Abstract
Epigenetic modifications, and particularly DNA methylation, have been studied in many tissues, both healthy and diseased, and across numerous developmental stages. The placenta is the only organ that has a transient life of 9 months and undergoes rapid growth and dynamic structural and functional changes across gestation. Additionally, the placenta is unique because although developing within the mother, its genome is identical to that of the foetus. Given these distinctive characteristics, it is not surprising that the epigenetic landscape affecting placental gene expression may be different to that in other healthy tissues. However, the role of epigenetic modifications, and particularly DNA methylation, in placental development remains largely unknown. Of particular interest is the fact that the placenta is the most hypomethylated human tissue and is characterized by the presence of large partially methylated domains (PMDs) containing silenced genes. Moreover, how and why the placenta is hypomethylated and what role DNA methylation plays in regulating placental gene expression across gestation are poorly understood. We review genome-wide DNA methylation studies in the human placenta and highlight that the different cell types that make up the placenta have very different DNA methylation profiles. Summarizing studies on DNA methylation in the placenta and its relationship with pregnancy complications are difficult due to the limited number of studies available for comparison. To understand the key steps in placental development and hence what may be perturbed in pregnancy complications requires large-scale genome-wide DNA methylation studies coupled with transcriptome analyses.
Introduction
Besides mediating maternal–foetal exchange throughout gestation, the placenta plays a major role in orchestrating maternal adaptation to pregnancy by secreting a variety of steroid and peptide hormones. These placental hormones stimulate maternal physiological changes that are essential for pregnancy success. The placenta is unique in several ways. First, although the placenta is a shared organ between mother and foetus, it is an extra-embryonic tissue and is therefore primarily regulated by the foetal genome. Secondly, the placenta separates from mother and foetus after birth, making it a truly transient organ. For these reasons, the epigenetic mechanisms involved in placental development and regulation of gene expression within this tissue may not be subject to the same lifetime epigenetic constraints as other organs that must function throughout an individual’s life.
In humans, from implantation of the blastocyst, the placenta invades the decidua colonizing and transforming the uterine spiral arterioles to sequester a maternal blood supply for efficient maternal–foetal exchange (Roberts 2010). Invading placental extravillous cytotrophoblasts employ molecular mechanisms that closely match those of a metastatic tumour (Murray & Lessey 1999); however, although this process is strictly controlled both spatially and temporally in the placenta, it is somewhat dysregulated in cancer. Such mechanisms are not fully understood but include complex interactions between both extravillous cytotrophoblasts and maternal endothelium and leucocytes (Graham & Lala 1992, Lyall 2002, Yu et al. 2015). Emerging evidence suggests that epigenetic regulation of the placental transcriptome is important for the molecular control of placental growth and differentiation. This review highlights some of the complexities of placental DNA methylation in humans and how this process may be disrupted in some pregnancy pathologies. The focus of this review is genome-wide DNA methylation studies on human placental tissues, what we have learnt from these studies and what remains to be discovered.
Epigenetics
Epigenetics is often defined as modifications that affect genome architecture and accessibility which can influence gene transcription, without altering the underlying DNA sequence. Such modifications include DNA methylation and histone modifications that, unlike changes to the DNA sequence, may be reversible. This review focuses on the most widely studied epigenetic modification: DNA methylation, which is the addition of a methyl group (–CH3) to cytosine bases, is a process catalyzed by DNA methyltransferases (DNMT1, DNMT3A and DNMT3B). DNMT1 maintains and repairs established DNA methylation, whereas DNMT3A and DNMT3B are involved in de novo DNA methylation (Goll & Bestor 2005, Denis et al. 2011). Typically, DNA methylation of gene regulatory regions is associated with repression of gene expression; however, many genome-wide DNA methylation studies have demonstrated that this is not always the case (Ke et al. 2010, Moarii et al. 2015, Becket et al. 2016). DNA methylation is important in genomic imprinting and X chromosome inactivation in females. It has been widely studied in diseases and has been used as a biomarker for predicting disease or environmental exposures (Portela & Esteller 2010).
DNA methylation in the placenta
DNA methylation plays a crucial role during cellular differentiation and development (Ji et al. 2010, Kim et al. 2010). Studying DNA methylation in the placenta is complicated by the presence of several different cell types. The majority of studies investigating this topic have used chorionic villi, which have a different DNA methylation profile than embryonic tissues, the maternal decidua or foetal membranes (amnion and chorion) (see Robinson & Price 2015). Chorionic villi are the site of maternal–foetal exchange and hormone production and contain a mixture of cell types with cells derived from both the trophectoderm and the inner cell mass. The cellular composition of individual placentas varies and this is most apparent in the presence of a pregnancy complication (Mayhew et al. 2004).
Hypomethylation of placental tissue
It has been known for some time that the genome of the placenta is hypomethylated compared with that in other healthy tissues (Ehrlich et al. 1982, Fuke et al. 2004). However, how or why the placental genome is hypomethylated remains unclear, but may reflect the heterogeneous nature of placental tissue and the corresponding different DNA methylation profiles of distinct cell populations. Shortly after fertilization, the embryonic DNA becomes largely demethylated (Smith et al. 2012). In the following days, the cells in the inner cell mass rapidly undergo de novo DNA methylation but the trophectoderm remains hypomethylated (reviewed in Robinson & Price 2015). However, this information has been obtained in the extensively and easily studied mouse model. Dissecting out the DNA methylation profiles in different cell types across development in human embryos is technically more difficult than in mice. Some data suggest that DNA methylation throughout embryo development in humans differs when compared with mice (Guo et al. 2014). Furthermore, it appears that the DNA methylation levels in the trophectoderm are marginally lower than those in the inner cell mass in humans, but these findings are based on observations in very few samples (Guo et al. 2014).
The chorionic villi of the placenta are composed of cells derived from both the trophectoderm (all populations of trophoblasts) and the inner cell mass (extra-embryonic mesoderm and endoderm progenitors comprising the villous stroma and blood vessels). The villous syncytiotrophoblast is the major cell type in the placenta and is derived from the trophectoderm via the villous cytotrophoblasts which fuse to form the syncytium (Gude et al. 2004). Therefore, hypomethylation of the placental genome may reflect the maintenance of the early hypomethylated state of the trophectoderm through development. It is also known that DNMT1 expression in the placenta is reduced with mono-allelic DNA methylation of the promoter region (Novakovic et al. 2010, Das et al. 2013), which may also contribute to the hypomethylated state of the placental methylome.
Partially methylated domains in the placenta
Hypomethylation within the placenta is not uniform but occurs in large domains (>100kb) called partially methylated domains (PMDs) which are regions of reduced DNA methylation that cover approximately 40% of the placental genome (Schroeder et al. 2013). PMDs are unique to a few different tissue types that include the placenta, some cultured cells and cancer (Lister et al. 2011, Schroeder & LaSalle 2013, Schroeder et al. 2013). Genes within placental PMDs are typically repressed, have tissue-specific functions and their methylation status is maintained throughout gestation (Schroeder et al. 2013). However, even though PMDs seem to be a characteristic trait of the placental methylome, the majority of studies published to date have largely ignored them; therefore, it remains unclear why the placental methylome is characterized by PMDs. It is also yet to be determined when PMDs are first established in the trophoblast or placenta, what roles PMDs and the genes located within them play and whether they are disrupted in pregnancy complications.
Foetal sex differences in DNA methylation
Although the placental genome contains fewer methylated cytosines than other tissues (Ehrlich et al. 1982, Fuke et al. 2004), a large study in 248 placentas has shown that there is a large range of DNA methylation of 2–5% (Dwi Putra et al. 2014), but the reason for this variation requires further investigation. The regions with reduced DNA methylation included long interspersed nuclear elements (LINE1) and Alu repeats, as well as CpG island promoters associated with X-linked genes (Cotton et al. 2009). By using an Illumina DNA methylation microarray, Cotton et al. (2009) assessed 84 sites within X chromosome-associated promoter CpG islands and found that overall DNA methylation of these sites was reduced in female placentas to a greater extent than in male placentas, suggesting that there was DNA methylation loss at the inactive X chromosome. This was further supported by pyrosequencing assays for CpG island-associated promoters on the X chromosome (Cotton et al. 2009).
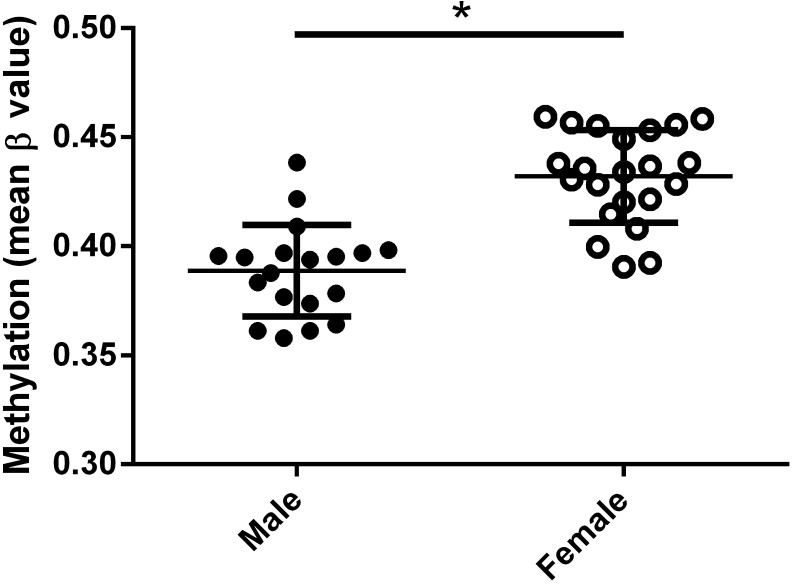
Our comparison of DNA methylation across 8346 CpG sites on the X chromosome using three datasets of placental tissue from uncomplicated term pregnancies (GSE44667, GSE54399 and GSE57767) (Blair et al. 2013, Anton et al. 2014) including 22 female and 19 male placentas indicated that the X chromosome from female placentas was more methylated than the X chromosome from male placentas (unpaired t-test, mean (female)=0.432; mean (male)=0.389, P-value=9.552e-08) (Fig. 1). When only the CpG sites within the 5′-UTR were analysed (2512 probes), DNA methylation on the X chromosome was higher in females than in males, as expected and seen for other organs (Yasukochi et al. 2010, Cotton et al. 2011, Hall et al. 2014, Joo et al. 2014).
Figure 1.
DNA methylation is higher on the X chromosome in placentas from female bearing pregnancies. DNA methylation levels were assessed at 8346 CpG sites on the X chromosome. DNA methylation of placental tissue from three publicly available data sets (GSE44667, GSE54399 and GSE57767) for a total of 19 male and 22 female term uncomplicated placentas was assessed. Probes that had missing values for samples were discarded, leaving 8346 X chromosome probes for all samples. Normalization was performed using the beta-mixture quantile normalization (BMIQ) method, which corrects for the two different designs of probes, followed by quantile normalization (Teschendorff et al. 2013). Batch effects were corrected using the Combat function implemented in the ChAMP Bioconductor package (Leek et al. 2012, Morris et al. 2014). Sample sex was identified using the minfi package in which the median value of theβvalues for probes that mapped uniquely for the X and Y chromosome, respectively, were first determined (Aryee et al. 2014). This resulted in the identification of 19 male and 22 female placentas. The overall DNA methylation for the X chromosome in each sample was calculated by taking the overall meanβvalue of all the probes that map to the X chromosome.
Cell-type differences in DNA methylation
As previously mentioned, the placenta is composed of several different cell types (Kanellopoulos-Langevin et al. 2003) that are likely to have their own unique corresponding methylome. Grigoriu et al. (2011) compared DNA from whole placenta with corresponding isolated fibroblasts and cytotrophoblasts separated from the same tissues using an enzymatic and magnetic bead separation methodology. By comparing DNA methylation profiles using the Illumina Infinium Human Methylation 27K BeadChip Array, the authors were able to identify 61 probes for genomic regions that were differentially methylated between whole placenta and cytotrophoblasts, 315 between whole placenta and fibroblasts and 442 between fibroblasts and cytotrophoblasts (Grigoriu et al. 2011). Interestingly, the cytotrophoblast DNA methylation profiles clustered with the whole placenta samples and the fibroblasts clustered on their own (Grigoriu et al. 2011). These results indicate that the methylome differs between the different cell types that make up the placenta and that this should be considered when performing DNA methylation studies in this organ. This is supported by other studies that have shown differential DNA methylation changes in distinct placental cell populations (Novakovic et al. 2008, 2011a).
Besides 5-methylcytosine, there are other derivatives, such as 5-hydroxymethylcytosine, that arise from the oxidation of 5-methylcytosine in the process of DNA demethylation (Hernando-Herraez et al. 2015). The precise role and function of 5-hydroxymethylcytosine require further study; however, it is important in development and disease (Tan & Shi 2012). In a recent study by Fogarty et al. (2015), differences in the levels of 5-methylcytosine and 5-hydroxymethylcytosine between syncytiotrophoblast and cytotrophoblasts were assessed using immunohistochemical quantification (Fogarty et al. 2015). This study showed higher levels of 5-methylcytsoine in cytotrophoblasts, whereas 5-hydroxymethylcytosine was more abundant in syncytiotrophoblast (Fogarty et al. 2015). The biological reason behind this difference is unclear but suggests that the methylomes may vary substantially between the different cell types that make up the placenta, which may be consistent with the differentiation state of the two villous trophoblast populations. The extent to which these epigenetic differences influence and define different cell populations in the placenta remains to be elucidated.
DNA methylation changes across gestation
One of the first studies to characterize DNA methylation across gestation used HPLC to show that global DNA methylation levels increase with gestational age (Fuke et al. 2004), which has been subsequently confirmed by others using different methodologies (Novakovic et al. 2011b, Price et al. 2012). Novakovic et al. (2011b) used Illumina arrays to show that DNA methylation increases from first trimester to third trimester with levels of DNA methylation very similar between second and third trimester. The authors suggest that the change in cell composition and differentiation of cells that occurs as gestation progresses may contribute to these differences, as well as the bias of this methodology with respect to the actual methylation sites assessed by the array probes (Novakovic et al. 2011b).
DNA methylation changes and pregnancy complications
The analysis of the placental methylome in pregnancy complications such as pre-eclampsia (PE), intrauterine growth restriction (IUGR), preterm birth (PTB) and gestational diabetes mellitus (GDM) has largely been performed using Illumina Infinium Human Methylation BeadChip Arrays (Banister et al. 2011, Lambertini et al. 2011, Jia et al. 2012, Blair et al. 2013, Ruchat et al. 2013, Anton et al. 2014, Chu et al. 2014, Liu et al. 2014, Finer et al. 2015, Hillman et al. 2015, Petropoulos et al. 2014) (Table 1). These studies identify differential methylation between placentas from controls and the pregnancy complication under investigation, with some studies showing overlap of differentially methylated sites in umbilical cord blood (Ruchat et al. 2013, Finer et al. 2015). However, Hillman et al. (2015) found no differences in DNA methylation in placenta, but observed differences in cord blood when comparing IUGR with controls. Interestingly, these differences in DNA methylation profiles in cord blood between healthy pregnant women and women diagnosed with pregnancy complications have also been reported in a preliminary study between healthy women and women diagnosed with either PE or PTB using maternal peripheral blood sampled at 15-week gestation, long before their diagnosis (Bianco-Miotto et al. 2015).
Table 1.
Summary of DNA methylation studies in placenta using genome-wide approaches.
| Reference | Method | Population | Sample (n) | Key findings |
|---|---|---|---|---|
| Finer et al. (2015) | Illumina HM450 | UK (South Asian origin) | Term placenta (25 GDM, 18 controls) Cord blood (27 GDM, 21 controls) |
More hypermethylated sites observed in both placenta and cord blood samples with GDM compared with the controls 4219 probes and 13,561 probes were differentially methylated between GDM and controls in placental tissue and cord blood respectively. 378 probes were common in placental tissue and cord blood |
| Hillman et al. (2015) | Illumina HM450 | UK | Placenta (23 IUGR, 22 controls) Cord blood (27 IUGR, 18 controls) |
No differentially methylated positions were observed in placental tissue between IUGR and controls. 839 differentially methylated regions were revealed in cord blood |
| Anton et al. (2014) | Illumina HM450 | USA (80% African American 20% Other) | Placenta (19 term PE, 12 PTB+PE, 14 controls) | 229 and 3411 loci were differentially methylated in PE and PTB+PE in comparison with the controls. Validation of four genes that were differentially methylated by qPCR confirmed altered mRNA expression |
| Chu et al. (2014) | Illumina HM27 | USA | Term placenta (24 PE, 24 controls) | PE samples were hypomethylated compared with controls. Clustering revealed that foetal sex is associated with DNA methylation irrespective of disease state |
| Liu et al. (2014) | Methylated-CpG island recovery assay (MIRA) | China | Placenta (27 PE, 28 GDM, 30 control) | 8191 (2140 genes) and 10,424 (2644 genes) differentially methylated regions were identified in PE and GDM compared with controls respectively 65% of the genes with different methylation revealed concordant changes in methylation between PE and GDM |
| Ou et al. (2014) | Illumina HM450 | China | First-trimester placenta and maternal blood (three of each) Term placenta and maternal blood (two of each) |
Identified 2944 and 5218 hypermethylated CpG sites that were foetal specific and found an overlap of 2613 differentially methylated sites between maternal blood and placenta tissue (present in both first- and third-trimester samples) |
| Petropoulos et al. (2014) | Illumina HM450 | Canada | Term placenta (seven GDM, seven controls) | 2021 CpG sites (981 genes) were differentially methylated between GDM and controls |
| Xiang et al. (2014) | MeDIP-Seq and Illumina HM450 | China | First-trimester placenta and maternal blood (14 of each) | Using both assays, 3759 CpG sites in 2188 regions were differentially methylated between maternal blood and placenta |
| Blair et al. (2013) | Illumina HM450 | Canada | Third-trimester placenta (20 EOPET, 20 controls) | 38,840 CpG sites were altered in EOPET vs controls. Gene expression microarray of a subset of samples (eight of each) showed negative correlation of gene expression changes with DNA methylation alterations |
| Ruchat et al. (2013) | Illumina HM450 | Canada | Term placenta and cord blood (30 GDM, 14 controls) | 3271 and 3758 genes were differentially methylated in controls vs GDM in placenta and cord blood, respectively, 25% common to both placenta and cord blood. The genes that were differentially methylated were involved in metabolic disease |
| Schroeder et al. (2013) | Illumina HM450 and MethylC-Seq & RNA-Seq | USA | Placenta (five first, ten second, 21 third trimester) for 450K Three term placentas for MethylC-Seq |
Identified partially methylated domains (PMDs) cover 37% of the placental genome. RNA-seq revealed that genes with PMDs are repressed. 450K data showed that PMDs are conserved throughout gestation |
| Gordon et al. (2012) | Illumina HM27 | Australia | Term placenta (eight MZ, seven DZ pairs) | MZ pairs showed greater similarity in intra-pair DNA methylation than DZ pairs |
| Jia et al. (2012) | MeDIP+NimbleGen human CpG island promoter microarray (385K) | China | Pooled term (3) compared with pooled PE (3)Validation (nine control, nine PE) | 3280 genes differentially methylated between controls and PE. Six genes (CAPN2, EPHX2, ADORA2B, SOX7, CXCL1, CDX1) were validated by bisulphite sequencing |
| Turan et al. (2012) | Illumina HM27+IlluminaHumanHT-12 v3 Expression BeadChip | USA | Term placenta (48) | Correlated DNA methylation levels with birth weight |
| Banister et al. (2011) | Illumina HM27 | USA | Term placenta (89 SGA, 117 controls) | Identified 22 differentially methylated loci that are associated with SGA |
| Novakovic et al. (2011b) | Illumina HM27 | Australia and Canada | Placenta (18 first, ten second, 14 third trimester) | An increase in overall genome methylation observed from first to third trimester. First-, second- and third-trimester cluster separately on a dendrogram |
| Lambertini et al. (2011) | MeDIP+Affymetrix Human Tiling Array 2.0R | USA | Term placenta (10 control, 7 IUGR) | Identified 113,020 genome-wide differentially methylated regions |
| Rakyan et al. (2008) | MeDIP+custom microarray | UK (European) | Term placenta (3) | Identified tissue-specific differentially methylated regions in the placenta |
Illumina HM27, Illumina Infinium Human Methylation 27K BeadChip; Illumina HM450, Illumina Infinium Human Methylation 450K BeadChip; MethylC-Seq, whole genome bisulphite sequencing; MeDIP, methylated DNA immunoprecipitation; DZ, dizygotic twins; MZ, monozygotic twins; EOPET, early-onset pre-eclampsia; GDM, gestational diabetes mellitus; IUGR, intrauterine growth restriction; PE, pre-eclampsia; SGA, small for gestational age.
Comparison between studies is made difficult due to different methodologies used; however, for studies in which comprehensive gene lists were available, we compared the overlap in differentially methylated genes. For GDM compared with uncomplicated pregnancies, we were able to compare two studies (Finer et al. 2015, Petropoulos et al. 2014) and found 91 genes that overlapped between the two studies. For PE, there appeared to be much less overlap with only two genes (DAPK3, PAPPA2) in common between the two studies that were compared (Blair et al. 2013, Chu et al. 2014). This is not surprising as Blair et al. (2013) assessed samples from women with early-onset pre-eclampsia and compared them with samples from gestational age-matched controls, whereas Chu et al. (2014) assessed samples from women with PE including 15 of 24 from women with term pre-eclampsia and compared them with term controls (Chu et al. 2014). Term PE has a very different aetiology to early-onset disease and preterm controls, although matched for gestational age could not be considered to be uncomplicated pregnancies (Andraweera et al. 2012). Despite this, Chu et al. (2014) found only one gene to be differentially methylated in early-onset versus term PE.
Of particular interest, only one study thus far has examined DNA methylation using methylC-seq (Schroeder et al. 2013), but was hampered by a small sample size (just three term placenta samples). MethylC-seq (Schroeder et al. 2013) uses sodium bisulphite treatment to convert all non-methylated cytosines to thymine, allowing single-base pair resolution of all 5mC sites within a sample. With sufficient coverage using high-throughput sequencing, the entire 5mC methylome can be identified in one methylC-seq library. This protocol is in stark contrast to the predominant use (driven by cost) of DNA methylation microarrays, which cover only ∼1–2% of the genome (Plongthongkum et al. 2014). Consequently, the majority of the placental methylome remains unexplored. This shortcoming is further compounded by the lack of studies quantifying both DNA methylation and gene expression in matched samples, which is crucial in linking altered DNA methylation to changes in gene regulation. There are few studies that have assessed gene expression and DNA methylation in the same samples and both of these only studied the term placenta (Turan et al. 2012, Schroeder et al. 2013). Without investigating gene expression together with DNA methylation, it is difficult to elucidate the role of DNA methylation in gene regulation in the placenta and how this may be disrupted in pregnancy complications. With reducing costs for next-generation sequencing, we anticipate more genome-wide placental DNA methylation studies in the future, which would increase our knowledge of placental DNA methylation and its effects on gene transcription in health and disease.
Conclusion
Although there are several studies of genome-wide DNA methylation profiling in the human placenta, our knowledge remains insufficient to draw well-supported conclusions about the role of DNA methylation in placental development, particularly since most studies have very small sample sizes. There are currently very few whole-genome bisulphite sequencing studies that provide comprehensive profiles of placental DNA methylation across gestation, and how these epigenetic modifications correlate with gene expression. What is also unclear is the role of other epigenetic modifications such as 5-hydroxymethylation and histone modifications in placental gene regulation. An integrated analysis of the placental epigenomic landscape may be required to begin elucidating the role of the placental epigenome in normal development and in pregnancy complications. Finally, as recently highlighted (Robinson & Price 2015), without understanding the distinct methylation profiles of the different cell types that make up the placenta, it is difficult to understand the role of DNA methylation in healthy placentas and what changes accompany pathology.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
C T R is supported by a National Health and Medical Research Council of Australia (NHMRC) Senior Research Fellowship (GNT1020749) and NHMRC project grant (GNT1059120) awarded to CTR and TB-M. SB is supported by a NHMRC and Australian Research Council (ARC) Dementia Research Development Fellowship (GNT1111206).
References
- Andraweera PH, Dekker GA, Laurence JA, Roberts CT. 2012. Placental expression of VEGF family mRNA in adverse pregnancy outcomes. Placenta 33 467–472. ( 10.1016/j.placenta.2012.02.013) [DOI] [PubMed] [Google Scholar]
- Anton L, Brown AG, Bartolomei MS, Elovitz MA. 2014. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS ONE 9 e100148. ( 10.1371/journal.pone.0100148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30 1363–1369. ( 10.1093/bioinformatics/btu049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. 2011. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics 6 920–927. ( 10.4161/epi.6.7.16079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becket E, Chopra S, Duymich CE, Lin JJ, You JS, Pandiyan K, Nichols PW, Siegmund KD, Charlet J, Weisenberger DJ, et al. 2016. Identification of DNA methylation-independent epigenetic events underlying clear cell renal cell carcinoma. Cancer Research 76 1954–1964. ( 10.1158/0008-5472.CAN-15-2622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco-Miotto T, Rodriguez Lopez C, Leemaqz S, Buckberry S, McCullough D, Zhuang Z, Dekker G, Wilkinson M, Roberts CT. 2015. DNA methylation biomarkers for predicting pregnancy complications. Placenta 36 A38–A39. ( 10.1016/j.placenta.2015.07.302) [DOI] [Google Scholar]
- Blair JD, Yuen RK, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. 2013. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Molecular Human Reproduction 19 697–708. ( 10.1093/molehr/gabib44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T, Bunce K, Shaw P, Shridhar V, Althouse A, Hubel C, Peters D. 2014. Comprehensive analysis of preeclampsia-associated DNA methylation in the placenta. PLoS ONE 9 e107318. ( 10.1371/journal.pone.0107318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Avila L, Penaherrera MS, Affleck JG, Robinson WP, Brown CJ. 2009. Inactive X chromosome-specific reduction in placental DNA methylation. Human Molecular Genetics 18 3544–3552. ( 10.1093/hmg/ddp299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Lam L, Affleck JG, Wilson IM, Penaherrera MS, McFadden DE, Kobor MS, Lam WL, Robinson WP, Brown CJ. 2011. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Human Genetics 130 187–201. ( 10.1007/s00439-011-1007-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Lee YK, Strogantsev R, Jin S, Lim YC, Ng PY, Lin XM, Chng K, Yeo G, Ferguson-Smith AC, et al. 2013. DNMT1 and AIM1 Imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. BMC Genomics 14 685. ( 10.1186/1471-2164-14-685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H, Ndlovu MN, Fuks F. 2011. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Reports 12 647–656. ( 10.1038/embor.2011.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwi Putra SE, Neuber C, Reichetzeder C, Hocher B, Kleuser B. 2014. Analysis of genomic DNA methylation levels in human placenta using liquid chromatography-electrospray ionization tandem mass spectrometry. Cellular Physiology and Biochemistry 33 945–952. ( 10.1159/000358666) [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Research 10 2709–2721. ( 10.1093/nar/10.8.2709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer S, Mathews C, Lowe R, Smart M, Hillman S, Foo L, Sinha A, Williams D, Rakyan VK, Hitman GA. 2015. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Human Molecular Genetics 24 3021–3029. ( 10.1093/hmg/ddv013) [DOI] [PubMed] [Google Scholar]
- Fogarty NM, Burton GJ, Ferguson-Smith AC. 2015. Different epigenetic states define syncytiotrophoblast and cytotrophoblast nuclei in the trophoblast of the human placenta. Placenta 36 796–802. ( 10.1016/j.placenta.2015.05.006) [DOI] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. 2004. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Annals of Human Genetics 68 196–204. ( 10.1046/j.1529-8817.2004.00081.x) [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry 74 481–514. ( 10.1146/annurev.biochem.74.010904.153721) [DOI] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, Alisch RS, Morley R, Visscher PM, Craig JM, Saffery R. 2012. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Research 22 1395–1406. ( 10.1101/gr.136598.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CH, Lala PK. 1992. Mechanisms of placental invasion of the uterus and their control. Biochemistry and Cell Biology 70 867–874. ( 10.1139/o92-135) [DOI] [PubMed] [Google Scholar]
- Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. 2011. Cell specific patterns of methylation in the human placenta. Epigenetics 6 368–379. ( 10.4161/epi.6.3.14196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gude NM, Roberts CT, Kalionis B, King RG. 2004. Growth and function of the normal human placenta. Thrombosis Research 114 397–407. ( 10.1016/j.thromres.2004.06.038) [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. 2014. The DNA methylation landscape of human early embryos. Nature 511 606–610. ( 10.1038/nature13544) [DOI] [PubMed] [Google Scholar]
- Hall E, Volkov P, Dayeh T, Esguerra JL, Salo S, Eliasson L, Ronn T, Bacos K, Ling C. 2014. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biology 15 522. (10.11862Fs13059-014-0522-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Herraez I, Garcia-Perez R, Sharp AJ, Marques-Bonet T. 2015. DNA methylation: insights into human evolution. PLoS Genetics 11 e1005661. ( 10.1371/journal.pgen.1005661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman SL, Finer S, Smart MC, Mathews C, Lowe R, Rakyan VK, Hitman GA, Williams DJ. 2015. Novel DNA methylation profiles associated with key gene regulation and transcription pathways in blood and placenta of growth-restricted neonates. Epigenetics 10 50–61. ( 10.4161/15592294.2014.989741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. 2010. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467 338–342. ( 10.1038/nature09367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia RZ, Zhang X, Hu P, Liu XM, Hua XD, Wang X, Ding HJ. 2012. Screening for differential methylation status in human placenta in preeclampsia using a CpG island plus promoter microarray. International Journal of Molecular Medicine 30 133–141. [DOI] [PubMed] [Google Scholar]
- Joo JE, Novakovic B, Cruickshank M, Doyle LW, Craig JM, Saffery R. 2014. Human active X-specific DNA methylation events showing stability across time and tissues. European Journal of Human Genetics 22 1376–1381. ( 10.1038/ejhg.2014.34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulos-Langevin C, Caucheteux SM, Verbeke P, Ojcius DM. 2003. Tolerance of the fetus by the maternal immune system: role of inflammatory mediators at the feto-maternal interface. Reproductive Biology and Endocrinology 1 121. ( 10.1186/1477-7827-1-121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke XS, Qu Y, Cheng Y, Li WC, Rotter V, Oyan AM, Kalland KH. 2010. Global profiling of histone and DNA methylation reveals epigenetic-based regulation of gene expression during epithelial to mesenchymal transition in prostate cells. BMC Genomics 11 669. ( 10.1186/1471-2164-11-669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467 285–290. ( 10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini L, Lee TL, Chan WY, Lee MJ, Diplas A, Wetmur J, Chen J. 2011. Differential methylation of imprinted genes in growth-restricted placentas. Reproductive Sciences 18 1111–1117. ( 10.1177/1933719111404611) [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 882–883. ( 10.1093/bioinformatics/bts034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 882–883. ( 10.1093/bioinformatics/bts034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471 68–73. ( 10.1038/nature09798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang X, Rong C, Rui C, Ji H, Qian YJ, Jia R, Sun L. 2014. Distinct DNA methylomes of human placentas between pre-eclampsia and gestational diabetes mellitus. Cellular Physiology and Biochemistry 34 1877–1889. ( 10.1159/000366386) [DOI] [PubMed] [Google Scholar]
- Lyall F. 2002. The human placental bed revisited. Placenta 23 555–562. ( 10.1053/plac.2002.0850) [DOI] [PubMed] [Google Scholar]
- Mayhew TM, Wijesekara J, Baker PN, Ong SS. 2004. Morphometric evidence that villous development and fetoplacental angiogenesis are compromised by intrauterine growth restriction but not by pre-eclampsia. Placenta 25 829–833. ( 10.1016/j.placenta.2004.04.011) [DOI] [PubMed] [Google Scholar]
- Moarii M, Boeva V, Vert JP, Reyal F. 2015. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics 16 873. ( 10.1186/s12864-015-1994-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S. 2014. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics 30 428–430. ( 10.1093/bioinformatics/btt682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MJ, Lessey BA. 1999. Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Seminars in Reproductive Endocrinology 17 275–290. [DOI] [PubMed] [Google Scholar]
- Novakovic B, Rakyan V, Ng HK, Manuelpillai U, Dewi C, Wong NC, Morley R, Down T, Beck S, Craig JM, et al. 2008. Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Molecular Human Reproduction 14 547–554. ( 10.1093/molehr/gan046) [DOI] [PubMed] [Google Scholar]
- Novakovic B, Wong NC, Sibson M, Ng HK, Morley R, Manuelpillai U, Down T, Rakyan VK, Beck S, Hiendleder S, et al. 2010. DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. Journal of Biological Chemistry 285 9583–9593. ( 10.1074/jbc.M109.064956) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Gordon L, Wong NC, Moffett A, Manuelpillai U, Craig JM, Sharkey A, Saffery R. 2011. a Wide-ranging DNA methylation differences of primary trophoblast cell populations and derived cell lines: implications and opportunities for understanding trophoblast function. Molecular Human Reproduction 17 344–353. ( 10.1093/molehr/gar005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. 2011b. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 12 529. ( 10.1186/1471-2164-12-529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Wang H, Qu D, Chen Y, Gao J, Sun H. 2014. Epigenome-wide DNA methylation assay reveals placental epigenetic markers for noninvasive fetal single-nucleotide polymorphism genotyping in maternal plasma. Transfusion 54 2523–2533. ( 10.1111/trf.12659) [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Guillemin C, Ergaz Z, Dimov S, Suderman M, Weinstein-Fudim L, Ornoy A, Szyf M. 2014. Gestational diabetes alters offspring DNA methylation profiles in human and rat: identification of key pathways involved in endocrine system disorders, insulin signaling, diabetes signaling, and ILK signaling. Endocrinology 156 2222–2238. ( 10.1210/en.2014-1643) [DOI] [PubMed] [Google Scholar]
- Plongthongkum N, Diep DH, Zhang K. 2014. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nature Reviews Genetics 15 647–661. ( 10.1038/nrg3772) [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. 2010. Epigenetic modifications and human disease. Nature Biotechnology 28 1057–1068. ( 10.1038/nbt.1685) [DOI] [PubMed] [Google Scholar]
- Price EM, Cotton AM, Penaherrera MS, McFadden DE, Kobor MS, Robinson W. 2012. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics 7 652–663. ( 10.4161/epi.20221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavare S, Beck S. 2008. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Research 18 1518–1529. ( 10.1101/gr.077479.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CT. 2010. IFPA Award in placentology lecture: complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta 31 (Supplement) S47–S53. ( 10.1016/j.placenta.2009.12.019) [DOI] [PubMed] [Google Scholar]
- Robinson WP, Price EM. 2015. The human placental methylome. Cold Spring Harbor Perspectives in Medicine 5 a023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchat SM, Houde AA, Voisin G, St-Pierre J, Perron P, Baillargeon JP, Gaudet D, Hivert MF, Brisson D, Bouchard L. 2013. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 8 935–943. ( 10.4161/epi.25578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, LaSalle JM. 2013. How has the study of the human placenta aided our understanding of partially methylated genes? Epigenomics 5 645–654. ( 10.2217/epi.13.62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, et al. 2013. The human placenta methylome. PNAS 110 6037–6042. ( 10.1073/pnas.1215145110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484 339–344. ( 10.1038/nature10960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. 2012. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139 1895–1902. ( 10.1242/dev.070771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29 189–196. ( 10.1093/bioinformatics/bts680) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan N, Ghalwash MF, Katari S, Coutifaris C, Obradovic Z, Sapienza C. 2012. DNA methylation differences at growth related genes correlate with birth weight: a molecular signature linked to developmental origins of adult disease? BMC Medical Genomics 5 10. ( 10.1186/1755-8794-5-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Maruyama O, Mahajan MC, Padden C, Euskirchen GM, Schulz V, Hirakawa H, Kuhara S, Pan XH, Newburger PE, et al. 2010. X chromosome-wide analyses of genomic DNA methylation states and gene expression in male and female neutrophils. PNAS 107 3704–3709. ( 10.1073/pnas.0914812107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Li Q, Zhou X, Wang T, Xu M, Xia S, Xing Q, Wang L, He L, Zhao X. 2014. DNA methylome profiling of maternal peripheral blood and placentas reveal potential fetal DNA markers for non-invasive prenatal testing. Molecular Human Reproduction 20 875–884. ( 10.1093/molehr/gau048) [DOI] [PubMed] [Google Scholar]
- Yu N, Yan W, Yin T, Wang Y, Guo Y, Zhou D, Xu M, Ding J, Yang J. 2015. HCG-Activated Human Peripheral Blood Mononuclear Cells (PBMC) Promote Trophoblast Cell Invasion. PLoS ONE 10 e0125589. ( 10.1371/journal.pone.0125589) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a