Abstract
Organ injuries caused by environmental chemical exposures or use of pharmaceutical drugs pose a serious health risk that may be difficult to assess because of a lack of non‐invasive diagnostic tests. Mapping chemical injuries to organ‐specific histopathology outcomes via biomarkers will provide a foundation for designing precise and robust diagnostic tests. We identified co‐expressed genes (modules) specific to injury endpoints using the Open Toxicogenomics Project‐Genomics Assisted Toxicity Evaluation System (TG‐GATEs) – a toxicogenomics database containing organ‐specific gene expression data matched to dose‐ and time‐dependent chemical exposures and adverse histopathology assessments in Sprague–Dawley rats. We proposed a protocol for selecting gene modules associated with chemical‐induced injuries that classify 11 liver and eight kidney histopathology endpoints based on dose‐dependent activation of the identified modules. We showed that the activation of the modules for a particular chemical exposure condition, i.e., chemical‐time‐dose combination, correlated with the severity of histopathological damage in a dose‐dependent manner. Furthermore, the modules could distinguish different types of injuries caused by chemical exposures as well as determine whether the injury module activation was specific to the tissue of origin (liver and kidney). The generated modules provide a link between toxic chemical exposures, different molecular initiating events among underlying molecular pathways and resultant organ damage. Published 2016. This article is a U.S. Government work and is in the public domain in the USA. Journal of Applied Toxicology published by John Wiley & Sons, Ltd.
Keywords: co‐expression modules, toxicogenomics, systems toxicology, nephrotoxicity, hepatotoxicity, histopathology, adverse outcome pathways
Short abstract
We proposed a protocol for selecting gene co‐expression modules associated with chemical‐induced injuries that classify 11 liver and eight kidney histopathology endpoints based on dose‐dependent activation of the identified modules. We showed that the activation of the modules for a particular chemical exposure correlated with the severity of histopathological damage and could distinguish different types of organ‐specific injuries. The generated modules provide a link between toxic chemical exposures, different molecular initiating events among underlying molecular pathways, and resultant organ damage.
Introduction
The release of toxic industrial chemicals in the environment, industrial accidents in manufacturing and transport, over‐use of pesticides and antibiotics in farming, as well as inappropriate use of pharmaceutical drugs, create an increasing chemical health hazard with the potential to cause both acute and long‐term adverse health effects. Accurately diagnosing chemical injuries through non‐invasive tests would allow for damage assessment, early intervention and treatment, and prediction of potential for recovery (Parkes et al., 2012). Efforts in elucidating the mechanism of toxicity and identifying potential biomarkers of exposure are key elements in starting to address these issues (Blomme et al., 2009; Permenter et al., 2013; Vinken et al., 2013; Hussainzada et al., 2014; Sturla et al., 2014; Madejczyk et al., 2015; Speir et al., 2015). Here, we present our efforts in identifying sets of genes (modules) that are characteristic of and specific to a wide variety of chemical exposure conditions causing liver and/or kidney injuries.
We used the Open Toxicogenomics Project‐Genomics Assisted Toxicity Evaluation System (TG‐GATEs) to generate sets of genes related to graded histopathology assessments of liver and kidney damage as manifestations of chemical exposure injuries. TG‐GATEs contains normalized, organ‐specific data on chemically induced gene expression changes coupled to graded histopathology assessments in male Sprague–Dawley rats (Igarashi et al., 2014). Ideally, the gene expression pattern in a module is transformed into an activation score that reaches statistical significance in those conditions for which a specific chemical/dose/exposure time combination causes an injury. Furthermore, the module activation score should correlate with the degree of observed injury in a dose‐dependent manner.
Many toxicogenomics studies have used gene expression microarrays to characterize differential transcriptional regulation resulting from chemical and toxicant insults (Blomme et al., 2009; Gresham and McLeod, 2009; Panagiotou and Taboureau, 2012; Smalley et al., 2010; Bai and Abernethy, 2013). Co‐expressed gene modules have been used to identify or classify genes specific to tumors of certain cancers (Segal et al., 2004), as well as for repurposing drugs as cancer therapeutics (Iskar et al., 2013). Computational methods to create these modules include bi‐clustering (Ihmels et al., 2002; Bergmann et al., 2003), in which the constituent genes share a correlated expression pattern across a subset of the chemical exposure conditions. A large dataset like TG‐GATEs – in which multiple conditions that vary in chemical, dose, and time are associated with an injury – is required to assess the specificity of modules to different types of cellular and tissue damage caused by different classes of toxicants.
In our previous work, we were able to conceptually connect molecular toxicity pathways to co‐expressed gene modules and link these pathways to specific injuries in the liver with the objective of identifying sensitive, specific and non‐invasive biomarkers for diseases and injuries (AbdulHameed et al., 2014; Tawa et al., 2014). We found that modules generated from the Iterative Signature Algorithm (ISA) performed satisfactorily in terms of generating modules that were specifically activated in response to chemical injuries (Tawa et al., 2014). However, the ISA model parameters that control selection and the association between gene sets and condition sets were not examined exhaustively. Furthermore, the selection of a suitable injury‐specific module was in part based on biological information and the presence of known biomarkers.
Here, we introduce a new and unbiased protocol for assigning gene modules for specific histopathology‐graded injuries based on co‐expression profile from a different, large‐scale and varied database containing multiple chemical exposure conditions ranging from 4 to 29 days in the Sprague–Dawley rat. We used multiple parameter sets for the generation of modules by the ISA and selected histopathology‐associated modules based on statistical metrics. This new protocol is applicable to any organ, e.g., liver and kidney, and does not require the input of biological information other than the gene expression data.
The proposed protocol associated chemical‐induced injury modules with 11 liver and eight kidney histopathology endpoints based on time‐ and dose‐dependent activation of the modules. We showed that the activation of the modules for a particular chemical exposure condition, i.e., chemical‐time‐dose combination, correlated with the onset, presence and severity of histopatological damage in a dose‐dependent manner. Furthermore, the modules could distinguish different types of injuries caused by chemical exposures as well as whether the injury‐module activation was specific to the tissue of origin (liver and kidney). The generated modules provide a link between toxic chemical exposures, different molecular‐initiating events among underlying molecular pathways, and resultant organ damage.
Methodology
Data
We used data from TG‐GATEs (Igarashi et al., 2014), a publicly available database that contains matched data associating chemical exposures with transcriptomic changes in the liver and kidney of male Sprague–Dawley rats along with graded histopathology assessments. TG‐GATEs contains repeated‐dose exposure of chemicals in low, middle and high dosages and four different time‐points (4, 8, 15 and 29 days), which we used for module generation. Chemical exposure conditions (or conditions, for brevity) are defined as a specific chemical‐time‐dose combination.
TG‐GATEs also provides histopathology data with corresponding severity or grades – minimal, slight, moderate and severe – associated with each chemical exposure condition. We considered one chemical exposure condition a positive instance of the histopathology endpoint if at least two animals in that condition showed a histopathology grade of at least 'minimal'. A histopathology assessment was considered for analyzes if at least two conditions were positive. Thirty‐five liver and 27 kidney histopathology endpoints satisfied the above requirement.
Data processing
We downloaded the liver and kidney microarray datasets on Affymetrix GeneChip Rat Genome 230 2.0 Array from TG‐GATEs (http://toxico.nibiohn.go.jp). The dataset contains whole genome microarray expression data for liver and kidney from 6 765 and 1 952 rats, respectively. According to our previous protocol (Tawa et al., 2014), we used the ArrayQualityMetrics (Kauffmann et al., 2009) Bioconductor package to assess the quality of the Robust Multi‐array Averaged (RMA) (Irizarry et al., 2003) pre‐processed data. In this process, we removed outlier arrays and renormalized the remaining data.
After array‐level filtering and normalization, we performed gene‐level filtering using the BioConductor package genefilter (Gentleman et al., 2004). Specifically, we removed genes without Entrez IDs or with low variance across conditions. We implemented the low variance criteria (Bourgon et al., 2010) by computing and sorting the expression variance of each gene over the complete condition set and removing the bottom half as low‐variance genes. We performed additional filtering using the default settings for the affy package from BioConductor to remove probe sets below a signal‐to‐noise threshold. The number of replicates for each condition that had a ’Present‘ call was determined for each probe set, and we retained probe sets for which at least 25% of the conditions had ’Present‘ calls for all replicates within a condition. As a result, the liver data comprised expression data for 9520 genes measured under 1679 distinct conditions. The kidney data contained fewer conditions and chemicals, with expression data for 9946 genes collected from 482 conditions. In addition, some chemicals do not necessarily have data collected across both organs or for all combinations of dose and time.
For the remaining genes and conditions, we calculated log ratios (LRs) for each gene as the difference between treatment and control RMA expression levels. We computed log2 expression values for treatment and control as averages over replicates. We assembled a log ratio matrix LR with rows defined by genes, columns defined by conditions and the matrix elements, LRi,j, defined as log ratios for genes i under conditions j. As a last step, we transformed the log ratios into Z‐scores (Tawa et al., 2014). The Z‐score of gene i under condition j is defined as the number of standard deviations this observation is above the average over all conditions for gene i.
Module/gene set generation
We used the R package eisa to generate ISA (Bergmann et al., 2003) co‐expression modules associated with the entire Z‐score matrix for the liver and the kidney. We first ran ISAIterate, which requires a starter gene set that is typically built using existing gene‐related knowledge; here we used ~200 starter gene sets from hierarchical clustering (Rinaldo et al., 2005). In line with our previous work (Tawa et al., 2014), random gene sets were added for ~15 000 starter gene sets. We used 25 combinations of tg (gene threshold) and tc (condition threshold) varying from 2.0 to 4.0 in 0.5 increments, i.e., 1) tg =2.0 and tc =2.0; 2) tg =2.5 and tc =2.0; … ; 25) tg = 4.0 and tc =4.0.
The two parameters individually control how similar the expression of genes and the subset of conditions are, with a higher number of either tg or tc being more restrictive, i.e., having a higher level of correlation of genes and conditions within the bi‐cluster. In order to avoid the creation of redundant modules, we pruned our results using the routine ISAUnique for every combination of tg and tc parameters, with the parameter cor.limit set to its default value.
To ensure that the gene sets were robust – i.e., the core module composition did not change when adding random genes – we used the routine ISAFilterRobust with default parameters. All gene modules with intra‐module correlation < 0.4 and with the number of genes >200 were filtered out. Using this procedure, we generated 891 liver modules and 303 kidney modules.
Module evaluation parameters
We used different metrics to select modules based on activation scores, data correlations and statistical significance associated with a specific histopathological outcome (injury). The activation score of module m associated with positive instances of histopathology endpoint p is the average of the absolute value of the Z‐score for all genes i in the module m across all conditions j in the histopathological outcome p.
To determine whether a module m is ’activated‘ for a particular histopathology endpoint p, the activation score must be greater than a threshold. The threshold was determined as having a P‐value < 0.025 from the distribution of all activation scores of all modules across all conditions. The activation threshold was 1.79 for the liver and 2.08 for the kidneys.
We also used the Matthews correlation coefficient (MCC) to evaluate the specificity of a module m to the histopathology endpoint p. The MCCm,p can be calculated from a confusion matrix as:
where TP, TN, FP and FN refer to the number of true positives, true negatives, false positives and false negatives, respectively. The MCC values range from −1.0 to +1.0, where the latter indicates a perfect correlation. In determining a true positive for a particular histopathology endpoint, the positive instance (condition j) of injury indicator p must have an activation score greater than the organ‐specific activation threshold. Positive instances of injury indicator p that is not activated (activation score less than organ‐specific threshold cutoff) were considered false negatives. In the same manner, we determined true negatives (non‐injury‐conditions j that did not activate a particular module m) and false positives (non‐injury‐causing conditions j that activated a module m).
To link module m with injury p, we chose the module with the highest MCCm,p for a particular injury or histopathology provided that the MCCm,p >0.4. In a 2 × 2 contingency table, the MCCm,p is related to the chi‐square (χ2) statistics (Powers, 2011). The false‐discovery‐rate‐corrected P‐value of each MCCm,p was determined from their corresponding χ2 values.
The statistical programming language R was used to perform principal component analyzes, linear regression, and heatmap clustering. To determine the overlap of the gene composition between two modules, we used the Sorensen‐Dice coefficient (Dice, 1945), where an overlap score of one means that the two modules are identical and zero means that the two modules have no genes in common.
External validation
We further evaluated the relevance of the modules for compatible histopathology assessments measured in DrugMatrix, a toxicogenomic database that contains organ‐specific gene expression data matched to dose‐dependent chemical exposures and histopathology assessments in Sprague–Dawley rats (Ganter et al., 2006). The animals were exposed to different chemicals for 0.25 to 7 days, typically at concentrations comparable to the high concentration in TG‐GATEs. This dataset utilized Affymetrix geneChip rat Genome 230 2.0 arrays. We used the same protocol for processing this data as discussed above. The activation of the modules from TG‐GATEs was determined from DrugMatrix gene expression data and assessed based on positive instances of histopathology endpoint in DrugMatrix.
Data availability
The gene composition of the selected liver and kidney modules are provided in the Supplemental Materials.
Results
Module generation and selection
We used the ISA to generate the modules for both liver and kidney. We systematically adjusted the gene threshold tg and condition threshold tc parameters of the ISA, used to control the correlation of the co‐expressed genes and conditions, respectively, from 2.0 to 4.0 (with 0.5 increments) for each parameter. Twenty‐five combinations of the ISA parameter were used, yielding 891 and 303 co‐expressed modules for the liver and kidney, respectively. In comparison, our previous efforts using one parameter set (tg = 3.5 and tc = 1.8) yielded 78 liver modules using DrugMatrix (Tawa et al., 2014).
We examined gene overlap among all generated modules (Supplementary Material Fig. S1 shows the results for the liver and kidney) and found instances of modules with identical gene composition (overlap score =1) as well as multiple, highly overlapping gene modules. As a consequence of this large gene overlap, the number of modules activated by an injury was also large. For example, for liver fibrosis as the histopathology endpoint, 243 modules had a statistically significant activation score , i.e., the average of the absolute value of the Z‐score for all genes in each of the 243 modules across the conditions that exhibited fibrotic histopathology was larger than 1.79.
In order to select a more manageable number of modules that would still be descriptive and specific to a particular histopathology endpoint, we needed to implement a module selection metric. One way of selecting modules is by choosing the module with the maximum activation score. In liver fibrosis, for example, the maximum activation score among all modules was 3.75 for module 538. This module correctly predicted fibrosis in 11 of the 13 fibrotic conditions, i.e., the module was activated and the injury was present. However, module 538 was also activated by 45 other conditions where no fibrosis was evident. Instead, we implemented selection of modules based on the highest MCCm,p as the specificity metric linking modules to a particular injury.
Histopathology‐specific modules
For each histopathology in the liver and kidney, we chose modules that scored the highest MCCm,p, provided that MCCm,p > 0.4. Of the 35 possible liver histopathology assessments in TG‐GATEs as determined in the Methodology Section, we were able to link 11 histopathology endpoints to their corresponding module (Table 1). All P‐values associated with the MCCm,p for the 11 modules were < 10−16. Figure 1 shows the MCCm,p of each histopathology endpoint with respect to the 11 modules. By design, the modules chosen have the highest MCCm,p for a particular histopathology endpoint p (highest MCCm,p in a column). However, some modules were activated by other injury conditions (column‐ and row‐wise comparison of MCCm,p). In addition, the modules with maximum MCCm,p (LM1 to LM11) were selected from different (tg, tc) parameter combinations of the ISA (Supplementary Material Table S1).
Table 1.
Modules associated with liver histopathology
| Module ID | Ng | MCC | TP | TN | FN | FP | Sen | Spc | PPV | NPV | BAc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alteration (cytoplasmic) | LM1 | 23 | 0.53 | 7 | 1654 | 0 | 18 | 1.00 | 0.99 | 0.28 | 1.00 | 0.99 |
| Alteration (nuclear) | LM2 | 131 | 0.42 | 9 | 1633 | 1 | 36 | 0.90 | 0.98 | 0.20 | 1.00 | 0.94 |
| Anisonucleosis | LM3 | 82 | 0.47 | 8 | 1649 | 2 | 20 | 0.80 | 0.99 | 0.29 | 1.00 | 0.89 |
| Cellular infiltration | LM4 | 31 | 0.44 | 20 | 1610 | 22 | 27 | 0.48 | 0.98 | 0.43 | 0.99 | 0.73 |
| Foci (cellular) | LM5 | 41 | 0.43 | 6 | 1647 | 0 | 26 | 1.00 | 0.98 | 0.19 | 1.00 | 0.99 |
| Granular degeneration (eosinophilic) | LM6 | 20 | 0.67 | 29 | 1622 | 12 | 16 | 0.71 | 0.99 | 0.64 | 0.99 | 0.85 |
| Fibrosis | LM7 | 56 | 0.56 | 12 | 1643 | 1 | 23 | 0.92 | 0.99 | 0.34 | 1.00 | 0.95 |
| Hematopoiesis | LM8 | 35 | 0.51 | 12 | 1636 | 1 | 30 | 0.92 | 0.98 | 0.29 | 1.00 | 0.95 |
| Proliferation (bile duct) | LM9 | 17 | 0.61 | 21 | 1631 | 9 | 18 | 0.70 | 0.99 | 0.54 | 0.99 | 0.84 |
| Proliferation (oval cell) | LM10 | 150 | 0.53 | 11 | 1640 | 0 | 28 | 1.00 | 0.98 | 0.28 | 1.00 | 0.99 |
| Single cell necrosis | LM11 | 13 | 0.53 | 19 | 1622 | 5 | 33 | 0.79 | 0.98 | 0.37 | 1.00 | 0.89 |
Ng = number of genes; MCC = Matthews correlation coefficient; TP = true positives; TN = true negatives; FN = false negatives; FP = false positives; Sen = sensitivity or TP/(TP + FN); Spc = specificity or TN/(FP + TN); PPV = positive predictive value or TP/(TP + FP); NPV = negative predictive value or TN/(TN + FN); BAc = Balanced Accuracy or ½ (Sen + Spc). The gene complement of each module is provided as an Excel file in the Supplementary Materials.
Figure 1.
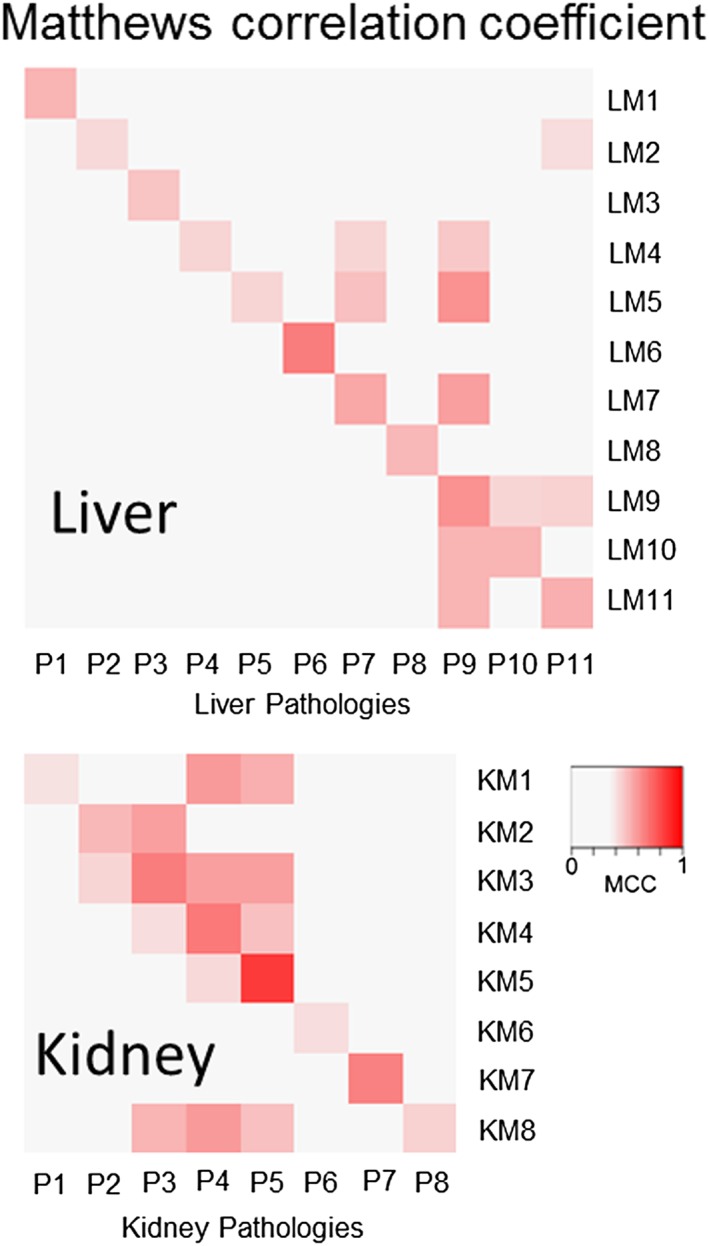
The module for the specific histopathology endpoints has the highest Matthews correlation coefficient (MCC). Shown are the MCCs for the liver histopathology assessments, ordered according to Table 1 from cytoplasmic alteration as P1 to single cell necrosis as P11, and 11 modules for the liver, LM1 to LM11. LM1 corresponds to the module for liver P1 and has the highest MCC for that histopathology. Also shown are the MCCs for the eight kidney injuries, ordered as listed in Table 2. As with the liver modules, KM1 is the module for kidney P1 and has the highest MCC for P1.
To illustrate the ability of the module to characterize injury conditions, we examined the chemicals in the dataset known to cause liver fibrosis. Figure 2 shows the agreement between the activation of the module for liver fibrosis (LM7) with the positive observation of the fibrosis histopathology. Of the six chemicals causing liver fibrosis, all activated module LM7. The activation of module LM7 correctly predicted 12 of the 13 fibrotic conditions (true positives), with the exception of high concentration of naphthyl isothiocyanate at 4 days. Overall, liver fibrosis module LM7 was associated with 12 TP, 1 643 TN, 1 FN, and 23 FP activations for an MCCLM7,liver fibrosis of 0.56.
Figure 2.
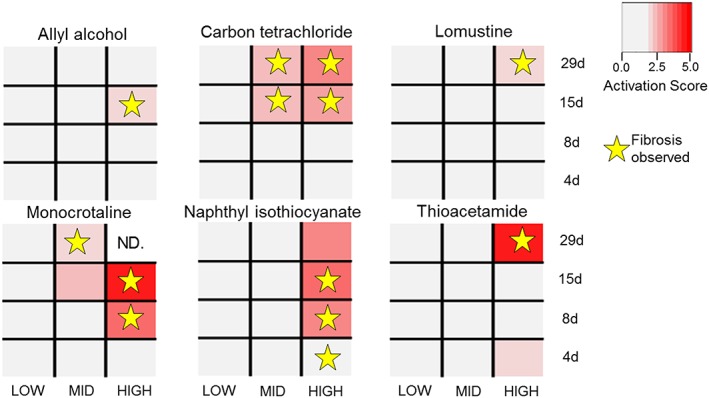
Positive instances of the liver fibrosis histopathology activate the liver fibrosis module. Conditions (chemical‐time‐dose combination) with activation scores above the activation threshold (corresponding to P‐value <0.025) are considered activated (in red). The activation score is defined as the average of the absolute value of the expression Z‐scores of the genes in the liver fibrosis module (LM7). Positive instances of liver fibrosis are marked with stars. Liver fibrosis is predicted if the histopathology endpoint is observed and LM7 is activated for the condition (red rectangles with stars). ND indicates no data are available.
In the kidney, we linked eight modules to a respective kidney histopathology (Fig. 1 and Table 2) with a maximum MCC P‐values of < 10−13. Figure 3 shows the case of module KM5 activation in response to chemical exposures linked to kidney fibrosis. In TG‐GATEs, two chemicals (allopurinol and puromycin) cause kidney fibrosis with five specific exposure conditions associated with identified kidney fibrosis histopathology – all of which are activated by module KM5. Overall, the kidney fibrosis module KM5 was associated with 5 TP, 475 TN, 0 FN and 2 FP activations for an MCCKM5,kidney fibrosis of 0.84. For the two FP instances, cisplatin and triamterene at the 29‐day exposure high‐concentration regimens activated KM5 but did not show positive histopathology for kidney fibrosis in TG‐GATEs.
Table 2.
Modules associated with kidney histopathology
| Histopathology | Module ID | Ng | MCC | TP | TN | FN | FP | Sen | Spc | PPV | NPV | BAc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cast (hyaline) | KM1 | 26 | 0.40 | 6 | 459 | 6 | 11 | 0.50 | 0.98 | 0.35 | 0.99 | 0.74 |
| Cellular infiltration | KM2 | 53 | 0.52 | 7 | 462 | 9 | 4 | 0.44 | 0.99 | 0.64 | 0.98 | 0.71 |
| Degeneration | KM3 | 72 | 0.66 | 12 | 458 | 9 | 3 | 0.57 | 0.99 | 0.80 | 0.98 | 0.78 |
| Dilatation | KM4 | 9 | 0.67 | 16 | 451 | 11 | 4 | 0.59 | 0.99 | 0.80 | 0.98 | 0.79 |
| Fibrosis | KM5 | 147 | 0.84 | 5 | 475 | 0 | 2 | 1.00 | 1.00 | 0.71 | 1.00 | 1.00 |
| Hypertrophy | KM6 | 18 | 0.41 | 5 | 463 | 5 | 9 | 0.50 | 0.98 | 0.36 | 0.99 | 0.74 |
| Intracytoplasmic inclusion body | KM7 | 46 | 0.65 | 7 | 467 | 1 | 7 | 0.88 | 0.99 | 0.50 | 1.00 | 0.93 |
| Necrosis | KM8 | 16 | 0.44 | 9 | 452 | 10 | 11 | 0.47 | 0.98 | 0.45 | 0.98 | 0.72 |
Ng = number of genes; MCC = Matthews correlation coefficient; TP = true positives; TN = true negatives; FN = false negatives; FP = false positives; Sen = sensitivity or TP/(TP + FN); Spc = specificity or TN/(FP + TN); PPV = positive predictive value or TP/(TP + FP); NPV = negative predictive value or TN/(TN + FN); BAc = Balanced Accuracy or ½ (Sen + Spc). The gene complement of each module is provided as an Excel file in the Supplementary Materials.
Figure 3.
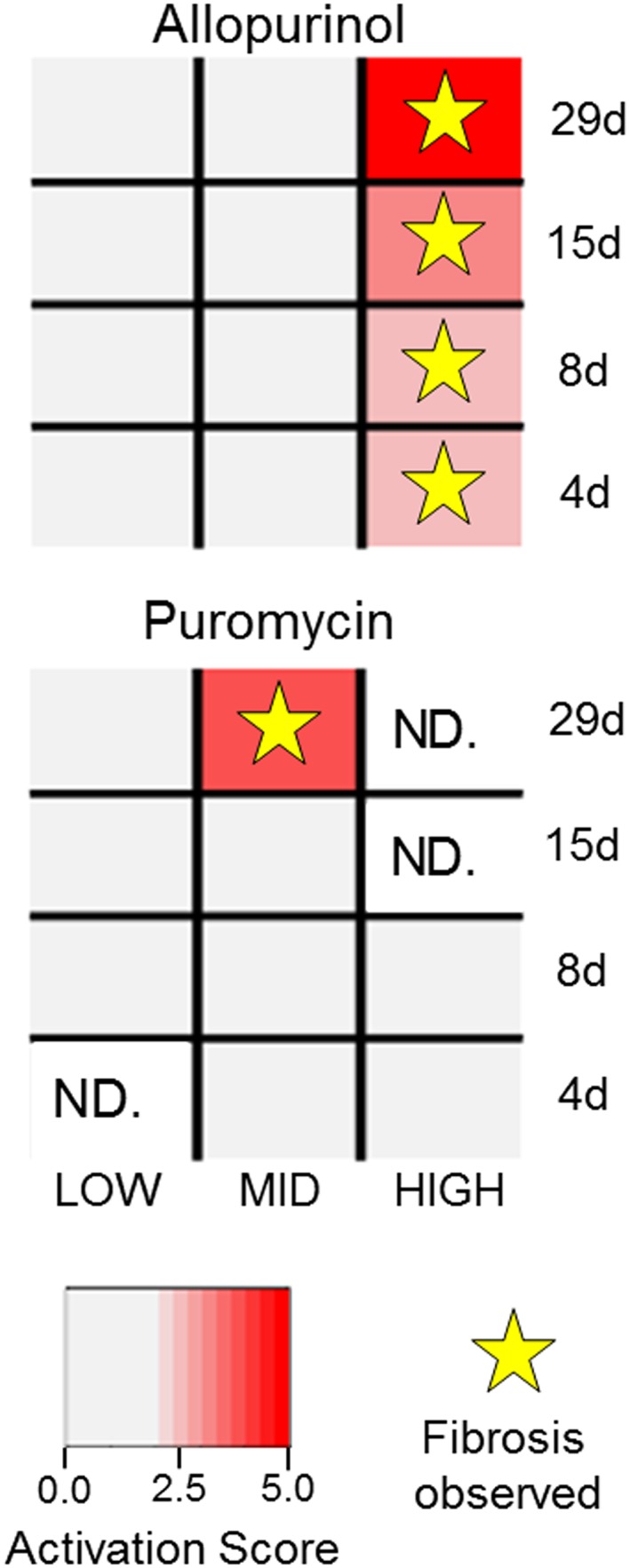
Positive instances of the kidney fibrosis histopathology activate the kidney fibrosis module. Conditions with KM5 activation scores above the activation threshold were considered activated (in red). Positive instances of kidney fibrosis are marked with stars. Liver fibrosis is predicted if the histopathology endpoint is observed and KM5 is activated for the condition (red rectangles with stars). ND indicates no data are available.
External validation of modules
We used DrugMatrix (Ganter et al., 2006) – a large‐scale toxicogenomics database with gene expression data and partly overlapping histopathology observations compared to TG‐GATEs – to validate the activation of the modules generated from TG‐GATEs in predicting the presence of an injury.
We used MCCm,p as a measure of how well the modules m from TG‐GATEs could predict the presence of histopathology endpoint p in DrugMatrix using the gene expression data of the latter database. Both kidney and liver tissues had three possible comparisons each, one of which was common to both tissue types (cellular infiltration). Table 3 summarizes the possible comparisons between these overlapping histopathology endpoints.
Table 3.
Verification of TG‐GATEs liver and kidney modules using DrugMatrix
| Histopathology | Module | Ng | MCCT | MCCDM | TP | TN | FN | FP | Sen | Spc | PPV | NPV | BAc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | |||||||||||||
| Cellular infiltration | LM4 | 31 | 0.44 | 0.24 | 22 | 405 | 211 | 1 | 0.09 | 1.00 | 0.96 | 0.66 | 0.55 |
| Fibrosis | LM7 | 56 | 0.56 | 0.57 | 8 | 619 | 2 | 11 | 0.80 | 0.98 | 0.42 | 1.00 | 0.89 |
| Proliferation (bile duct) | LM9 | 17 | 0.61 | 0.60 | 11 | 614 | 3 | 12 | 0.79 | 0.98 | 0.48 | 1.00 | 0.88 |
| Kidney | |||||||||||||
| Cast (hyaline) | KM1 | 26 | 0.40 | 0.43 | 3 | 349 | 0 | 13 | 1.00 | 0.96 | 0.19 | 1.00 | 0.98 |
| Cellular infiltration | KM2 | 53 | 0.52 | 0.16 | 1 | 352 | 2 | 10 | 0.33 | 0.97 | 0.09 | 0.99 | 0.65 |
| Necrosis | KM8 | 16 | 0.44 | 0.52 | 9 | 341 | 7 | 8 | 0.56 | 0.98 | 0.53 | 0.98 | 0.77 |
Ng = number of genes; MCCT = Matthews correlation coefficient in TG‐GATEs; MCCDM = Matthews correlation coefficient in DrugMatrix; TP = true positives; TN = true negatives; FN = false negatives; FP = false positives; Sen = sensitivity or TP/(TP + FN); Spc = specificity or TN/(FP + TN); PPV = positive predictive value or TP/(TP + FP); NPV = negative predictive value or TN/(TN + FN); BAc = Balanced Accuracy or ½ (Sen + Spc).
Among the liver modules, fibrosis (LM7) and bile duct proliferation (LM9) were specifically activated in their corresponding histopathology endpoints in DrugMatrix with MCCs of 0.57 and 0.60, respectively. These correlation values were similar to the values from the TG‐GATEs data. LM7 was activated in eight of the 10 liver fibrosis conditions in DrugMatrix (true positives) and in 11 conditions not associated with fibrosis (false positives). The negative predictive rate, i.e., the fraction of true negatives among all conditions predicted to be negative, was > 0.99 for both of these modules. For the kidney histopathology assessments found in both TG‐GATEs and DrugMatrix, the kidney modules from TG‐GATEs could be used to predict the histopathology in DrugMatrix for the hyaline cast and necrosis with an MCCm,p >0.4. These predictions were similarly associated with similar positive and negative predictive rates to the liver modules. Modules assigned to cellular infiltration for both liver and kidney tissues have significantly lower MCC values for predicting positive instances of this condition in DrugMatrix as compared with TG‐GATEs.
Module properties
The creation of gene modules represents a systems toxicology effort to organize the transcriptional response around specific injury endpoints, here chosen as graded histopathology assessments of liver and kidney tissues. Next, we examined the properties of these modules with respect to activation and classification as a response to chemical toxicant exposures.
Modules classify and quantify injuries
Module activation can be used to separate exposure conditions that are associated with injury from those that are not. Using principal component analysis (PCA) based on the activation score for the histopathology‐related modules; Fig. 4A shows the separation of the 1679 conditions in the liver for TG‐GATEs into those that cause an adverse histopathology outcome versus those conditions that do not cause any injury. The first three principal components of module activation in the liver accounted for 87% of the variance. In contrast, the PCA of all conditions based on individual gene activation – here limited to genes found in the modules – did not distinguish between conditions associated with adverse and normal histopathology outcomes (Fig. 4B). The first three principal components of the Z‐scores of genes in the liver accounted for 41% of the variance.
Figure 4.
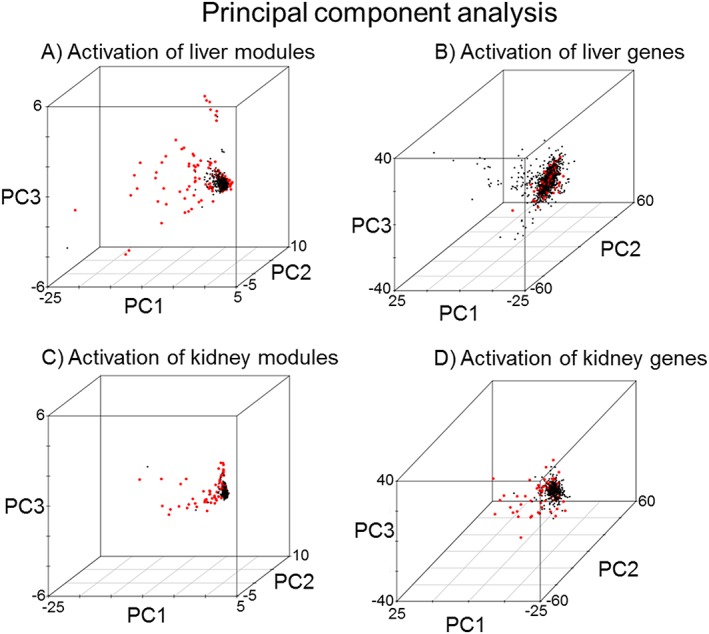
Module activation can predict histopathology‐causing conditions. Principal component analysis of the activation of the modules by histopathology‐graded chemical exposure conditions (red dots) for modules (A and C) and for genes (B and D). A condition was considered injury causing if at least one of the 11 liver (A and B) or 8 kidney (C and D) histopathology endpoints was positive. All other conditions not associated with histopathology graded damage are marked with black dots.
The PCA of all 482 conditions involving the kidney in TG‐GATEs based on the activation score for the eight kidney modules showed a separation between adverse histopathology conditions from those devoid of any injury (Fig. 4C). The first three principal components of module activation in the kidney accounted for 93% of the variance. In contrast, gene‐level PCA did not clearly separate out injury versus non‐injury conditions (Fig. 4D). The first three principal components of Z‐scores of genes (involved in modules) in the kidney accounted for only 50% of the variance.
We also examined the dose‐dependent relationship between the module activation score and the histopathology assessment, i.e., whether heightened activation correlated with increased severity of the histopathology. The histopathology grades were converted to numerical values based on the severity of the injury, ranging from 1 for ’minimal‘ to 4 for ’severe’, and averaged across the animals studied for a particular exposure conditions. Figure 5A shows the general trend for all injury conditions as a function of increasing activation score. The overall coefficient of determination (R2) as a measure of the goodness of the linear fit for all liver histopathology scores and activation of the appropriate injury module was 0.45. Table 4 shows R2 for each of the 11 individual histopathologies, ranging from the best in cytoplasmic alteration (R2 = 0.90) to absence of correlation in hematopoiesis (R2 = 0.09). The corresponding data are shown in Supplementary Material Fig. S2. The kidney modules also showed increased injury severity as a function of activation (Fig. 5B); albeit the R2 for the linear relationship was lower than in the liver (0.36). Among the eight kidney histopathology endpoints, the correlation of histopathology grade versus activation score was best for the presence of intracytoplasmic inclusion bodies (0.65) and worst (0.04) in hypertrophy (Table 4 and Supplemental Material Fig. S2).
Figure 5.
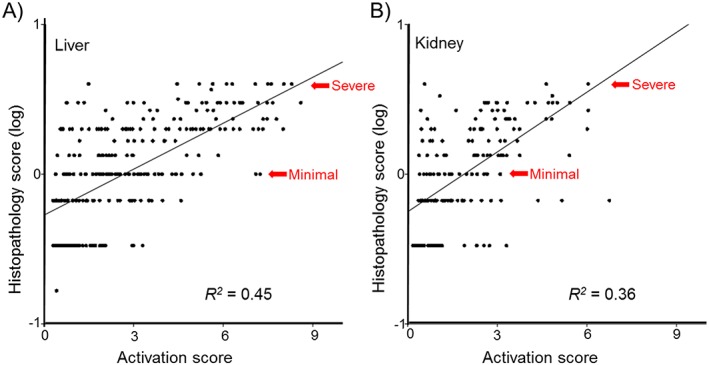
Injury severity is associated with correspondingly higher activation scores. In general, as the severity of the histopathology increases, the activation score of the module increases. The histopathology assessments were converted to numbers (with minimal = 1, slight = 2, moderate = 3, severe = 4) and the histopathology score was determined as the average over all replicates. Only conditions with at least one replicate having an observed histopathology were considered for the linear regression. The goodness of the fit was measured as the coefficient of determination (R 2).
Table 4.
Linear regression of the activation score relative to the histopathology grade or severity
| Histopathology | Module ID | R 2 |
|---|---|---|
| Liver | ||
| Alteration (cytoplasmic) | LM1 | 0.90 |
| Alteration (nuclear) | LM2 | 0.73 |
| Anisonucleosis | LM3 | 0.15 |
| Cellular infiltration | LM4 | 0.56 |
| Foci (cellular) | LM5 | 0.43 |
| Granular degeneration (eosinophilic) | LM6 | 0.23 |
| Fibrosis | LM7 | 0.57 |
| Hematopoiesis | LM8 | 0.09 |
| Proliferation (bile duct) | LM9 | 0.34 |
| Proliferation (oval cell) | LM10 | 0.68 |
| Single cell necrosis | LM11 | 0.36 |
| Kidney | ||
| Cast (hyaline) | KM1 | 0.26 |
| Cellular infiltration | KM2 | 0.63 |
| Degeneration | KM3 | 0.16 |
| Dilatation | KM4 | 0.62 |
| Fibrosis | KM5 | 0.64 |
| Hypertrophy | KM6 | 0.04 |
| Intracytoplasmic inclusion body | KM7 | 0.65 |
| Necrosis | KM8 | 0.20 |
R2 = coefficient of determination. The histopathology assessments were converted to scores (minimal = 1, slight = 2, moderate = 3, and severe = 4) and averaged over all replicates in a condition. The R2 was determined through linear regression of the averaged histopathology grade and the activation of the module by the positive instances of the injury.
A unique module describes each histopathology
One of the advantages of our module selection protocol was the assignment of unique modules for each histopathology endpoint although some endpoints are closely related. For example, the four liver histopathology endpoints – cellular infiltration, fibrosis, bile duct proliferation and single cell necrosis – share common exposure conditions with the same chemical causing multiple abnormal histopathologies. Using the full set of 891 liver modules, 61 modules were activated by all four endpoints. Figure 6A shows that all 61 modules shared common genes and had high overlap scores, including overlap scores of 1.0 indicating that two modules have the same gene composition. However, in our proposed protocol, the MCCm,p metric was used to link a unique module to each of these closely related endpoints. Figure 6B shows that the selected modules for the four closely related liver endpoints have very limited gene membership overlap. The limited gene overlap extended to all 11 liver and eight kidney modules (Supplementary Material Table S2).
Figure 6.
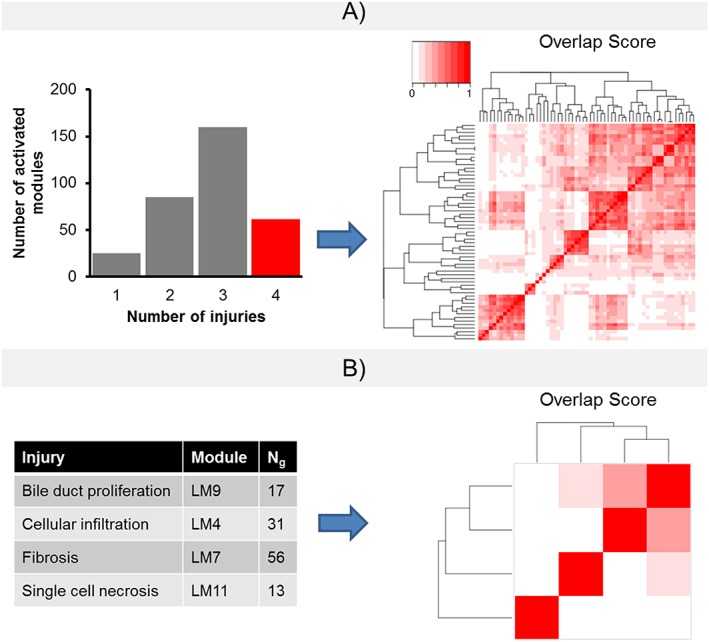
The module selection assigns unique modules to closely related histopathology endpoints. In TG‐GATEs, chemical exposure conditions may cause a number of injuries. Four closely related histopathology assessments (cellular infiltration, fibrosis, bile duct proliferation and single cell necrosis) share a number of common chemicals. (A) All four injuries activate 61 modules in the liver, with the 61 modules having overlapping genes, as determined by the Sorensen‐Dice coefficient (Dice, 1945). (B) Using our module selection protocol, the number of overlapping genes among the modules is limited.
Modules capture organ‐specific injuries
For a number of chemical exposure conditions, the TG‐GATEs data provide matched liver and kidney data for the same set of animals. Figure 7 shows the histopathology assessment for liver and kidney fibrosis and the global module activation pattern for two nephrotoxicants and two hepatotoxicants. Allopurinol and puromycin, two chemicals that cause kidney fibrosis, activated a number of kidney modules but no liver modules. Similarly, the known hepatotoxicants thioacetamide and carbon tetrachloride primarily activated liver modules and not kidney modules. The corresponding data for allyl alcohol, lomustine, monocrotaline, and naphthyl isothiocyanate are shown in Supplementary Material Fig. S3.
Figure 7.
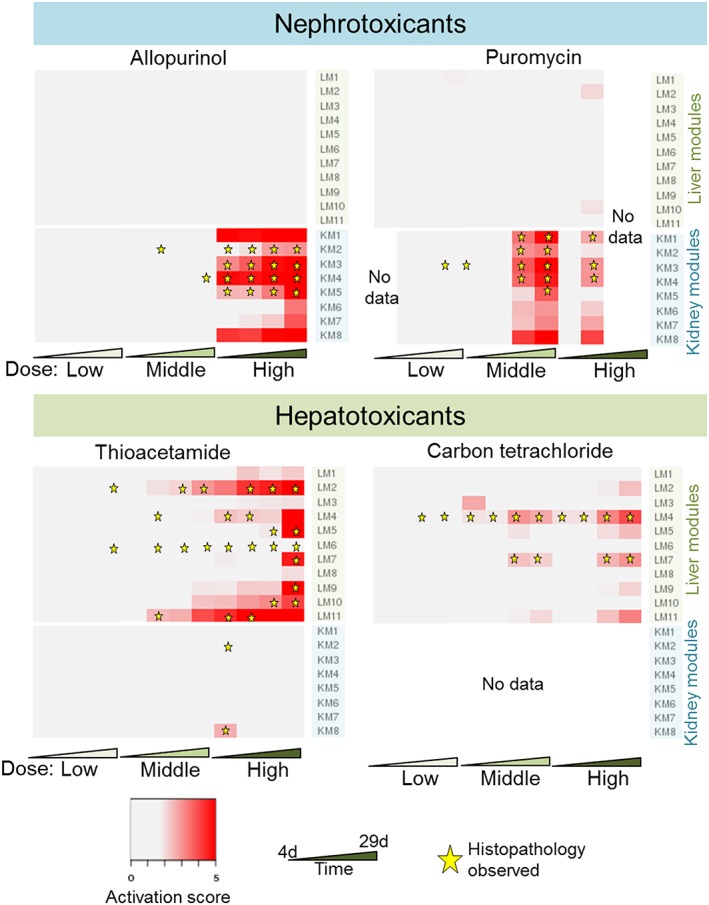
The activation of the modules is organ‐specific. Allopurinol and puromycin, two nephrotoxicants that cause a number of kidney injuries, including kidney fibrosis, generally activate kidney modules. Conversely, thiocetamide, a hepatotoxicant that causes liver fibrosis among other liver injuries, selectively activates liver injury modules. Also shown is carbon tetrachloride, a known hepatotoxicant. The four other chemicals causing liver fibrosis are shown in Supplementary Material Fig. S3.
Discussion
Module generation and selection
In this study, we proposed a comprehensive scheme for creating and selecting modules associated with different histopathologies. Our protocol provides for an exhaustive module generation and a metric for module selection applicable to any organ or tissue type. Here, we implemented this scheme for assessing graded histopathology for both liver and kidney using TG‐GATEs data.
The module generation scheme was designed to essentially be both non‐parametric and unsupervised. We consider the module generation as non‐parametric because we systematically varied the gene threshold tg and condition threshold tc in the ISA to generate the modules. Twenty‐five sets of ISA parameters were used to generate the modules. For the most restrictive parameter set (tg = 4.0 and tc = 4.0), 14 liver modules were generated containing an average of ~37 genes each. As either tg and tc were relaxed to lower values, the number of modules as well as the number of genes and conditions in a module increased. The comprehensive module generation identified 891 modules for the liver and 303 modules for the kidney. As we filtered out modules that had >200 genes, no module from parameter sets where tg = 2.0 was part of the modules used in the analyzes. The module generation was also unsupervised, in the sense that no prior knowledge was required beyond the expression data and no biological information or pathway knowledge was used to determine the subset of genes and conditions in a module. We emphasize that the histopathology is not an input of the ISA protocol and does not per se influence the composition of the subset of conditions composing the co‐expression module.
The module generation process has the advantage that the modules are diverse in terms of how correlated the genes and conditions are within a co‐expression module. Thus, there is no a priori assumption on how correlated a biological process needs to be in order to be considered part of a molecular toxicity pathway. However, with more partially overlapping modules, a biological effect as gauged by the positive instances of a histopathology assessment might activate multiple non‐independent modules. This creates a problem for prioritizing and selecting modules. Hence, we used the MCCm,p (MCC of module m for histopathology outcome p) – a balanced measure of true and false positives and negatives – to address two issues with the comprehensive module generation, namely (i) to determine which modules are more predictive of the presence of an injury and (ii) to identify which set of ISA parameters creates modules that are specific to injuries.
By selecting modules with the highest MCCm,p, we chose modules with better chances of predicting the presence of the injury. In our liver fibrosis exemplar, 12 of the 13 conditions with observable liver fibrosis histopathology also activated (above the threshold cut‐off) the module LM7 (Fig. 2). This module is not necessarily the same as the module with the highest activation score (module 538 in liver fibrosis). Such highly activated modules can be influenced by outliers, i.e., positive instances of the injury that have an abnormally high average expression Z‐score of the genes in the module. Module 538 activated 85% of the fibrotic conditions (versus 92% for module LM7) and had a higher number of false positive conditions than LM7 (45 versus 23).
Linking histopathology endpoints with modules, we found that no single set of ISA parameters produced modules that consistently scored the highest MCCm,p for the 11 liver and eight kidney histopathology endpoints (Supplementary Material Table S1). As the ISA parameter controls the coherence of the co‐expression within a module, and with the assumption that modules with the highest MCCm,p are the most predictive module for a histopathology, then differing tg and tc thresholds generated the most consistently linked modules to the histopathology. In comparison, for the entire set of 11 liver injuries, our previously published ISA parameter set of tg = 3.5 and tc = 1.8 (Tawa et al., 2014) can only describe one injury better than the comprehensive module generation presented here, underscoring the importance of generating and examining additional modules.
From the module selection protocol, 11 modules were directly linked to the same number of histopathology assessments in the liver (Table 1) and, similarly, eight modules for the kidney (Table 2). The module selection is based on statistical metrics applied to co‐expressed genes rather than using biological information, e.g., specific cellular pathways or presence of known genes associated with a particular injury.
Correlated prediction of histopathology with module activation
Two of the histopathology outcomes we linked using module activation were liver and kidney fibrosis. We showed a strong correlation between activation and abnormal histopathology for chemicals known to cause liver fibrosis in TG‐GATEs, where 12 of the 13 fibrotic conditions activated module LM7 (Fig. 2 and Supplementary Material Fig. S4). Similarly, a lack of module activation correctly predicted a lack of fibrosis in 1 643 out of 1 666 non‐fibrotic exposure conditions. Module LM7 was activated in 23 conditions that did not have observed fibrosis. The 23 false positive cases listed in Table 1 involved seven different chemicals under multiple exposure conditions. Three of these chemicals caused fibrosis at different exposure time‐points in TG‐GATEs (Fig. 2), whereas the remaining four chemicals (acetamidofluorene, methapyrilene, methylene dianiline and nitrosodiethylamine) had multiple exposure conditions activating the LM7 module but without observed liver fibrosis. However, all of these chemicals are known to cause or promote liver fibrosis under certain conditions (Fukushima et al., 1979; Nakazato et al., 2010; Chobert et al., 2012; Probert et al., 2014).
For kidney fibrosis, activation of KM5 (the module for kidney fibrosis) predicted all five positive instances of the histopathology assessment in a dose‐dependent manner (Supplementary Material Fig. S4). For two conditions involving cisplatin and triamterene, KM5 was activated but did not show fibrotic histopathology even though cisplatin is known to induce kidney fibrosis (Yuasa et al., 2014) whereas triamterene is an acute nephrotoxicant known to cause nephropathy (Nasr et al., 2014).
TG‐GATEs modules cross‐validated using an external database
We further verified whether the module LM7 was activated in fibrotic conditions in the DrugMatrix, an external dataset of the same scope and size as TG‐GATEs. Eight of the 10 conditions with observed liver fibrosis were correctly predicted by the activation of module LM7 (Table 3), including three conditions of naphthyl isothiocyanate and one condition of lomustine – two chemicals with observed liver fibrosis in TG‐GATEs. In addition, two conditions of methylene dianiline (a false positive chemical in TG‐GATEs) caused fibrosis and activated module LM7 using DrugMatrix. Differences in experimental setup and histopathological assessment from the two studies may in part explain why the methylene dianiline in TG‐GATEs (100 mg kg−1 from 4 to 29 days) did not cause fibrosis, but the same chemical in DrugMatrix (81 mg kg−1 from 3 to 5 days) did. Nitrosodimethylamine, which is chemically related to the TG‐GATEs compound nitrosodiethylamine that was labeled a false positive, causes fibrosis in DrugMatrix and activated module LM7 for this dataset.
Six hundred and thirty conditions in DrugMatrix did not cause liver fibrosis. Most of these conditions also did not activate LM7 (619 true negatives). However, 11 conditions were considered as false positives where these conditions activated the TG‐GATEs module LM7 without the presence of liver fibrosis in DrugMatrix. Most of the false‐positive conditions are related to liver fibrosis. In our previous study, a liver fibrosis module was generated from a combination of differentially expressed genes, co‐expression clustering, pathway enrichment analyzes and protein–protein interaction networks using the DrugMatrix database (AbdulHameed et al., 2014). Seventeen conditions, either with observed fibrosis in DrugMatrix or known to cause fibrosis, clustered together based on the activation of the previous DrugMatrix‐based fibrosis module (AbdulHameed et al., 2014). Of the 11 false positive conditions of LM7 in DrugMatrix, 10 conditions were part of the previously identified cluster of 17 conditions associated with liver fibrosis (AbdulHameed et al., 2014). Thus, the false positives in the verification dataset are also connected to fibrosis, including vinblastine, allyl alcohol, carbon tetrachloride and lipopolysaccharide (Liedtke et al., 2013).
Given that the module construction was geared towards identifying sets of co‐expressing genes, partly reflecting the underlying transcriptional program that needs to be in place to cause fibrosis, module activation may be present before the histological manifestation of fibrosis is evident. Thus, activation of LM7 may be a true pre‐fibrotic signal indicative of the early onset of liver fibrosis, implying that our protocol has the potential to generate modules that can predict the early onset of initiating events in molecular toxicity pathways that can lead to histological damage.
The modules derived from TG‐GATEs were able to predict conditions that cause the same or similar histopathology endpoints in DrugMatrix. The comparison using liver fibrosis for module LM7 was discussed above and in addition, module LM9 has good predictive power for the presence of bile duct proliferation or hyperplasia in DrugMatrix. In the kidney, KM1 for the hyaline cast and KM8 for necrosis were verified through the DrugMatrix external dataset (Table 3). For these endpoints, the MMC values of the classification in DrugMatrix were comparable to the values in TG‐GATEs.
The chemical exposure conditions in DrugMatrix and TG‐GATEs do not overlap to a great extent, and, thus, they represent both similar and different aspects of chemical toxicity. DrugMatrix conditions typically include high doses for a short period of time (0.25 to 7 days). In TG‐GATEs, the chemical exposure conditions ranged from 4 to 29 days with high concentrations that are usually lower than those used in DrugMatrix. In spite of the differences in experimental setup, etc., the cross‐validation of modules generated in TG‐GATEs indicated the potential general applicability of these modules in detecting chemical injuries and activation of molecular toxicity pathways.
In contrast, cellular infiltration in the liver and the kidney in DrugMatrix were not satisfactorily predicted by modules generated from TG‐GATEs. A relatively large number of conditions were annotated in DrugMatrix as having observable cellular infiltration in the liver as compared to in TG‐GATEs (233 vs. 42). This is partly due to the large number of control animals, i.e., those that were not exposed to any chemical, also showing positive histopathology for cellular infiltration in the DrugMatrix data. For the kidney, only three instances of cellular infiltration of the kidney were observed and only one of these was correctly predicted by TG‐GATEs module using DrugMatrix data.
Biological significance of the modules
The construction of gene co‐expression modules that broadly characterize injuries can be used to select specific gene signatures that may be proposed as genes and proteins for future development as clinical biomarkers. Although biological information was not used in the selection of modules, the modules (Tables 1 and 2) may still be related to the biological pathways involved in injury generation or progression. Using fibrosis as an example, both liver and kidney fibrosis modules contain known biomarkers for fibrosis (Adams, 2011), including tissue inhibitor of metalloproteinase (Timp1), type IV collagens (Col4a1, Col4a2) and laminin (LM7 has Lamb1 and KM5 has Lama4). Other known biomarkers (or related genes) such as Timp2, fibronectin Fn1 gene and matrix metalloproteinase 2 (Mmp2) were part of the kidney fibrosis module KM5. In addition, both liver and kidney fibrosis modules also contained Col1a1, Col6a2 and fibrillin. The extracellular matrix‐receptor pathway was the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway that was most significantly overrepresented pathway in both modules, consistent with the excessive scarring associated with fibrosis.
We reviewed the Comparative Toxicogenomics Database (Davis et al., 2011) for genes associated with liver and kidney injuries. The 11 liver modules have on average ~5 genes related to general liver injury while the eight kidney modules have ~7 genes related to general kidney injury. The kidney modules included some of the known biomarkers (Dieterle et al., 2010) for the kidney, namely hepatitis A virus cellular receptor 1 (Havcr1), also known as kidney injury molecule‐1 (Kim‐1), and clusterin (Clu).
Module properties
We examined properties of the modules and their behavior in response to different chemical exposures. First, from the PCA, the activation of the modules could be used to differentiate conditions that are associated with injurious exposure conditions from those that are not. In contrast, just using individual genes, even those that are contained in the modules associated with the injuries, the PCA could not separate out conditions related to injuries (Fig. 4). However, even using modules, this analysis is somewhat limited, as the PCA could not distinguish between injuries, in part because they are closely related, i.e., the same condition can cause two or more distinct histopathology conditions.
We also showed that when the histopathology outcome is positive, the severity of the injury correlated with the activation score (Fig. 5), a property which could be exploited in identifying biomarkers or gene signatures indicative of early detection and damage prognosis of chemical exposures. While in general we see that severe injuries lead to higher activation scores, some injuries have a poor correlation between severity and module activation. However, in injuries where the linear regression has a poor correlation, the positive instances of the injury still activated the module. In liver hematopoiesis, associated with a low R2 of 0.09 for LM8 (Table 4), high‐dose iproniazid exposures have activation scores of 1.84 and 3.65 with severity grades of 2.0 and 2.3 at 8 and 15 days, respectively. High doses of naproxen activate LM8 at 8 and 15 days with activation scores of 2.74 and 4.18, albeit with a 2.0 histopathology grade for both conditions. Taken together, the modules with lower values of R2 may be associated with non‐linear dose–response relationships, and, hence, less likely to be informative as a source of early detection biomarkers, yet would still be good indicators of the presence of the injury.
We further showed that the genes have limited or no overlap among modules (Supplementary Material Table S2). The selection of the module was solely based on using the MCCm,p as a metric, thus the uniqueness of the composition of genes in the module was unexpected. Even modules associated with closely related injuries have genes with limited overlap among them (Fig. 6B). In some cases, genes in one module may appear in the other modules as genes may be part of multiple response pathways that are commonly activated in response to different injuries. However, having unique modules with limited overlap in gene composition is important for selecting specific biomarkers for a given particular injury.
Lastly, we showed that differential organ toxicity and specificity for nephrotoxicants and hepatotoxicants was also captured by the generated modules (Fig. 7 and Supplementary Material Fig. S3). Examining activation of the modules, the same chemical exposure conditions administered to the animal could specifically affect the kidney, i.e., activate kidney modules, but not the liver for nephrotoxicants. Hepatotoxicants also activated liver modules, but not kidney modules, in animals exposed to the same chemical. This toxicant specificity might be utilized in developing assays based on the gene/protein composition of these modules to identify organ‐specific molecular toxicity pathways for predicting adverse effects of a given chemical.
Limitations of current protocol and prospects for use
The module methodology is based on identifying co‐expressed genes by the ISA bi‐clustering algorithm and dependent on the availability of comprehensive, large‐scale in vivo datasets. Inclusion of additional chemical exposure conditions – especially from chemical exposures that cause multiple, closely related injuries (e.g., cellular infiltration, fibrosis, bile duct proliferation, and single cell necrosis) as well as chemical exposures that are very specific to a particular endpoint – should further improve module generation for distinguishing these and other endpoints from one another.
Despite these general limitations, the elucidation of co‐expressed modules associated with histopathology outcomes can provide insight into the underlying molecular mechanism of the injury. We showed that gene co‐expression modules could characterize chemically induced liver and kidney injuries and may provide a rational basis for identifying and developing potential biomarkers for diagnosis or prognosis. For example, the liver fibrosis module LM7 could predict the presence of the injury and may predict the onset of fibrosis based on dose‐dependent activation of the module. Even for the exposures that did not develop fibrosis yet activated the LM7 module, the chemicals themselves are known to cause fibrosis under different conditions. Hence, LM7 activation may constitute a prognostic module for the early onset of fibrosis. If so, the modules may serve as a foundation for creating diagnostic tests for monitoring adverse health effects.
Conclusion
Here, we used TG‐GATEs to identify co‐expressed genes (modules) specific to injury endpoints in the liver and kidney. Our implemented method associated chemical‐induced injury modules with 11 liver and eight kidney histopathology endpoints based on time‐ and dose‐dependent activation of the modules. We showed that the activation of the modules for a particular chemical exposure condition, i.e., chemical‐time‐dose combination, correlated with the onset and presence of histopathological damage in a dose‐dependent manner. We showed that the liver fibrosis‐linked module (LM7) was activated in 92% of liver fibrotic conditions; similarly, the kidney fibrosis‐linked module (KM5) was activated in 100% of the kidney fibrotic conditions. The false‐positive conditions, where the module was activated without the presence of injury, were also identified to be related to the injury. In general, the activation of the module can be verified and validated by positive instances of the same or similar histopathology endpoints in DrugMatrix, an external toxicogenomics database. Furthermore, the modules could distinguish different types of injuries caused by chemical exposures as well as whether the injury‐module activation was specific to the tissue of origin (liver and kidney). The generated modules provide a link between toxic chemical exposures, different molecular initiating events among underlying molecular pathways and resultant organ damage.
Conflict of interest
The authors did not report any conflict of interest.
Supporting information
Supporting info item
Acknowledgments
The authors were supported by the U.S. Army Medical Research and Materiel Command (Fort Detrick, MD) as part of the U.S. Army's Network Science Initiative, and by the Defense Threat Reduction Agency grant CBCall14‐CBS‐05‐2‐0007. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or of the U.S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Te, J. A. , AbdulHameed, M. D. M. , and Wallqvist, A. (2016) Systems toxicology of chemically induced liver and kidney injuries: histopathology‐associated gene co‐expression modules. J. Appl. Toxicol., 36: 1137–1149. doi: 10.1002/jat.3278.
References
- AbdulHameed MDM, Tawa GJ, Kumar K, Ippolito DL, Lewis JA, Stallings JD, Wallqvist A. 2014. Systems level analysis and identification of pathways and networks associated with liver fibrosis. PLoS One 9 e112193. DOI:10.1371/journal.pone.0112193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams LA. 2011. Biomarkers of liver fibrosis. J. Gastroenterol. Hepatol. 26: 802–809. DOI:10.1111/j.1440-1746.2010.06612.x. [DOI] [PubMed] [Google Scholar]
- Bai JP, Abernethy DR. 2013. Systems pharmacology to predict drug toxicity: integration across levels of biological organization. Annu. Rev. Pharmacol. Toxicol. 53: 451–473. DOI:10.1146/annurev-pharmtox-011112-140248. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Ihmels J, Barkai N. 2003. Iterative signature algorithm for the analysis of large‐scale gene expression data. Phys. Rev. E 67: 031902 DOI:10.1103/PhysRevE.67.031902. [DOI] [PubMed] [Google Scholar]
- Blomme EA, Yang Y, Waring JF. 2009. Use of toxicogenomics to understand mechanisms of drug‐induced hepatotoxicity during drug discovery and development. Toxicol. Lett. 186: 22–31. DOI:10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. 2010. Independent filtering increases detection power for high‐throughput experiments. Proc. Natl. Acad. Sci. 107: 9546–9551. DOI:10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chobert M‐N, Couchie D, Fourcot A, Zafrani E‐S, Laperche Y, Mavier P, Brouillet A. 2012. Liver precursor cells increase hepatic fibrosis induced by chronic carbon tetrachloride intoxication in rats. Lab. Invest. 92: 135–150. DOI:10.1038/labinvest.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, King BL, Mockus S, Murphy CG, Saraceni‐Richards C, Rosenstein M, Wiegers T, Mattingly CJ. 2011. The comparative toxicogenomics database: update 2011. Nucleic Acids Res. 39: D1067–D1072. DOI:10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR. 1945. Measures of the amount of ecologic association between species. Ecology 26: 297–302. DOI:10.2307/1932409. [Google Scholar]
- Dieterle F, Sistare F, Goodsaid F, Papaluca M, Ozer JS, Webb CP, Baer W, Senagore A, Schipper MJ, Vonderscher J. 2010. Renal biomarker qualification submission: a dialog between the FDA‐EMEA and Predictive Safety Testing Consortium. Nat. Biotechnol. 28: 455–462. DOI:10.1038/nbt.1625. [DOI] [PubMed] [Google Scholar]
- Fukushima S, Shibata M, Hibino T, Yoshimura T, Hirose M, Ito N. 1979. Intrahepatic bile duct proliferation induced by 4, 4'‐diaminodiphenylmethane in rats. Toxicol. Appl. Pharmacol. 48: 145–155. DOI:10.1016/S0041-008X(79)80017-4. [DOI] [PubMed] [Google Scholar]
- Ganter B, Snyder RD, Halbert DN, Lee MD. 2006. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class‐dependent effects using the DrugMatrix® database. Pharmacogenomics 7: 1025–1044. DOI:10.2217/14622416.7.7.1025. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80 DOI:10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham V, McLeod HL. 2009. Genomics: applications in mechanism elucidation. Adv. Drug Delivery Rev. 61: 369–374. DOI:10.1016/j.addr.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussainzada N, Lewis JA, Baer CE, Ippolito DL, Jackson DA, Stallings JD. 2014. Whole adult organism transcriptional profiling of acute metal exposures in male Zebrafish. BMC Pharmacol. Toxicol. 15: 15 DOI:10.1186/2050-6511-15-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H. 2014. Open TG‐GATEs: a large‐scale toxicogenomics database. Nucleic Acids Res. 43: D921–927. DOI:10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31: 370–377. DOI:10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer‐Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. DOI:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iskar M, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH, Runz H, Gavin AC, Pepperkok R, van Noort V. 2013. Characterization of drug‐induced transcriptional modules: towards drug repositioning and functional understanding. Mol. Syst. Biol. 9: 662 DOI:10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416. DOI:10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C, Luedde T, Sauerbruch T, Scholten D, Streetz K, Tacke F, Tolba R, Trautwein C, Trebicka J, Weiskirchen R. 2013. Experimental liver fibrosis research: update on animal models, legal issues and translational aspects. Fibrogenesis Tissue Repair 6: 1–25. DOI:10.1186/1755-1536-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madejczyk MS, Baer CE, Dennis WE, Minarchick VC, Leonard SS, Jackson DA, Stallings JD, Lewis JA. 2015. Temporal changes in rat liver gene expression after acute cadmium and chromium exposure. PLoS One 10 e0127327. DOI:10.1371/journal.pone.0127327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato K, Takada H, Iha M, Nagamine T. 2010. Attenuation of N‐nitrosodiethylamine‐induced liver fibrosis by high‐molecular‐weight fucoidan derived from Cladosiphon okamuranus. J. Gastroenterol. Hepatol. 25: 1692–1701. DOI:10.1111/j.1440-1746.2009.06187.x. [DOI] [PubMed] [Google Scholar]
- Nasr SH, Milliner DS, Wooldridge TD, Sethi S. 2014. Triamterene crystalline nephropathy. Am. J. Kidney Dis. 63: 148–152. DOI:10.1053/j.ajkd.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Panagiotou G, Taboureau O. 2012. The impact of network biology in pharmacology and toxicology. SAR QSAR Environ. Res. 23: 221–235. DOI:10.1080/1062936X.2012.657237. [DOI] [PubMed] [Google Scholar]
- Parkes J, Guha IN, Harris S, Rosenberg WM, Roderick PJ. 2012. Systematic review of the diagnostic performance of serum markers of liver fibrosis in alcoholic liver. Comp. Hepatol. 11: 5 DOI:10.1186/1476-5926-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permenter MG, Dennis WE, Sutto TE, Jackson DA, Lewis JA, Stallings JD. 2013. Exposure to cobalt causes transcriptomic and proteomic changes in two rat liver derived cell lines. PLoS One 8 e83751. DOI:10.1371/journal.pone.0083751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers DM. 2011. Evaluation: from precision, recall and F‐measure to ROC, informedness, markedness and correlation. J Mach. Learn. Tech. 2: 37–63. [Google Scholar]
- Probert PM, Ebrahimkhani MR, Oakley F, Mann J, Burt AD, Mann DA, Wright MC. 2014. A reversible model for periportal fibrosis and a refined alternative to bile duct ligation. Toxicol. Res. 3: 98–109. DOI:10.1039/C3TX50069A. [Google Scholar]
- Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K. 2005. Characterization of multilocus linkage disequilibrium. Genet. Epidemiol. 28: 193–206. DOI:10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. 2004. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 36: 1090–1098. DOI:10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Smalley JL, Gant TW, Zhang S‐D. 2010. Application of connectivity mapping in predictive toxicology based on gene‐expression similarity. Toxicology 268: 143–146. DOI:10.1016/j.tox.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Speir RW, Stallings JD, Andrews JM, Gelnett MS, Brand TC, Salgar SK. 2015. Effects of valproic acid and dexamethasone administration on early bio‐markers and gene expression profile in acute kidney ischemia‐reperfusion injury in the rat. PLoS One 10 e0126622. DOI:10.1371/journal.pone.0126622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturla SJ, Boobis AR, FitzGerald RE, Hoeng J, Kavlock RJ, Schirmer K, Whelan M, Wilks MF, Peitsch MC. 2014. Systems toxicology: from basic research to risk assessment. Chem. Res. Toxicol. 27: 314–329. DOI:10.1021/tx400410s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa GJ, AbdulHameed MDM, Yu X, Kumar K, Ippolito DL, Lewis JA, Stallings JD, Wallqvist A. 2014. Characterization of chemically induced liver injuries using gene co‐expression modules. PLoS One 9 e107230. DOI:10.1371/journal.pone.0107230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M, Maes M, Vanhaecke T, Rogiers V. 2013. Drug‐induced liver injury: mechanisms, types and biomarkers. Curr. Med. Chem. 20: 3011–3021. [DOI] [PubMed] [Google Scholar]
- Yuasa T, Yano R, Izawa T, Kuwamura M, Yamate J. 2014. Calponin expression in renal tubulointerstitial fibrosis induced in rats by cisplatin. J. Toxicol. Pathol. 27: 97–103. DOI:10.1293/tox.2013-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item
Data Availability Statement
The gene composition of the selected liver and kidney modules are provided in the Supplemental Materials.


