCS2+C-Flag, pCS2+HA, and pCS2+GFP, respectively. FAM92-6E mutants were generated by PCR-based site-directed mutagenesis and subcloned into pCS2+Flag. A cDNA for Cby1 lacking the N-terminal half (amino acids [aa] 1 to 63) (Cby1-ΔN) or for GFP was amplified by PCR and ligated into pCS2+Flag. A His-FAM92A bacterial expression plasmid was constructed with pET28a (Novagen). Lentiviral expression constructs for Flag-tagged FAM92 were created by inserting PCR-amplified cDNAs into the lentiviral transfer vector pEF1α-IRES-EGFP (6, 18).
Cell culture, transfection, and lentiviral production.
HEK293T, U2OS, RPE1, and GFP-centrin1-expressing RPE1 (14) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) or DMEM-GlutaMAX with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin. Transfections were performed by using polyethyleneimine (PEI; Sigma). Lentiviruses were produced by transient transfection of HEK293T cells according to standard protocols.
siRNA-mediated knockdown of CEP164 and FAM92A was performed as described previously (6, 14). Briefly, U2OS or RPE1 cells seeded onto coverslips in a 12-well plate were transfected with or without 40 pmol siRNA duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Mock-transfected controls were treated identically except for the omission of siRNA. RPE1 cells were serum starved for 48 h to induce ciliogenesis. siRNAs were synthesized by Sigma. The CEP164 siRNA sequence was described previously (6). The human FAM92A siRNA target sequences were (i) 5′-GGATCAACAAGCAGAAGAT-3′ (nucleotide positions 801 to 819) and (ii) 5′-GCAGAAACGGAATTACAGA-3′ (nucleotide positions 439 to 457). KD of Cby1 in U2OS cells was achieved by transient transfection of a human Cby1-specific SureSilencing shRNA (SABiosciences) as described previously (14), and a scrambled shRNA was used as a negative control.
TAP and mass spectrometry of Cby1-interacting proteins.
A HEK293 stable cell line expressing TAP-Cby1 or TAP-GFP was generated by using the HEK293 Flp-In T-Rex system (Invitrogen). Cells were seeded into 20 15-cm dishes and grown to 60% confluence. Protein expression was then induced with 0.02 μg/ml tetracycline, which yielded TAP-Cby1 expression levels similar to those of the endogenous protein. After 48 h of induction, cells were washed three times with ice-cold phosphate-buffered saline (PBS), and TAP was performed as described previously (32). In brief, these cells were collected by scraping and resuspended in 10 ml lysis buffer containing 125 mM KCl, and Cby1-interacting proteins were purified by using 150 μl IgG-Sepharose beads (Sigma) and 150 μl calmodulin affinity resin (Agilent Technologies). Final eluates were run 1.5 to 2 cm into a NuPAGE Novex 8% Tris-acetate midigel (Invitrogen). The gel was then silver stained, and the entire stained protein area was excised, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Proteomics Center at Stony Brook University.
MBP pulldown assays.
MBP- and His-tagged recombinant proteins were expressed in Escherichia coli strains BL21 and BL21(DE3) and purified by using amylose beads (New England BioLabs) and Ni-nitrilotriacetic acid (Ni-NTA) His-Bind resin (Novagen), respectively, according to the manufacturers' instructions. MBP pulldown assays were performed as previously described (33, 34).
Co-IP and Western blotting.
Coimmunoprecipitation (co-IP) assays and immunoblotting were performed as described previously (28, 33). Briefly, for co-IP assays, transfected HEK293T cells were harvested in ice-cold lysis buffer (20 mM Tris-HCl [pH 8.0], 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and 10% glycerol) with a protease inhibitor cocktail (Sigma) and incubated for 20 min on ice with intermittent agitation. Cell lysates were cleared by centrifugation at 12,000 rpm for 30 min at 4°C. The supernatants were then incubated with 1 μg of primary antibody as indicated for 1 h at 4°C, followed by the addition of protein A/G beads (Sigma) and rotation for 1 h. The beads were collected and washed twice with 1 ml of ice-cold lysis buffer before SDS-PAGE. The primary antibodies used for co-IP assays and Western blotting were as follows: mouse Flag M2 (Sigma), rat HA (Roche), rat HA-peroxidase (Roche), rabbit FAM92A (Proteintech), rabbit FAM92B (Sigma), rabbit Cby1 (30), rabbit CEP164 (Sigma), MBP (New England BioLabs), rabbit GFP (generated by Open Biosystems), and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Meridian Life Science). Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Jackson ImmunoResearch.
Immunofluorescence (IF) microscopy.
RPE1 or U2OS cells were seeded onto coverslips in 12-well plates and fixed with cold methanol. MTECs on supported membranes were fixed with methanol-acetone (1:1), cut from supports into quarters, and processed for immunostaining as described previously (6, 18, 27). After permeabilization and blocking, the fixed samples were incubated with primary antibodies overnight at 4°C, followed by secondary antibodies conjugated with cyanine 3 (Cy3) and Cy5, fluorescein isothiocyanate (FITC), Texas Red, and DyLight 488 and 549 (Jackson ImmunoResearch) or Alexa Fluor 647 (Invitrogen) fluorescent dye. The primary antibodies used were as follows: mouse Flag M2 (Sigma), mouse HA (Proteintech), rabbit FAM92A (Proteintech), rabbit FAM92B (Sigma), mouse Cby1 8-2 (35), mouse γ-tubulin (G-tub; Sigma), mouse CEP135 (36), mouse centrobin (Abcam), rabbit CEP164 (5, 20), and mouse acetylated α-tubulin (A-tub; Sigma). For double staining with rabbit FAM92A and rabbit CEP164 antibodies as shown in Fig. 3C, RPE1 cells were first labeled with FAM92A antibody, followed by incubation with Cy3-conjugated secondary antibody. After images of FAM92A staining were taken, the same cells identified on a grid dish were labeled with primary CEP164 and secondary Cy5 antibodies. No FAM92A staining with Cy5 was detectable after incubation of Cy3-labeled FAM92A with Cy5-conjugated secondary antibody alone. Labeled samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei before mounting with Fluoromount-G (Southern Biotechnology).
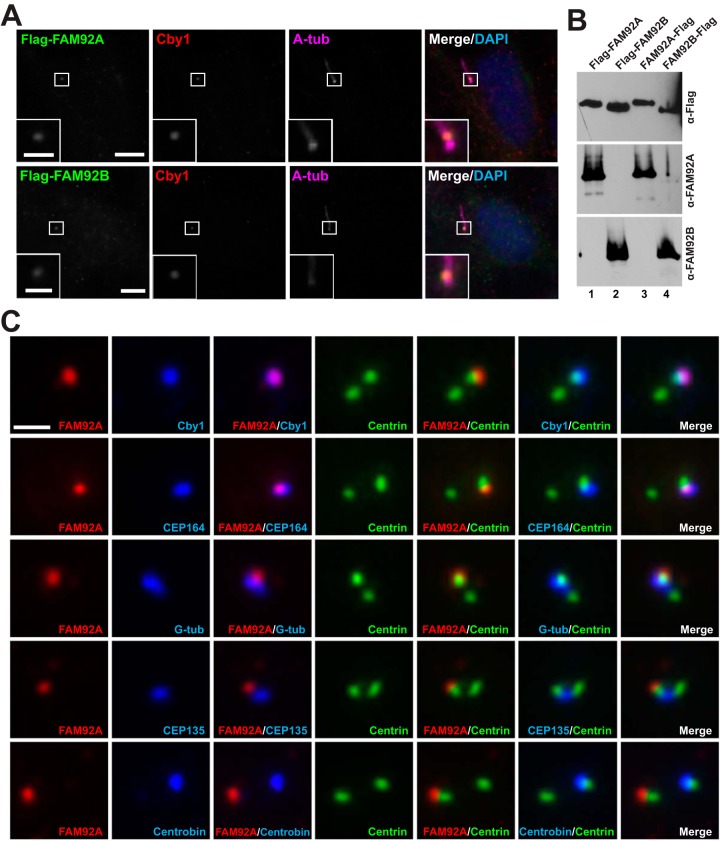
FIG 3.
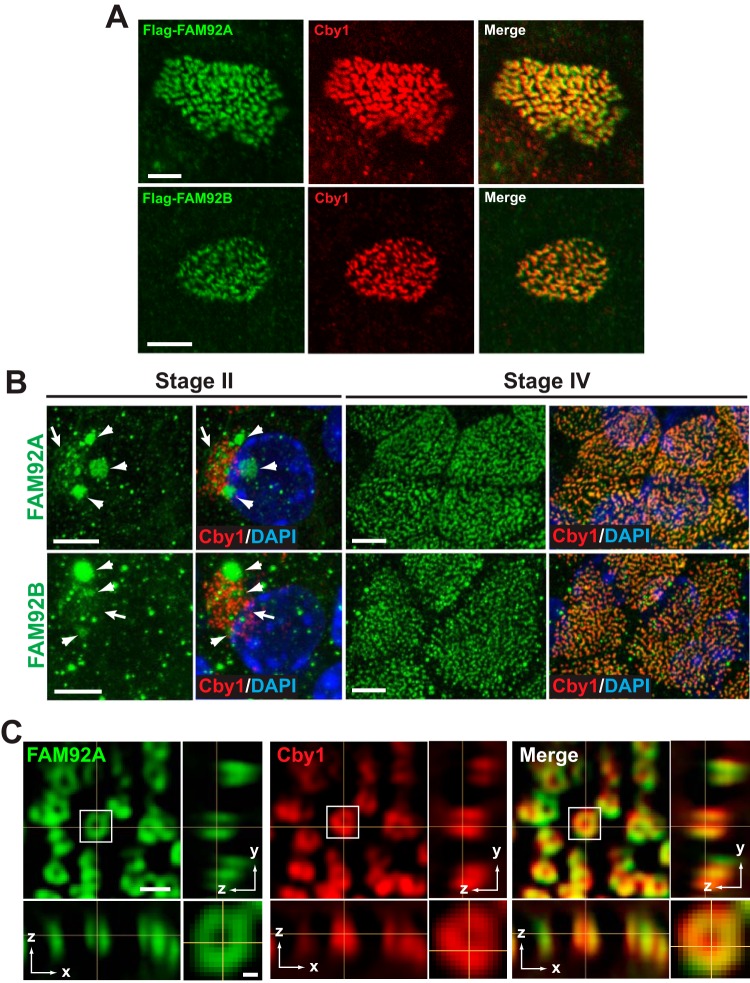
FAM92 proteins colocalize with Cby1 at mother centrioles/basal bodies. (A) RPE1 cells were infected with lentiviruses expressing Flag-tagged FAM92A or -92B and fixed after 48 h of serum starvation to induce ciliogenesis. These cells were triple stained with antibodies against the Flag tag, Cby1, and the centriolar/ciliary axoneme marker A-tub. Merged images are shown with DAPI nuclear stain. The insets show enlarged views of the boxed areas. Bars, 10 μm; bars in the insets, 3 μm. (B) Specificity of the FAM92A and -92B antibodies. Cell lysates were prepared from HEK293T cells expressing Flag-tagged FAM92A or -92B and subjected to Western blotting with FAM92A or -92B antibody as shown. Both N- and C-terminally tagged proteins were used to ensure that tagging did not interfere with antibody binding. The blot was also probed with Flag antibody to confirm equal loading of Flag-tagged proteins. (C) Cycling RPE1 cells expressing GFP-tagged centrin1 were costained for FAM92A and Cby1, G-tub (centrosomes), CEP135 (proximal ends of centrioles), centrobin (daughter centrioles), or CEP164 (distal appendages of mother centrioles). Bar, 1 μm.
Samples were viewed at room temperature with an LSM510 confocal microscope (Zeiss) with a 63×/1.4-numerical-aperture (NA) or a 100×/1.4-NA objective or with a DMI6000B epifluorescence microscope (Leica) with an HCX PL Fluotar 100×/1.3-NA oil objective. The LSM510 and DMI6000B microscopes were equipped with an AxioCam MRm digital camera with AxioVision software (Carl Zeiss) and a DFC300FX camera with Application Suite software (Leica), respectively. For structured illumination microscopy (SIM) imaging, specimens were analyzed by using a Nikon SIM (N-SIM) instrument with a 100×/1.49-NA objective. The N-SIM instrument was equipped with an Andor iXon3 897 electron multiplying charge-coupled device (EMCCD) camera (Andor Technology) with NIS-Elements image analysis software (Nikon).
Statistical analysis.
Two-tailed Student's t test was used for data analysis, and a P value of <0.05 was considered significant.
RESULTS
Identification of FAM92A as a Cby1-interacting protein.
To elucidate the role of Cby1 in ciliogenesis, we attempted to isolate novel Cby1-interacting proteins using TAP. The TAP tag consists of two IgG-binding domains of Staphylococcus aureus protein A and a calmodulin-binding peptide (37). Stable tetracycline-inducible HEK293 cells for TAP-Cby1 or control TAP-GFP were grown in the presence of tetracycline. Cell lysates were then subjected to TAP, and Cby-binding candidates in the final eluates were identified by proteomic analysis as described in Materials and Methods.
All identified proteins are listed in Table S1 in the supplemental material. Thirty-seven candidate Cby1 interactors were specifically purified with TAP-Cby1 but not with TAP-GFP. Among them were multiple isoforms of 14-3-3 proteins, which are known Cby1-binding partners (28), thereby validating our approach. Interestingly, cilium-related proteins such as tubulin family members and DZIP1/Iguana were specifically purified with TAP-Cby1. DZIP1 has been shown to localize to mother centrioles and to play an essential role in primary cilium formation (38–40). It also binds to the Gli3 transcription factor to regulate Hedgehog signaling (38–40). Further inspection of the list led us to focus on FAM92A for the following reasons: (i) its role in ciliogenesis is unknown; (ii) it contains a putative lipid-binding BAR domain, which is consistent with the localization and functional association of Cby1 with membranes (6); and (iii) it colocalizes with Cby1 at mother centrioles (see Fig. 3).
FAM92A is a 33-kDa protein and harbors a putative BAR domain that is known to bind to membranes and induce their curvature (Fig. 1A) (23–26). Similarly to Cby1, FAM92A is highly conserved in all ciliated unikont organisms (except for nematodes) but missing in most bikonts. BAR domain-containing proteins are implicated in a diverse set of biological processes such as endocytosis, vesicle fission and fusion, and regulation of the actin cytoskeleton (23–26). In Xenopus laevis, FAM92A is expressed throughout embryonic development, and its overexpression or morpholino oligonucleotide (MO)-mediated depletion in embryos causes severe developmental defects (41). Studies from that same laboratory also demonstrated that ectopic expression of FAM92A affects the growth of human cultured cells (42, 43). The other family member, FAM92B, has a predicted molecular mass of 35 kDa and displays 52% amino acid identity and 75% similarity to FAM92A within the BAR domain, although the rest of the protein is diverged. FAM92B is conserved only in vertebrates, and its biological function is currently unknown.
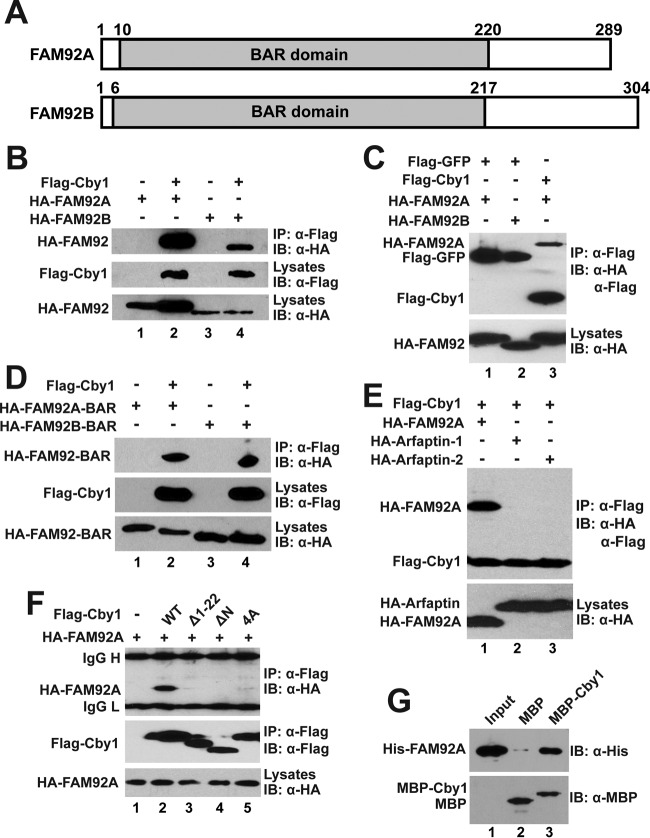
FIG 1.
Interactions between Cby1 and FAM92 proteins. (A) Schematic representation of the domain structures of human FAM92A and -92B. The conserved BAR domains are shaded with the amino acid positions indicated above. (B to F) HEK293T cells were transfected with the indicated plasmids, and cell lysates were immunoprecipitated with Flag antibody and detected with HA antibody. Cell lysates and immunoprecipitates were also probed with the indicated HA and Flag antibodies to confirm stable protein expression. (B) FAM92A and -92B interact with Cby1. (C) FAM92 proteins specifically bind to Cby1 but not to GFP. (D) The BAR domains of FAM92A and -92B are sufficient for Cby1 binding. (E) Cby1 does not interact with the BAR domain-containing proteins arfaptin-1 and arfaptin-2. (F) Both the N- and C-terminal regions of Cby1 are involved in FAM92 binding. (G) FAM92A directly binds to Cby1. Bacterially produced MBP or an MBP-Cby1 fusion was incubated with His-tagged FAM92A. The protein complex was then pulled down with amylose resin and subjected to Western blotting with FAM92A antibody. The input lane was loaded with 1/10 of the amount of His-FAM92A used in the binding reaction mixture. Western blotting with MBP antibody indicated that similar amounts of MBP and MBP-Cby1 were pulled down by amylose resin. IB, immunoblotting.
FAM92A and -92B bind to Cby1 through their BAR domains.
To confirm the interaction between FAM92A and Cby1 and examine if FAM92B binds to Cby1, we performed co-IP assays. Cell lysates from HEK293T cells expressing Flag-Cby1 and HA-tagged FAM92A or -92B were immunoprecipitated with Flag antibody and analyzed by Western blotting using an HA antibody. As shown in Fig. 1B, both FAM92A and -92B specifically interacted with Cby1. FAM92A and -92B did not interact with the GFP control under the same conditions (Fig. 1C). Next, we tested whether the BAR domains of FAM92 proteins are sufficient for Cby1 binding. As determined by co-IP assays, the BAR domains of both FAM92A and -92B bound to Cby1 (Fig. 1D). In contrast, the BAR domain proteins arfaptin-1 and arfaptin-2 (31) failed to interact with Cby1 (Fig. 1E), indicating the selective nature of Cby1 binding to the BAR domains of FAM92 proteins. To determine which region of Cby1 is responsible for FAM92 binding, we tested the interaction of FAM92A with various Cby1 mutants using co-IP assays (Fig. 1F). Deletion of the N-terminal 22 aa (Δ1–22) or the N-terminal half (aa 1 to 63) (ΔN) of Cby1 abolished the interaction with FAM92A. The Cby1-4A mutant, which contains alanine substitutions for four leucine residues in the C-terminal coiled-coil motif and is deficient in homodimerization (29), was also unable to interact with FAM92A. Thus, it appears that both the N-terminal region and the C-terminal coiled-coil motif of Cby1 are involved in FAM92 binding. Furthermore, we investigated if FAM92A and Cby1 directly interact using bacterially expressed and purified recombinant proteins. In vitro MBP pulldown assays using His-FAM92A demonstrated that FAM92A directly interacts with MBP-Cby1 but not with the negative-control MBP tag alone (Fig. 1G). These results suggest that the BAR domains of FAM92A and -92B directly bind to Cby1.
FAM92 proteins form homo- and heterodimers via their BAR domains.
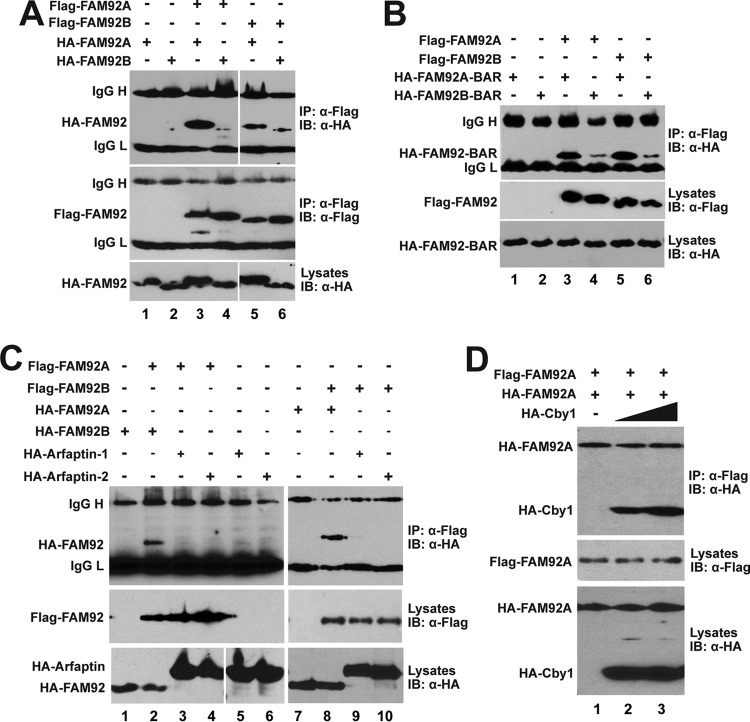
The BAR domain consists of three extended α-helices in an antiparallel arrangement (23–26). BAR domain proteins typically homo- and/or heterodimerize to form crescent-shaped structures that wrap around membranes. Heterodimers are most commonly formed among closely related family members such as endophilin family members (44, 45). To examine whether FAM92A and -92B have the ability to dimerize, Flag-tagged FAM92A and -92B were tested for their interaction with HA-tagged versions by using co-IP assays. As shown in Fig. 2A, FAM92A and -92B exhibited the capacity to homo- as well as heterodimerize. Next, we sought to determine if their BAR domains are sufficient for dimerization. Indeed, HA-tagged BAR domains of FAM92A and -92B were coimmunoprecipitated with Flag-tagged full-length proteins (Fig. 2B). FAM92A and -92B did not form heterodimers with arfaptin-1 and arfaptin-2 (Fig. 2C). To investigate if Cby1 influences FAM92 dimerization, we conducted co-IP assays for FAM92A homodimerization in the presence of Cby1. As shown in Fig. 2D, overexpression of Cby1 did not have significant effects on FAM92A homodimerization, and both HA-FAM92A and HA-Cby1 were coprecipitated with Flag-FAM92A. This suggests that the contact surfaces of FAM92 proteins for dimerization and Cby1 binding are different. Taken together, our results indicate that FAM92A and -92B homo- and heterodimerize through their BAR domains and that FAM92 dimers form a stable complex with Cby1.
FIG 2.
FAM92A and -92B homo- and heterodimerize through their BAR domains. (A to C) HEK293T cells were cotransfected with the indicated Flag-FAM92 and HA-FAM92 (A), HA-FAM92 BAR domain (B), or HA-arfaptin (C) expression constructs for co-IP assays. IgG H and IgG L denote IgG heavy and light chains, respectively. (D) Overexpression of Cby1 does not interfere with FAM92A homodimer formation. HEK293T cells were cotransfected with constant amounts of Flag-FAM92A and HA-FAM92A and increasing amounts of HA-Cby1, and cell lysates were immunoprecipitated with Flag antibody and detected with HA antibody.
FAM92A colocalizes with Cby1 at the distal end of mother centrioles and the ciliary base in RPE1 cells.
To study the subcellular localization of FAM92 proteins, we employed human retinal pigment epithelial (RPE1) cells that assemble primary cilia upon serum starvation. First, we examined the localization of ectopically expressed proteins. RPE1 cells were infected with lentiviruses expressing Flag-tagged FAM92A or -92B and serum starved for 48 h to induce ciliogenesis. Fixed cells were then triple labeled with antibodies for Flag, Cby1, and the centriolar/ciliary axoneme marker acetylated A-tub. Excitingly, both FAM92A and -92B colocalized with Cby1 at the ciliary base (Fig. 3A). In addition to the centriolar localization, FAM92 proteins, albeit weak, also showed a punctate vesicular distribution throughout the cytoplasm. To detect endogenous proteins, we verified the specificity of FAM92A and -92B antibodies by Western blotting using cell lysates from HEK293T cells expressing Flag-tagged FAM92 proteins (Fig. 3B). Indeed, these antibodies were highly specific, with each antibody recognizing its own family member. Next, cycling RPE1 cells stably expressing GFP-centrin1 as a marker for the distal lumen of centrioles were fixed and subjected to immunofluorescence (IF) staining with antibodies for FAM92A, Cby1, and various centriole markers (Fig. 3C). FAM92A showed extensive colocalization with Cby1 at mother centrioles while displaying a partial overlap with CEP164, which localizes to the distal appendages of mother centrioles (5, 6, 14, 15, 20). Furthermore, the results from costaining with different centrosome/centriole markers, including G-tub (centrosomes), CEP135 (proximal ends of centrioles), and centrobin (daughter centrioles), support the notion that FAM92A localizes to the distal end of mother centrioles. On the other hand, no clear centriolar localization of endogenous FAM92B was observed in RPE1 cells. Western blotting revealed that FAM92B was detectable in RPE1 cell lysates after a long exposure of the blots (see Fig. 5A), suggesting that the FAM92B protein is present at low levels, while FAM92A is the predominant family member in RPE1 cells.
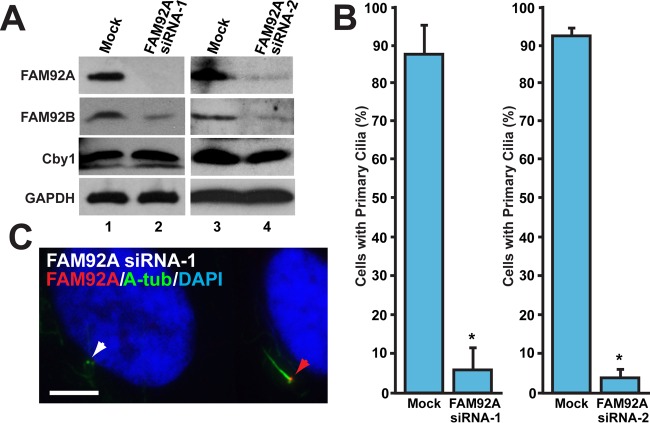
FIG 5.
FAM92A is required for primary cilium formation. RPE1 cells were either mock transfected or transfected with FAM92A siRNA-1 or FAM92A siRNA-2 and serum starved for 48 h to induce ciliogenesis. (A) Western blot analysis verified the efficient depletion of FAM92A by siRNA. Interestingly, the levels of the FAM92B protein were decreased, while those of the Cby1 protein were unchanged. GAPDH was used a loading control. (B) Quantification of the number of cells with primary cilia after siRNA treatment. RPE1 cells were immunostained for FAM92A and A-tub. FAM92A-positive cells for mock-treated controls and FAM92A-negative cells for siRNA-treated samples were counted in 4 (siRNA-1) or 12 (siRNA-2) independent experiments (a total of over 1,000 cells per siRNA). Data are presented as means ± standard deviations. *, P < 0.05. (C) Representative image of FAM92A siRNA-1-treated cells that were immunostained for FAM92A and A-tub. The red arrowhead indicates a FAM92A-positive cell with a primary cilium, whereas the white arrowhead indicates a FAM92A-depleted cell with no primary cilium. Nuclei were visualized by DAPI staining. Bar, 5 μm.
FAM92A and -92B colocalize with Cby1 at basal bodies in airway multiciliated cells.
Primary cultures of mouse tracheal epithelial cells (MTECs) provide an excellent model to study centriole formation and ciliogenesis (6, 27, 46). We previously demonstrated that Cby1 localizes to centrioles/basal bodies throughout the differentiation of multiciliated cells in MTEC cultures (6). In agreement with the results from RPE1 cells (Fig. 3A), lentivirally expressed Flag-tagged FAM92A and -92B colocalized with Cby1 at the ciliary base in fully differentiated multiciliated cells (Fig. 4A). Next, we examined whether the endogenous proteins colocalize with Cby1 in multiciliated cells using IF staining. At early stage II, when centrioles start to replicate en masse, FAM92A appeared in a weak and diffuse pattern around clusters of newly formed Cby1-positive centrioles (Fig. 4B, arrows). In addition, a more intense FAM92A signal was observed in patches of various sizes, with some overlapping these Cby1-positive centrioles (Fig. 4B, arrowheads). The nature of these structures, however, is currently unknown. In fully differentiated multiciliated cells at stage IV, FAM92A extensively colocalized with Cby at the base of cilia. In striking contrast to RPE1 cells, FAM92B was clearly detectable in multiciliated cells and demonstrated distribution patterns similar to those of FAM92A at both stages II and IV.
FIG 4.
FAM92A and -92B colocalize with Cby1 at the ciliary base in multiciliated cells. (A) MTECs were infected with lentiviruses expressing Flag-tagged FAM92A or -92B and costained with Flag and Cby1 antibodies. Bars, 5 μm. (B) MTECs were colabeled at the indicated differentiation stages with antibodies for Cby1 and FAM92A or -92B. Top-down views are shown. Arrows denote the areas of centriole replication in stage II ciliated cells, where FAM92 and Cby1 partially colocalized. Arrowheads indicate larger puncta or clusters of FAM92 signals. Merged images are shown with DAPI nuclear stain. Bars, 5 μm. (C) Fully differentiated ciliated cells in ALI day 14 MTEC cultures were costained for FAM92A and Cby1 and imaged by superresolution 3D-SIM. Top-down (x-y) (single section) and side (y-z and x-z) (maximum projection) views are shown. The squared area is shown at a higher magnification at the bottom right corner. Bar, 0.5 μm; bar in the high-magnification image, 0.1 μm.
Using superresolution microscopy, we previously reported that Cby1 clusters at the base of cilia in ring-shaped patterns in multiciliated cells, characteristic of distal appendage/transition fiber proteins (6). We therefore used three-dimensional structured illumination microscopy (3D-SIM) to examine FAM92A localization at the superresolution level (Fig. 4C). FAM92A also showed a ring-shaped distribution extensively overlapping Cby1 at the ciliary base in both the lateral and axial dimensions. Based on these results, we concluded that FAM92A and -92B are bona fide Cby1-interacting proteins that might play crucial roles in ciliogenesis.
FAM92A is required for ciliogenesis.
Our data so far are consistent with the idea that FAM92 proteins physically interact with Cby1, localize to mother centrioles, and hence play a role in ciliogenesis. To directly assess this, RPE1 cells were transfected with FAM92A siRNA-1 or -2 or mock transfected and serum starved to induce primary cilia, followed by IF staining for FAM92A and A-tub. As shown in Fig. 5A, both FAM92A siRNAs efficiently depleted endogenous FAM92A in RPE1 cells. Intriguingly, FAM92A KD significantly reduced FAM92B protein levels but had no effects on Cby1 protein levels. This supports the notion that FAM92A heterodimerizes with FAM92B and stabilizes it. Depletion of FAM92A effectively suppressed the assembly of primary cilia (Fig. 5B and C). These data suggest that FAM92A plays a crucial role in ciliogenesis.
Centriolar targeting of FAM92 proteins depends on Cby1.
The colocalization of FAM92A and -92B with Cby1 at mother centrioles/basal bodies raises the possibility that Cby1 facilitates the recruitment of FAM92 proteins to centrioles/basal bodies or vice versa. To explore this, we employed U2OS cells as a model system, as Cby1 is present at mother centrioles and as siRNA-mediated gene KD can be effectively performed (6). U2OS cells were transfected with an expression plasmid for either Cby1 shRNA or control scrambled shRNA. Western blotting confirmed the efficient depletion of the endogenous Cby1 protein (Fig. 6A). We then conducted IF staining for Cby1 and FAM92A and counted the numbers of cells with FAM92A at centrioles. In scrambled shRNA-transfected cells, 83.1% ± 3.2% of cells showed FAM92A localization at the mother centriole. In direct contrast, the vast majority of Cby1 shRNA-transfected cells, where the lack of Cby1 staining at centrioles was apparent, lost centriolar FAM92A, with only 10.2% ± 5.2% having detectable but reduced levels of FAM92A at centrioles. Importantly, Western blot analysis demonstrated that Cby1 depletion did not significantly affect the steady-state levels of the FAM92A protein. Similarly to our findings for RPE1 cells, a clear centriolar localization of FAM92B was not observed in U2OS cells by IF staining (data not shown). Nonetheless, as in RPE1 cells, the FAM92B protein was detectable by Western blotting after extended exposure times, and the steady-state levels of FAM92B were not altered by Cby1 depletion (Fig. 6A).
FIG 6.
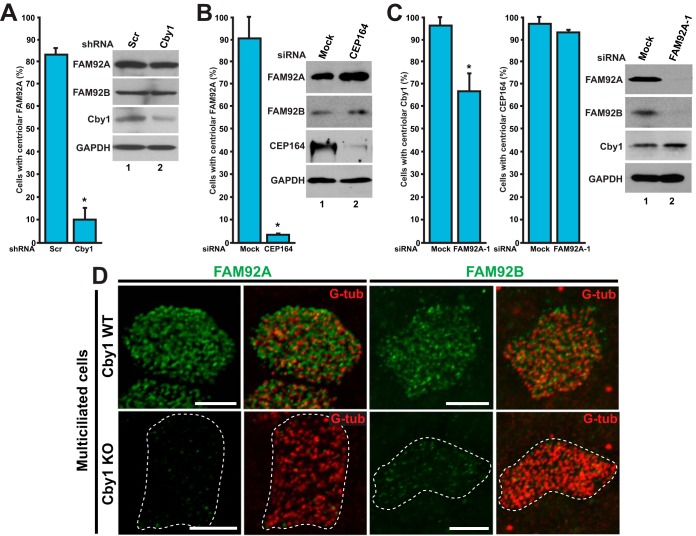
Cby1 is required for proper targeting of FAM92A and -92B to centrioles/basal bodies. (A) U2OS cells were transfected with an expression vector for either Cby1 shRNA or control scrambled shRNA (Scr) and immunostained for Cby1 and FAM92A to examine the centriolar recruitment of FAM92A. Cby1-positive cells for scrambled shRNA-transfected controls and Cby1-negative cells for Cby1 shRNA-transfected samples were counted in three independent experiments (a total of over 500 cells per shRNA). Data are presented as means ± standard deviations. *, P < 0.05. Cell lysates were subjected to Western blotting as indicated. GAPDH served as a loading control. (B) U2OS cells were transfected with CEP164 siRNA or mock transfected and immunostained for FAM92A and G-tub to assess the effects of CEP164 depletion on FAM92A localization at centrioles. At least 200 cells were counted in three independent experiments under each condition. Data are presented as means ± standard deviations. *, P < 0.05. (C) U2OS cells were transfected with FAM92A siRNA-1 or mock transfected and immunostained for Cby1 and FAM92A or CEP164 and G-tub to examine whether FAM92A depletion influences the centriolar recruitment of Cby1 and CEP164. For Cby1, FAM92A-positive cells for mock-transfected controls and FAM92A-negative cells for FAM92A siRNA-1-transfected samples were counted in 15 independent experiments (a total of over 2,000 cells under each condition). For CEP164, at least 200 cells were counted in three independent experiments. Data are presented as means ± standard deviations. *, P < 0.05. Note that FAM92A depletion substantially reduced FAM92B protein levels. (D) Cby1-WT and -KO MTECs at ALI day 14 were colabeled for FAM92A or -92B and the basal body marker G-tub. Bars, 5 μm.
The above-described data suggest that Cby1 plays a critical role in localizing FAM92A to centrioles. We previously reported that Cby1 is recruited to centrioles through its physical interaction with the distal appendage protein CEP164 (6). We therefore asked whether the centriolar localization of FAM92A depends on CEP164. To address this, we depleted CEP164 using siRNA in U2OS cells, followed by IF staining for FAM92A and G-tub. In mock-treated cells, 90.8% ± 10.0% of cells displayed FAM92A localization at their centrioles, whereas only 3.8% ± 0.1% of CEP164-depleted cells showed a normal centriolar localization of FAM92A (Fig. 6B). Western blot analysis revealed that CEP164 siRNA efficiently depleted the endogenous protein, while no significant changes in the steady-state levels of FAM92A and -92B were noted.
Next, we performed the reciprocal experiments using FAM92A siRNA-1 to determine if the centriolar localization of Cby1 and CEP164 depends on FAM92A in U2OS cells. Interestingly, FAM92A KD diminished the number of cells with Cby1 at the mother centrioles by about 30% (67.2% ± 7.6% of FAM92A siRNA-treated cells versus 97.0% ± 3.4% of mock-treated cells) (Fig. 6C), suggesting that, to at least some extent, FAM92A and Cby1 centriolar localizations are mutually dependent. Consistent with the idea that FAM92A lies downstream of CEP164, FAM92A KD showed no major influences on the centriolar localization of CEP164. A clear dependency of FAM92A centriolar localization on CEP164 was also observed in RPE1 cells (data not shown). FAM92A KD significantly reduced FAM92B protein levels without any detectable effects on Cby1 protein levels in U2OS cells. FAM92A siRNA-2 yielded similar trends, with statistically significant results (for centriolar Cby1, 97.7% ± 1.4% for mock treatment and 86.1% ± 3.3% for siRNA-2 treatment [n = 15] [P < 0.05]; for centriolar CEP164, 98.4% ± 1.5% for mock treatment and 98.9% ± 0.9% for siRNA-2 treatment [n = 15] [P < 0.05]).
To confirm that FAM92A centriolar localization is reliant on Cby1 and to examine if Cby1 is required for the basal body localization of FAM92B, we prepared MTECs from the tracheas of Cby1-WT and -KO mice and differentiated them under air-liquid interface (ALI) conditions for 14 days. In fully differentiated multiciliated cells, the basal body localization of FAM92A as well as FAM92B was lost or severely reduced in ciliated Cby1-KO cells (Fig. 6D). Collectively, these data suggest that Cby1 is required for centriole/basal body targeting of both FAM92A and -92B and that FAM92 proteins also play a role in the centriolar targeting of Cby1.
Coexpression of FAM92 proteins and Cby1 induces deformed membrane-like structures containing Rab8.
BAR domain proteins possess the ability to bind to lipid membranes and facilitate membrane remodeling into various shapes in vivo and in vitro (23–26). For example, overexpression of amphiphysin or arfaptin induces the formation of extensive membrane tubules in mammalian cultured cells (31, 47), while ectopic expression of sorting nexin 1 (SNX1) or SNX4 predominantly induces globular structures (48, 49). To gain insight into the biological significance of the FAM92-Cby1 interaction, we ectopically expressed GFP-tagged FAM92A or -92B with or without Flag-tagged Cby1 in U2OS cells. Either GFP-FAM92A or GFP-FAM92B alone showed diffuse/punctate distribution patterns, with occasional localization to small globular compartments (Fig. 7A, top, arrowheads). Flag-Cby1 alone also showed a similar diffuse/punctate distribution throughout the cytoplasm. In marked contrast, coexpression of GFP-FAM92A with Cby1 induced the formation of larger globular domains in which both proteins colocalized, particularly in cells with high levels of protein expression (Fig. 7A, middle, arrowheads). Strikingly, ectopic expression of GFP-FAM92B and Cby1 promoted the formation of membrane tubule-like structures (Fig. 7A, bottom, arrowheads). FAM92B and Cby1 proteins extensively colocalized in these tubular structures. Similar results were obtained by using COS7 cells (data not shown). Whether these FAM92-Cby1-decorated structures are composed of membranes awaits further investigation by using TEM.
FIG 7.
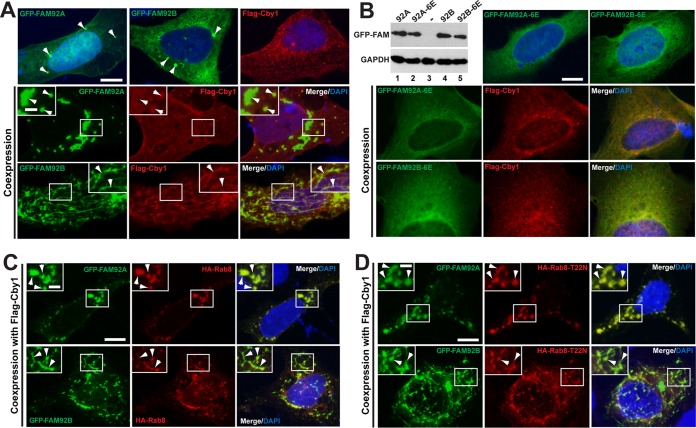
Coexpression of FAM92 and Cby1 induces membrane globule- and tubule-like structures containing Rab8. (A) U2OS cells were transfected with GFP-FAM92A, GFP-FAM92B, or Flag-Cby1 alone (top) or in combination, as indicated, and immunostained with Flag antibody. Nuclei were visualized by DAPI staining. When expressed individually, all proteins demonstrated a diffuse punctate distribution throughout the cytoplasm. GFP-tagged FAM92A and -92B occasionally localized to larger puncta (arrowheads in top panels). GFP-FAM92A and Flag-Cby1, when coexpressed, localized to large globular compartments (arrowheads in the inset). In contrast, ectopic expression of GFP-FAM92B and Cby1 induced extensive tubule-like structures where both proteins were present (arrowheads in the inset). Bar, 10 μm; bar in the inset, 3 μm. (B) U2OS cells were transfected with GFP–FAM92A-6E (K107E/R110E/K114E/R132E/R134E/R136E) or GFP–FAM92B-6E (K105E/R108E/K112E/K130E/R132E/K134E), which carries glutamic acid substitutions for putative lipid-binding residues, either individually or in combination with Flag-Cby1 as indicated, and immunostained with Flag antibody. Nuclei were visualized by DAPI staining. These mutants mainly showed a diffuse cytoplasmic distribution in the presence or absence of Cby1. Protein levels of the mutants were tested by Western blotting with GFP antibody. A minus sign indicates the nontransfected negative control. GAPDH was used a loading control. Bar, 10 μm. (C and D) Rab8 localizes to FAM92-Cby1-decorated membrane-like structures. U2OS cells were transfected with GFP-FAM92A or GFP-FAM92B, Flag-Cby1, and HA-Rab8 (C) or HA-Rab8-T22N (D) as indicated and immunostained with HA antibody. Arrowheads indicate colocalization of FAM92 and Rab8. Nuclei were visualized by DAPI staining. Bar, 10 μm; bar in the inset, 3 μm.
Protein structural modeling between FAM92 BAR domains and known BAR domains using the Phyre2 server (50) predicted that K107, R110, K114, R132, R134, and R136 on FAM92A and K105, R108, K112, K130, R132, and K134 on FAM92B are exposed on their concave surfaces and potentially involved in direct lipid binding. Hence, we generated the FAM92A-6E and FAM92B-6E mutants, carrying glutamic acid substitutions at these 6 basic amino acids. Upon coexpression with Cby1, the FAM92-6E mutants failed to induce globular or tubular structures, although they were stably expressed (Fig. 7B). Of note, the centriolar localization of the FAM92-6E mutants was severely compromised (data not shown).
We previously demonstrated that Cby1 promotes the recruitment of Rab8 to facilitate CV formation during ciliogenesis (6). We therefore investigated if the FAM92-Cby1-decorated membrane-like structures contain Rab8 by coexpressing GFP-FAM92A or GFP-FAM92B, Flag-Cby1, and HA-Rab8 in U2OS cells. As shown in Fig. 7C, indeed, Rab8 was recruited to FAM92-positive structures. In contrast, Rab11 and ARF6 small GTPases were not detectable in these structures (data not shown). To determine whether Rab8 plays a direct role in this process, we coexpressed the dominant negative Rab8-T22N mutant and found that FAM92-positive globular and tubular structures were able to form and contained Rab8-T22N (Fig. 7D), suggesting that Rab8 activity is not essential for the formation of these structures. Taken together, our data demonstrate that, consistent with the membrane-remodeling functionality of BAR domain proteins, FAM92A and -92B in cooperation with Cby1 facilitate the formation of globular and tubular membrane-like structures containing Rab8.
DISCUSSION
In this study, we identified FAM92A and -92B of the BAR domain family as novel Cby1-interacting proteins using TAP and mass spectrometry. The BAR domains of FAM92 proteins are responsible for their homo- and heterodimer formation and binding to Cby1. Both FAM92A and -92B colocalize with Cby1 at the mother centrioles/basal bodies of cilia. We furthermore found that the centriolar targeting of FAM92 proteins depends on Cby1 and CEP164. Notably, KD of FAM92A in RPE1 cells results in impaired ciliogenesis. Consistent with the established BAR domain functions, FAM92 proteins in concert with Cby1 exert apparent membrane-remodeling activities in mammalian cultured cells.
We previously demonstrated that Cby1 localizes to the distal appendages/transition fibers of centrioles/basal bodies throughout the differentiation of airway ciliated cells (6). Consistent with this, FAM92A and -92B are expressed from early differentiation stages of multiciliated cells, partially colocalizing with Cby1 at nascent centrioles (Fig. 4B). At later stages, the centriolar localization of FAM92A and -92B becomes more discrete. SIM imaging revealed that FAM92A colocalizes with Cby1 in a ring-shaped pattern at the base of mature cilia (Fig. 4C). These observations indicate that FAM92A and -92B represent new distal appendage/transition fiber proteins. It is noteworthy that FAM92B was undetectable at centrioles in RPE1 and U2OS cells, most likely due to its low expression levels, but was highly expressed in multiciliated cells, suggesting its tissue-restricted expression pattern. In support of this, according to data from gene expression databases, including the FANTOM5 database (51) and the Human Protein Atlas (52), FAM92B expression is restricted to certain tissues, most prominently multiciliated tissues such as tracheas, lungs, and fallopian tubes, while FAM92A is ubiquitously expressed in multiple different tissues. Additionally, a recent expression profiling study identified the FAM92B gene as a gene highly expressed in the lung (53).
We found that the centriolar localization of FAM92A depends largely on Cby1 (Fig. 6A). Interestingly, FAM92A KD decreased the number of cells with centriolar Cby1 (Fig. 6C). One possible explanation is that FAM92A KD is incomplete and variable among siRNA-treated cells, even though FAM92A is undetectable by IF staining, and a small amount of the residual protein is sufficient to target Cby1 to centrioles. Another possibility is that although FAM92B protein levels are significantly reduced upon FAM92A KD, the residual FAM92B protein is able to recruit Cby1 to centrioles. In these scenarios, Cby1 and FAM92 proteins collaborate with each other in trafficking to centrioles. Another likely scenario is that a fraction of Cby1 may be targeted to centrioles in a FAM92-independent manner. Further investigation is therefore required to clarify the molecular mechanisms for the centriolar recruitment of Cby1 and FAM92 proteins.
BAR domain family members are categorized into several subfamilies, including the N-, F-, and I-BAR proteins, based on sequence homologies and structural similarities (23–26). The BAR domains of FAM92A and -92B display only limited homologies to other BAR domains, and elucidation of the subfamily to which they belong awaits structural characterization. BAR domain proteins bind negatively charged phospholipids such as phosphoinositides via positively charged residues on their concave surface to induce membrane deformation in vivo and in vitro (23–26). We found that coexpression of FAM92A or -92B with Cby1 provokes robust membrane-remodeling activity (Fig. 7). What might be the mechanistic basis for this? N-BAR proteins such as amphiphysin and endophilin contain an N-terminal amphipathic α-helix (H0) that inserts into membranes and facilitates the anchoring of BAR domains to membranes (54–56). Some other BAR domain proteins harbor a lipid-binding domain such as the pleckstrin homology (PH) domain, which is important for membrane binding and the generation of membrane curvature (47, 57). However, both FAM92A and -92B lack such domains. Thus, it is tempting to speculate that Cby1 may either directly or indirectly associate with membranes to promote the local recruitment of FAM92 proteins for their higher-order assembly and subsequent membrane remodeling.
The functions of BAR domain proteins in ciliogenesis remain poorly defined. ASAP1 is an Arf4 GTPase-activating protein (GAP) containing a BAR domain. Although it is dispensable for ciliogenesis, ASAP1 serves as a scaffold for the recruitment and activation of the Rab11-Rabin8-Rab8 cascade to facilitate the ciliary targeting of rhodopsin in retinal photoreceptor cells (58). The F-BAR protein syndapin I localizes around the base of cilia, and its KD in zebrafish leads to defects in left-right body asymmetry and sensory hair cells (59), both of which depend on the proper functions of cilia. Missing in metastasis (MIM) is a member of the I-BAR subfamily (60). It localizes to the base of primary cilia and plays a crucial role in ciliogenesis and Hedgehog signaling. However, the precise molecular functions of syndapin I and MIM during ciliogenesis remain to be elucidated. What might be the functions of FAM92 proteins during ciliogenesis? In agreement with the membrane-remodeling activities of BAR domains, coexpression of FAM92A and -92B with Cby1 induces the formation of membrane globule- and tubule-like structures, respectively, which contain Rab8 (Fig. 7). Besides its localization to pericentriolar vesicles and ciliary membranes (61), it has been shown that Rab8 localizes to an endocytic recycling tubular compartment and regulates clathrin-independent endocytosis and recycling (62). Thus, FAM92-Cby1-positive structures may result from the successive fusion of endocytic vesicles in the cytoplasm. We speculate that FAM92 proteins in cooperation with Cby1 may be involved in the recruitment and fusion of small vesicles of endosomal origin at the distal appendages during early stages of ciliogenesis. Another possibility, but not mutually exclusive, is that FAM92 proteins function as a protein coat to shape or stabilize the underlying membranes of small vesicles that are targeted to the ciliary base by Cby1. Somewhat surprisingly, FAM92-positive structures are observed in the presence of the dominant negative Rab8-T22N mutant (Fig. 7D). A likely explanation is that Cby1 may be involved in the recruitment of other small GTPases that are able to compensate for the loss of Rab8 function in the formation of these structures and possibly CVs. In conclusion, our study identifies BAR domain-containing FAM92A and -92B as bona fide Cby1-binding partners that localize to mother centrioles/basal bodies and highlights the importance of dynamic membrane deformation and remodeling during ciliogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erich Nigg and Gislene Pereira for the CEP164 antibodies, Kazuhisa Nakayama and Hye-Won Shin for the HA-tagged arfaptin-1 and arfaptin-2 expression constructs, Miguel Garcia-Diaz for help with 3D structural modeling of FAM92 BAR domains using the Phyre2 server, and Antonius Kohler at the Stony Brook Proteomics Center for help with mass spectrometry.
This work was supported by an NSF grant (MCB1140033) to R.K. and an NIH-NHLBI grant (R01HL107493) to K.-I.T. S.S.S. was supported by Stony Brook Medical Scientist Training Program grant T32 GM008444-23. The purchase of the Nikon SIM instrument was supported in part by an NIH Shared Instrumentation grant (S10OD016405-01).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00160-16.
REFERENCES
- 1.Hildebrandt F, Benzing T, Katsanis N. 2011. Ciliopathies. N Engl J Med 364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. 2010. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigg EA, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Horani A, Ferkol TW, Dutcher SK, Brody SL. 11 September 2015. Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol 199:1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke MC, Li FQ, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O'Connell CB, Love D, Westlake CJ, Reynolds SD, Kuriyama R, Takemaru K. 2014. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol 207:123–137. doi: 10.1083/jcb.201406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Q, Ling K, Hu J. 2015. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol 35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter JF, Blacque OE, Leroux MR. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiao YC, Tuz K, Ferland RJ. 2012. Trafficking in and to the primary cilium. Cilia 1:4. doi: 10.1186/2046-2530-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmer BT, Maric D, Engman DM. 2010. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci 123:529–536. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbaum JL, Witman GB. 2002. Intraflagellar transport. Nat Rev Mol Cell Biol 3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 12.Voronina VA, Takemaru K, Treuting P, Love D, Grubb BR, Hajjar AM, Adams A, Li FQ, Moon RT. 2009. Inactivation of Chibby affects function of motile airway cilia. J Cell Biol 185:225–233. doi: 10.1083/jcb.200809144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love D, Li FQ, Burke MC, Cyge B, Ohmitsu M, Cabello J, Larson JE, Brody SL, Cohen JC, Takemaru K. 2010. Altered lung morphogenesis, epithelial cell differentiation and mechanics in mice deficient in the Wnt/beta-catenin antagonist Chibby. PLoS One 5:e13600. doi: 10.1371/journal.pone.0013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steere N, Chae V, Burke M, Li FQ, Takemaru K, Kuriyama R. 2012. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS One 7:e41077. doi: 10.1371/journal.pone.0041077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YL, Sante J, Comerci CJ, Cyge B, Menezes LF, Li FQ, Germino GG, Moerner WE, Takemaru K, Stearns T. 2014. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol Biol Cell 25:2919–2933. doi: 10.1091/mbc.E14-02-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, Soulavie F, Kernan MJ, Laurencon A, Durand B. 2012. Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol 197:313–325. doi: 10.1083/jcb.201109148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J, Zhao Y, Galati D, Winey M, Klymkowsky MW. 2014. Chibby functions in Xenopus ciliary assembly, embryonic development, and the regulation of gene expression. Dev Biol 395:287–298. doi: 10.1016/j.ydbio.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siller SS, Burke MC, Li FQ, Takemaru K. 2015. Chibby functions to preserve normal ciliary morphology through the regulation of intraflagellar transport in airway ciliated cells. Cell Cycle 14:3163–3172. doi: 10.1080/15384101.2015.1080396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li FQ, Siller SS, Takemaru KI. 2015. Basal body docking in airway ciliated cells. Oncotarget 6:19944–19945. doi: 10.18632/oncotarget.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorokin SP. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3:207–230. [DOI] [PubMed] [Google Scholar]
- 23.Daumke O, Roux A, Haucke V. 2014. BAR domain scaffolds in dynamin-mediated membrane fission. Cell 156:882–892. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Suetsugu S, Kurisu S, Takenawa T. 2014. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev 94:1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- 25.Qualmann B, Koch D, Kessels MM. 2011. Let's go bananas: revisiting the endocytic BAR code. EMBO J 30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. 2006. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev 70:37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Y, Richer EJ, Huang T, Brody SL. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 28.Li FQ, Mofunanya A, Harris K, Takemaru K. 2008. Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J Cell Biol 181:1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mofunanya A, Li FQ, Hsieh JC, Takemaru K. 2009. Chibby forms a homodimer through a heptad repeat of leucine residues in its C-terminal coiled-coil motif. BMC Mol Biol 10:41. doi: 10.1186/1471-2199-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. 2003. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 31.Man Z, Kondo Y, Koga H, Umino H, Nakayama K, Shin HW. 2011. Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J Biol Chem 286:11569–11578. doi: 10.1074/jbc.M110.201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen GI, Gingras AC. 2007. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 33.Li FQ, Mofunanya A, Fischer V, Hall J, Takemaru K. 2010. Nuclear-cytoplasmic shuttling of Chibby controls beta-catenin signaling. Mol Biol Cell 21:311–322. doi: 10.1091/mbc.E09-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takemaru KI, Moon RT. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol 149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cyge B, Fischer V, Takemaru K, Li FQ. 2011. Generation and characterization of monoclonal antibodies against human Chibby protein. Hybridoma (Larchmt) 30:163–168. doi: 10.1089/hyb.2010.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. 2002. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol 156:87–99. doi: 10.1083/jcb.200108088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Low WC, Liu A, Wang B. 2013. Centrosomal protein DZIP1 regulates Hedgehog signaling by promoting cytoplasmic retention of transcription factor GLI3 and affecting ciliogenesis. J Biol Chem 288:29518–29529. doi: 10.1074/jbc.M113.492066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HR, Richardson J, van Eeden F, Ingham PW. 2010. Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol 8:65. doi: 10.1186/1741-7007-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay SY, Yu X, Wong KN, Panse P, Ng CP, Roy S. 2010. The iguana/DZIP1 protein is a novel component of the ciliogenic pathway essential for axonemal biogenesis. Dev Dyn 239:527–534. doi: 10.1002/dvdy.22199. [DOI] [PubMed] [Google Scholar]
- 41.Ruan XZ, Yang HS, Yao SH, Ma FX, Zhao XY, Yan F, Wang CT, Lai ST, Deng HX, Wei YQ. 2007. Isolation and characterization of a novel Xenopus gene (xVAP019) encoding a DUF1208 domain containing protein. Mol Reprod Dev 74:1505–1513. doi: 10.1002/mrd.20739. [DOI] [PubMed] [Google Scholar]
- 42.Ruan XZ, Yan F, Zhao XY, Wang CT, Song M, Yang HS, Deng HX, Wei YQ. 2007. Identification and characterization of two novel variants of the DUF1208 protein FAM92A1. Mol Cells 23:391–397. [PubMed] [Google Scholar]
- 43.Liang S, Gong F, Zhao X, Wang X, Shen G, Xu Y, Yang H, Ruan X, Wei Y. 2009. Prokaryotic expression, purification of a new tumor-relative protein FAM92A1-289 and its characterization in renal cell carcinoma. Cancer Lett 276:81–87. doi: 10.1016/j.canlet.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 44.Ralser M, Nonhoff U, Albrecht M, Lengauer T, Wanker EE, Lehrach H, Krobitsch S. 2005. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum Mol Genet 14:2893–2909. doi: 10.1093/hmg/ddi321. [DOI] [PubMed] [Google Scholar]
- 45.Pierrat B, Simonen M, Cueto M, Mestan J, Ferrigno P, Heim J. 2001. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics 71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 46.Vladar EK, Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol 178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 48.Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. 2007. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol 9:1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- 49.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol 14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 50.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 51.FANTOM Consortium and RIKEN PMI and CLST (DGT), Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, et al. 2014. A promoter-level mammalian expression atlas. Nature 507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, et al. 2005. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Lindskog C, Fagerberg L, Hallstrom B, Edlund K, Hellwig B, Rahnenfuhrer J, Kampf C, Uhlen M, Ponten F, Micke P. 2014. The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. FASEB J 28:5184–5196. doi: 10.1096/fj.14-254862. [DOI] [PubMed] [Google Scholar]
- 54.Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R. 2010. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J Biol Chem 285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isas JM, Ambroso MR, Hegde PB, Langen J, Langen R. 2015. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure 23:873–881. doi: 10.1016/j.str.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. 2006. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang X, Fan J, Zhang Y, Zhang K, Gao B, Ma J, Li J, Deng Y, Zhou Q, Egelman EH, Hsu VW, Sun F. 2014. A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature. Dev Cell 31:73–86. doi: 10.1016/j.devcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deretic D. 2013. Crosstalk of Arf and Rab GTPases en route to cilia. Small GTPases 4:70–77. doi: 10.4161/sgtp.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. 2013. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J Cell Sci 126:196–208. doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- 60.Bershteyn M, Atwood SX, Woo WM, Li M, Oro AE. 2010. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell 19:270–283. doi: 10.1016/j.devcel.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, Sheffield VC, Scheller RH, Jackson PK. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A 108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grant BD, Donaldson JG. 2009. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol 10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.