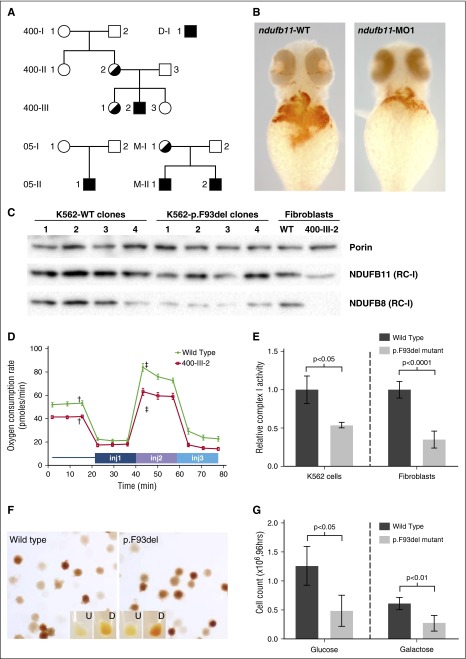
Figure 1.
NDUFB11 mutations in CSA. (A) NDUFB11 CSA pedigrees. Black shading indicates the genotype. The genotype of M-I-1 is inferred on the basis of her 2 sons. (B) Zebrafish embryos stained for hemoglobin with o-dianisidine at 48 hours postfertilization (hpf) following microinjection with water control or antisense morpholino reagents targeting the fish ortholog of human NDUFB11. (C) Total mitochondrial protein was analyzed for the RC-I proteins NDUFB11 (middle) and NDUFB8 (bottom) in K562 cells and patient fibroblasts by western blot. Equivalent loading of mitochondrial lysates was confirmed by immunoblot analysis for porin (VDAC1) (top). (D) Mitochondrial function was measured using oxygen consumption rate (ocr) using the Seahorse Mitostress test. Patient and control fibroblasts were treated with 2 µM oligomycin injection 1 (inj1), 0.5 µM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) (inj2), and 1 µM rotenone/antimycin A (inj3). Error bars are ± standard error of the mean (SEM). Basal OCR was measured before adding treatments (†) and maximum OCR was measured after the addition of 0.5 µM FCCP (‡). The difference in basal and maximum OCR is plotted in supplemental Figure 6. (E) RC-I function was analyzed in K562 cell lines (n = 4, clones as in panel C) and control and patient fibroblasts (n = 6, technical replicates) and normalized to citrate synthase activity. Data are expressed relative to the wild-type (WT) value, whose mean was defined as 1.0. Ratios are ± standard deviation (SD). (F) Representative WT and p.F93del K562 cells (clones K562-WT3 and K562-p.F93del1) stained with o-dianisidine show normal hemoglobinization. Undifferentiated (U) and differentiated (D) K562 cells cultured in the presence of 6.0 mM sodium butyrate for differentiation. (G) Growth of undifferentiated K562 cells (n = 4 clones) in glucose or galactose after 96 hours. Cells were initially plated at 4 × 104 cells per mL and analyzed in triplicate.