Abstract
Melanocortin receptor accessory protein 2 (MRAP2) is a transmembrane accessory protein predominantly expressed in the brain. Both global and brain-specific deletion of Mrap2 in mice results in severe obesity. Loss-of-function MRAP2 mutations have also been associated with obesity in humans. Although MRAP2 has been shown to interact with MC4R, a G protein-coupled receptor with an established role in energy homeostasis, appetite regulation and lipid metabolism, the mechanisms through which loss of MRAP2 causes obesity remains uncertain. In this study, we used two independently derived lines of Mrap2 deficient mice (Mrap2tm1a/tm1a) to further study the role of Mrap2 in the regulation of energy balance and peripheral lipid metabolism. Mrap2tm1a/tm1a mice have a significant increase in body weight, with increased fat and lean mass, but without detectable changes in food intake or energy expenditure. Transcriptomic analysis showed significantly decreased expression of Sim1, Trh, Oxt and Crh within the hypothalamic paraventricular nucleus of Mrap2tm1a/tm1a mice. Circulating levels of both high-density lipoprotein and low-density lipoprotein were significantly increased in Mrap2 deficient mice. Taken together, these data corroborate the role of MRAP2 in metabolic regulation and indicate that, at least in part, this may be due to defective central melanocortin signalling.
Keywords: obesity, melanocortin, accessory protein, metabolism, MC4R, MRAP2, SIM1, OXT, AVP, CRH, TRH
Introduction
Melanocortin receptor accessory protein (MRAP) and its paralogue MRAP2 are a recently identified class of small, single-pass transmembrane domain accessory proteins (Chan et al. 2009, Novoselova et al. 2013). Both MRAP and MRAP2 have been shown to interact with the melanocortin receptors (MCRs), a family of G protein-coupled receptors (GPCRs) with diverse physiological function stimulated by pro-opiomelanocortin (POMC) derived peptide agonists such as adrenocorticotropin hormone (ACTH) and α-MSH (Cone 2005, Chan et al. 2009). Of the five MCRs (MC1R–MC5R), only the function of the melanocortin 2 receptor (MC2R) is clearly recognized to be facilitated by MRAPs (Metherell et al. 2005, Chan et al. 2009), although in-vitro data suggests a broader role in conjunction with all the MCRs (Chan et al. 2009, Sebag & Hinkle 2009, 2010).
MRAP is highly expressed in the adrenal gland and is essential for MC2R function. Mutations in MRAP are associated with familial glucocorticoid deficiency (OMIM#607398) (Metherell et al. 2005). MRAP2 is predominantly expressed in the central nervous system and hypothalamus, in particular within the paraventricular nucleus (PVN), a region known to have a role in energy homeostasis (Chan et al. 2009). Mice with global and brain-specific Mrap2 deletion developed marked obesity and rare loss-of-function or missense heterozygous variants in MRAP2 were also identified in humans with severe early-onset obesity (Asai et al. 2013). This work demonstrated that MRAP2’s role in the control of body composition and growth is via MC4R signalling (Asai et al. 2013). Further evidence for a link with MC4R signalling came from a study on the role of Mrap2 in zebrafish feeding and growth (Sebag et al. 2013).
Given these data, the phenotype observed in Mrap2-deficient mice is likely, at least in part, to be driven by disruption of central melanocortin signalling. However, some areas of uncertainty remain. In particular, the paradoxical observation that the mutant mice become obese without detectable changes in food intake or energy balance (Asai et al. 2013) requires exploration, as does the potential role of MRAP2 in peripheral cholesterol and lipid metabolism, a function known to be regulated by melanocortins (Nogueiras et al. 2007, Perez-Tilve et al. 2010). In this study, we have used an independently derived line of Mrap2-deficient mice (Mrap2tm1a/tm1a) on two different genetic backgrounds to further study the role of MRAP2 in the regulation of energy homeostasis and the control of cholesterol and lipid metabolism.
Materials and methods
Generation of Mrap2-deficient mouse
Mice carrying the knockout-first conditional-ready allele Mrap2tm1a(EUCOMM)Wtsi (abbreviated to Mrap2tm1a) were generated on a C57BL/6N background as part of the Sanger Mouse Genetics Project (MGP) (Fig. 1A). Mice carrying the same Mrap2tm1a allele were generated separately on a 129S5/SvEvBrdWtsi;129P2/OlaHsdWtsi background (abbreviated to 129/Sv). Detailed description of the Sanger Mouse Genetics Project methodology has been reported (Skarnes et al. 2011). Briefly, a promoter-containing cassette (L1L2_Bact_P) was introduced upstream of the critical Mrap2 exon 4 at position 87175333 of Chromosome 9, Build GRCm38 (Fig. 1A). The vectors containing Mrap2tm1a were electroporated into C57BL/6N derived JM8F6 and 129P2/OlaHsd derived E14Tg2a embryonic stem (ES) cells. Correct ES cell gene targeting was confirmed by long-range PCR and quantitative PCR. Targeted ES cells were microinjected into blastocysts and used to generate chimeras. Germ-line transmission was confirmed by genotyping PCR analyses (http://www.knockoutmouse.org/kb/25/). Mice obtained from heterozygous intercross were genotyped for the Mrap2tm1a allele by PCR (Supplementary Table 1, see section on supplementary data given at the end of this article).
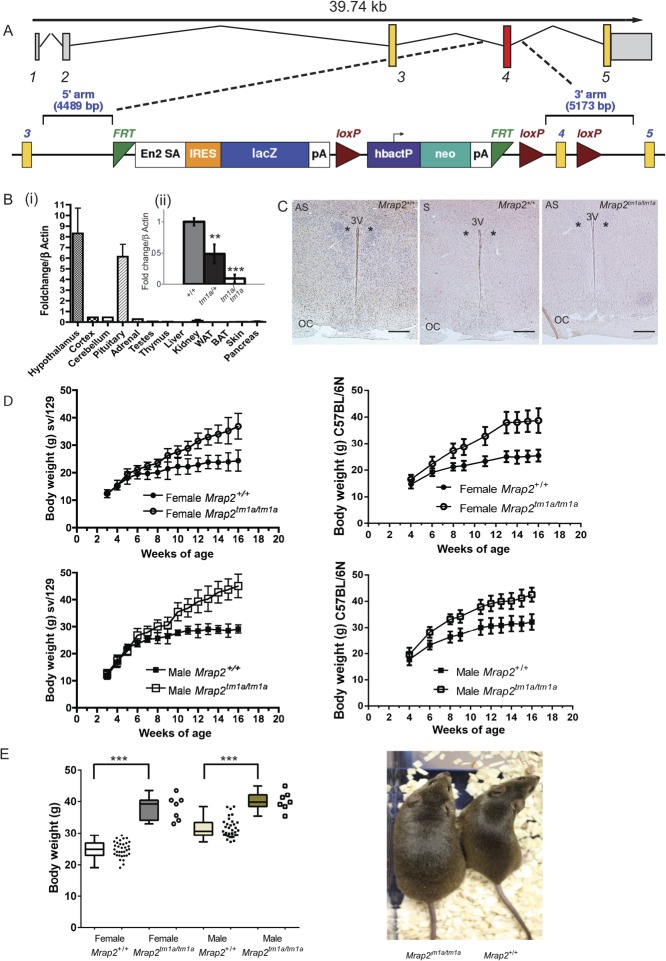
Figure 1.
Mrap2 gene disruption results in weight gain (A) Schematic of knockout-first strategy for Mrap2. A promoter driven cassette including lacZ and neo genes are inserted upstream of critical exon 4. (B) Q-RT-PCR in 129/Sv mice demonstrating (i) Mrap2 expression in a range of wild-type mouse tissues showing the highest expression levels in the hypothalamus, n=3. (ii) reduced hypothalamic Mrap2 transcript in Mrap2tm1a/+and Mrap2tm1a/tm1a compared with Mrap2+/+ mice (n=3 per genotype); mean plotted ± s.e.m.; **P<0.005; ***P<0.0005). (C) Expression of Mrap2 in the hypothalamus of the wild-type 129/Sv Mrap2+/+ and Mrap2tm1a/tm1a mice as shown by in situ hybridization using coronal brain sections (approx. bregma – 0.6mm). AS, antisense probe, S, sense probe as a negative control. Third ventricle indicated as 3 V, asterisk indicates position of the PVN, OC, optic chiasm; scale bars=200μm. (D) Weight curves of Mrap2tm1a/tm1a in both genders and genetic backgrounds illustrated. 129/Sv Mrap2tm1a/tm1a n > 8 per genotype and gender, C57BL/6N Mrap2tm1a/tm1a n=7 of each gender and genotype. (E) Total body weight gain of C57BL/6N Mrap2tm1a/tm1a mice by the age of 16weeks, n=7 for each gender/genotype (left) and appearance of the mutant mice compared with the wild type (right). Data is presented as both box-and-whiskers plot (showing min-mean-max values, with the box representing the 25th and 75th percentiles), and as a scatter dot plot for individual values. P-values presented on graphs are either global P-values for genotype adjusted for multiple correction testing, or (in the cases of sexual dimorphism) the P-value is the impact of genotype for that sex.
Animal husbandry
The care and use of all animals were carried out in accordance with the UK Home Office regulations, UK Animals (Scientific Procedures) Act 1986. Mice were kept under a standard 12h light:12h darkness cycle with food and water ad libitum unless otherwise stated. 129/Sv background mice were maintained in a facility at 22°C and fed a standard chow (SDS RM3, Essex, UK). Mice on a C57BL/6N background were maintained at 21°C±2°C, humidity 55%±10% and fed a standard rodent chow (LabDiets 5021-3, IPS, Richmond, VA, USA).
Metabolic phenotyping
Metabolic phenotyping was undertaken at two independent centres. In accordance to the 3R (replacement, reduction and refinement) principles of humane experimental technique and based on scientific objectives, not all procedures were performed on both lines. The genetic background of Mrap2tm1a mice used in each experiment have been specified previously.
Phenotyping using C57BL/6N mice was performed at the Wellcome Trust Sanger Institute as part of the MGP (White et al. 2013), whilst studies using 129/Sv background mice were carried out at the University of Cambridge Metabolic Research Laboratories (MRL). For data arising from the MGP, a cumulative baseline was generated from controls of the same genetic background, age and sex. Seven male and seven female Mrap2tm1a mice were processed in five batches for each sex (one to three mice per batch) and were phenotyped unblinded as part of a larger mixed genotype group that included weekly wild-type controls, with the individual mouse as the experimental unit. Animals for testing were randomly assigned to test sessions and operators. Mice were group housed to a minimum density of three per cage.
Body composition of 14-week-old anaesthetized C57BL/6N Mrap2tm1a mice were determined by dual-energy X-ray absorptiometry (DEXA) using a Lunar PIXImus2 mouse densitometer (General Electric Medical Systems, Fitchburg, WI, USA).
After overnight fasting (approximately 16h), intraperitoneal glucose tolerance tests (IPGTT) were carried out on 13-week-old mice. After taking a baseline glucose measurement, mice were given a single glucose injection (2g/kg) and blood glucose measured at 15, 30, 60 and 120min (Accu-Chek Aviva, Roche).
Blood for plasma biochemistry was collected from 16-week-old C57BL/6N animals into lithium-heparin tubes. Animals were not fasted unless otherwise indicated. Clinical blood chemistry was carried out using an Olympus AU400 chemistry analyser (Olympus). Insulin levels were measured by Mesoscale Discovery array technology platform.
Additional data relating to the C57BL/6N Mrap2tm1a/tm1a line can be found at http://www.mousephenotype.org/data/genes/MGI:3609239. For studies at MRL, individual experiments were matched for age and sex of mice. The body weight and length were measured weekly since weaning. Food intake was carried out on 8-week-old single-housed acclimatized animals. Response to fasting was measured after mice were moved into clean cages, and food was removed at 07:00 for 24h.
Energy expenditure was determined at 8 weeks of age using indirect calorimetry. Animals were placed in a custom-built monitoring system based on their home cages (Ideas Studio, Cambridge, UK). Oxygen consumption and carbon dioxide production was measured, and samples were taken at 18min intervals for 48h. Energy expenditure was calculated using indirect calorimetry with the Elia and Livesey constants for respiratory quotient (Elia & Livesey 1992). Activity was assessed by beam breaks (beams 1.25cm apart) and measurements were taken as total, rather than consecutive beam breaks.
Behavioural tests
Open field assessment was used to quantify spontaneous locomotor behaviour in a novel environment. The open field, custom-designed-walled, infra-red backlit arena 75cm2 (Tracksys Ltd., Nottingham, UK) was subdivided into a centre zone (42 cm2) with the remainder designated as border zone. Twenty-week-old 129/Sv background mice were recorded for a 20min period using Noldus Ethovision-3 video tracking software. The position of the centre-point of the mouse within the open field was recorded. A mouse was considered to begin moving when its velocity surpassed 2 cm/s and stop moving when below 1.75 cm/s. Mrap2tm1a/tm1a and their Mrap2+/+ controls were littermates housed in single-sex groups of three to five.
Histology, non-radioactive in-situ hybridization, immunohistochemistry and PVN stereotaxic counts
For haematoxilin-eosin (H&E) staining, tissues were fixed in 4% paraformaldehyde (PFA) (Sigma), washed, dehydrated and embedded into paraffin before sectioning to 7µm. For Oil-Red-O staining, flash frozen liver was embedded into OCT (VWR), 10µm cryosections were adhered onto slides (Thermo Fisher) and stained with Oil-Red-O (Sigma). Both staining techniques were performed according to standard protocols. Ucp1 immunohistochemistry was carried out using brown fat paraffin sections anti-Ucp1 antibody (1/500) according to the manufacturer’s instructions (ab10983, Abcam) followed by detection using anti-rabbit HRP antibody (Thermo-Fisher) with DAB staining (Vector).
To generate riboprobes for in-situ hybridization (ISH), RNA was extracted from hypothalamus and cDNA prepared. Full-length Mrap2 cDNA fragment (898 bp) was PCR amplified (Supplementary Table 1), ligated into pGEM-T easy vector (Promega), sequenced and then linearized with EcoRI or NotI (Promega). Digoxigenin (DIG)-labelled antisense and sense cRNA probes were synthesized by in-vitro transcription with T7 or SP6 RNA polymerases (Roche). Dissected brains were embedded into OCT and frozen in liquid nitrogen, 20µm cryosections were cut onto slides and fixed with ice-cold 4% PFA for 20min. Slides were then subjected to ISH as described previously (Gorrigan et al. 2011).
To determine the PVN neuron counts, brains of Mrap2tm1a/tm1a and wild-type littermates (three brains per group) were fixed in 4% PFA, cryoprotected with 20% sucrose and cryosectioned 20μm each, starting from −0.58mm to −1.22mm to bregma (Franklin & Paxinos 2012). After Nissl staining, the slides were visualized and images taken using Zeiss Axio Scope A1. The neurons within the PVN were then counted using ImageJ software (http://imagej.nih.gov/ij/).
Laser-captured microdissection and RNA isolation
Mouse brains from 9-week-old, 129/Sv Mrap2tm1a/tm1a and Mrap2+/+ mice were dissected, immediately embedded into OCT and frozen in liquid nitrogen. Coronal sections (20μm) covering the region from −0.58mm to −1.22mm caudal to bregma (Franklin & Paxinos 2012) were cut on a cryostat and mounted on Superfrost Plus slides (Thermo-Fisher). Frozen sections were fixed for 40s in 95% ethanol and then rehydrated (75% and 50% ethanol, 30s each). The slides were stained with 1% cresyl violet in 75% ethanol (w/v) for 45s, dehydrated in a graded ethanol series (50, 75, 95, 100% for 30s each), in 100% ethanol for 5min and air-dried. Laser microdissection was performed using a P.A.L.M. MicroBeam (Zeiss). The PVN was collected into AdhesiveCap tubes (Zeiss). Total RNA was immediately isolated using the RNAqueous-Micro Kit (Ambion). Quality and quantity of the total RNA samples were determined by the Agilent BioAnalyzer using PicoChip. RNAse free technique and RNAase free reagents were used throughout.
RNA microarray hybridization and analysis
Fifteen nanograms of isolated RNA with the RNA Integrity Number of at least 6.5 (n=4 for Mrap2+/+; n=3 for Mrap2tm1a/tm1a) was converted into cDNA using Ovation PICO SL System V2 (NuGEN), which was then fragmented and labelled using Encore BiotinIL Module (NuGEN). About 1500ng of each labelled product was then hybridized with MouseRef-8v2.0 Expression BeadChip Kit according to the manual and scanned using iScan (Illumina, San Diego, CA, USA). Raw image data were converted to bsc format using Illumina GenomeStudio 2011.1 software. Bonferroni correction with Family-Wise Error Rate (FWER) of 0.05 was applied to identify statistical significance of gene expression changes. Pathway analysis was performed using DAVID6.7 (http://david.abcc.ncifcrf.gov/tools.jsp) and STRING 10 (http://string-db.org/).
Quantification of RNA by real-time quantitative PCR (Q-RT-PCR)
Dissected tissues were immediately frozen in liquid nitrogen, homogenized using Precellys24 (Precellys, Bertin Technologies, Paris, France) into RPL buffer (Qiagen) and the RNA was extracted with RNeasy Mini Kit (Qiagen). cDNA was produced with SuperScriptII (Life Technologies) and 50ng of cDNA was used for RT-Q-PCR with TaqMan Universal MasterMix II and gene-specific TaqMan probes (Life technologies, Supplementary Table 2). The fold change related to Actin-b was calculated using 2-ΔΔTh method (Livak & Schmittgen 2001).
Protein quantification
White and brown fat tissues were homogenized using Precellys24 in ice-cold RIPA buffer (Sigma) containing phosphotase (Roche) and protease inhibitors cocktail (Sigma). Lysates were centrifuged for 20min at 4°C before separation of the lipid layer. The SDS–PAGE samples were prepared with 2× Sample buffer (Sigma), heated at 95°C for 5min, centrifuged for 20min at 4°C to separate samples from residual lipids and subjected to western blotting. The membrane was blocked with 5% bovine serum albumin in TBS (Life Technologies) for 1h at 22°C followed by incubation at 4°C overnight with the primary antibody: anti-ACTB antibody 1/10,000 (Abcam), anti-UCP1 1/5000 (Abcam) and antibodies for Fatty Acid and Lipid Metabolism and Lipolysis Activation (8334, 8335 Cell Signaling Technology). After three washes, the membranes were probed with anti-mouse 680 and anti-rabbit 800 IRDye antibodies (LI-COR). The band intensities were quantified using Odyssey software.
Statistical analysis
All data were generated from the MGP utilized statistical analysis with RStudio running R version 3.1.2 and Phenstat package version 2.0.1. This uses a mixed-model framework (Karp et al. 2012) to assess the impact of genotype on phenotype. The analysis was performed by loading the model without body weight, therefore analysing the absolute differences between genotypes whilst accounting for sex, using the model: Y=Genotype+Sex+Genotype×Sex. Multiple correction testing was performed on the global P-values using the Hochberg correction. Data is presented as both box-and-whiskers plot (showing min-mean-max values, with the box representing the 25th and 75th percentiles), and as a scatter dot plot for individual values. P-values presented on graphs are either global P-values for genotype adjusted for multiple correction testing, or (in the cases of sexual dimorphism) the P-value is the impact of genotype for that sex.
For other data, males and females were assessed independently and the effect of genotype compared with wild-type controls was statistically tested using a two-tailed Student’s t-test. For calorimetry data, multiple linear regression analysis (ANCOVA) was used. Data is plotted as mean±s.e.m. and analysed using Microsoft excel and GraphPad Prism.
Results
Production of Mrap2-deficient mice
Mice carrying the mutant Mrap2tm1a allele were viable with expected homozygous mutant offspring born from heterozygous matings (21% C57BL/6N Mrap2tm1a/tm1a and 23% 129/Sv Mrap2tm1a/tm1a). Both female and male Mrap2tm1a/tm1a mice were fertile and did not exhibit any changes in skin or hair colour/appearance. The introduction of the knockout-first Mrap2tm1a allele resulted in targeted disruption of the critical exon 4 encoding the transmembrane domain of the protein. The predicted outcome would be a premature stop codon, thus producing a short 132bp transcript that, if translated, would produce a 44 amino acid protein (predicted MW 5 kDa). Previous work demonstrated that such a protein was unlikely to be translated (Asai et al. 2013). However, generation of hypomorphic mice have been demonstrated previously using the ‘knockout-first’ strategy targeting other genes (McIntyre et al. 2012, Chen et al. 2013, White et al. 2013). We therefore determined the expression of Mrap2 by Q-RT-PCR analysis using a TaqMan probe spanning exons 4–5. cDNA generated from whole hypothalamus extracted from mice on an 129/Sv background revealed a low but detectable residual Mrap2 transcript of Mrap2tm1a/tm1a within homozygous mice (13%, range 11–16%), whilst heterozygotes Mrap2tm1a/+ mice had approximately half of the Mrap2 transcript expression compared with Mrap2+/+ (Fig. 1B).
Mrap2 is predominantly expressed in the paraventricular nucleus of the hypothalamus
Mrap2 RNA expression was studied in wild-type mice tissues (Fig. 1B). The highest expression level was detected in the hypothalamus with substantial expression observed in the pituitary gland. Mrap2 expression was also detected in the cortex, cerebellum and adrenal gland. Kidney, testes, thymus and pancreas had very low expression levels whilst expression in white fat, liver, brown fat and skin was undetectable (Fig. 1B). ISH using a full-length Mrap2 probe showed visible Mrap2 RNA expression in the PVN of Mrap2+/+ mice on a 129/Sv background, which was absent in Mrap2tm1a/tm1a mice (Fig. 1C).
Mrap2 deficiency results in obesity in both C57BL/6N and 129/Sv background
Before weaning, there was no difference in body weight between wild-type and Mrap2tm1a/tm1amice (Fig. 1D). However, in both genetic backgrounds and in both sexes, Mrap2tm1a/tm1a mice had a significant increase in body weight from 6 weeks of age. By 16 weeks of age on a C57BL/6N background, mean body weight in male wild-type mice was 32.2g, compared with 42.5g in Mrap2tm1a/tm1a mice; the corresponding weights in females were 25.4g and 38.7g, respectively (Fig. 1E). Similarly, on a 129/Sv background, mean body weight in male wild-type mice was 29.1g, compared with 45.1g in Mrap2tm1a/tm1a mice; the corresponding weights in females were 24.4g and 36.9g, respectively.
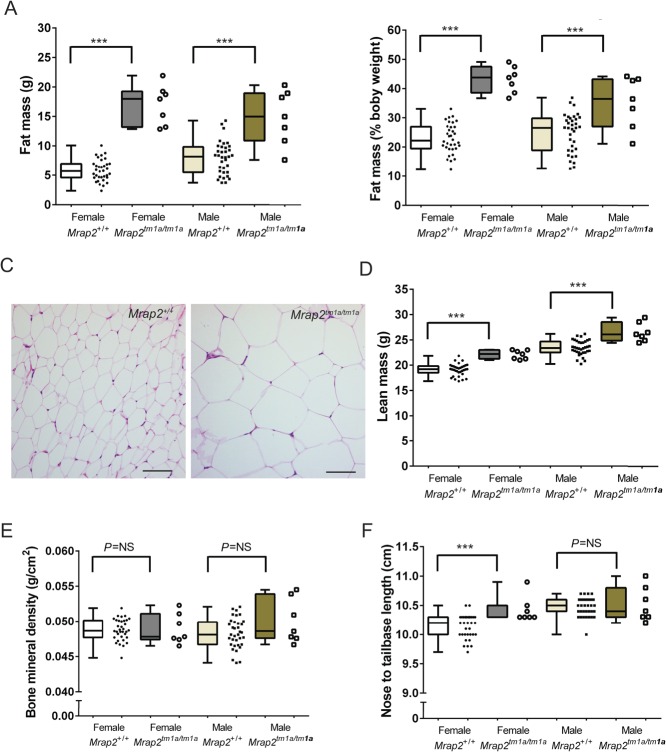
In C57BL/6N, this increase in body weight was as a result of a significant increase in both fat and lean mass (Fig. 2A and D). C57BL/6N Mrap2tm1a/tm1a females had twice the fat/body weight ratio of Mrap2+/+ controls and Mrap2tm1a/tm1a males displayed a 1.5-fold increase (Fig. 2A and B) with clear increase in adipocyte size macroscopically (Fig. 2C). No difference in bone mineral content or density was observed in either sex (Fig. 2E). Body length was significantly increased in female, but not male Mrap2tm1a/tm1a mice (Fig. 2F). On the 129/Sv background, a significant increase in fat mass was recorded in both male and female Mrap2tm1a/tm1a mice compared with wild-type controls with no change in lean mass observed (data not shown).
Figure 2.
Increased fat content and lean mass in Mrap2tm1a/tm1a mice at 14 weeks. (A) Increased fat mass in mutant C57BL/6N mice. (B) Fat mass represented as a percentage of body weight. (C) Adipocyte hypertrophy as demonstrated by H&E histology of peripheral fat (representative image of inguinal white fat from 129/Sv mice). Scale bars=200μm. (D) Increased lean mass and (E) no difference in bone mineral density or content (data not shown) in Mrap2tm1a/tm1a mice (C57BL/6N) compared with Mrap2+/+. (F) Increased body length in the female mutant mice only (C57BL/6N). n=7 for each Mrap2tm1a/tm1a group, 34 for female Mrap2+/+ and 35 for Mrap2+/+ controls gender/genotype. P-values presented on graphs are either global P-values for genotype adjusted for multiple correction testing, or (in the cases of sexual dimorphism) the P-value is the impact of genotype for that sex. ***P=0.0005; NS, not significant.
Mrap2-deficient mice display little difference in food intake and energy expenditure compared with wild-type control mice
Activation of the melanocortin system has a role in both feeding behaviour in both ad libitum conditions and in re-feeding after fasting. To determine if loss of Mrap2 affects feeding behaviour in either situation, the food intake and body weight of 8-week-old male and female 129/Sv mice were monitored over a period of 60h. For the first 24h, food was freely available, after which mice were fasted for 24h followed by reintroduction of food (Fig. 3A, B, C and D).
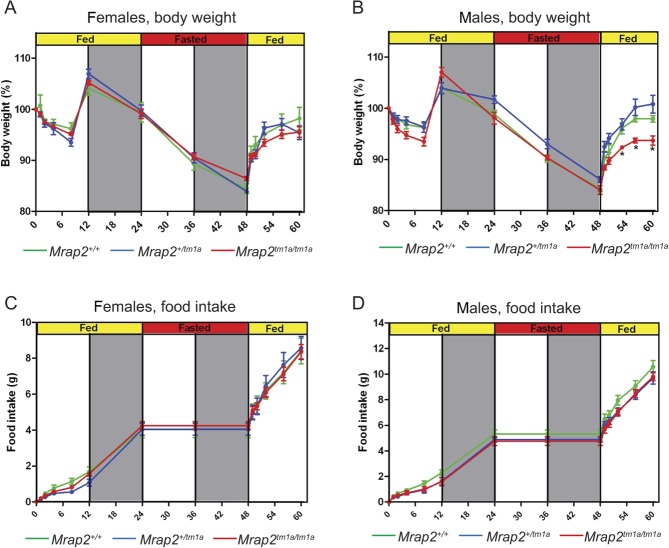
Figure 3.
Food intake and energy expenditure balance in Mrap2+/+, Mrap2tm1a/+ and Mrap2tm1a/tm1a mice. Body weight dynamics of female (A) Mrap2+/+ n=6; Mrap2tm1a/+ n=4; Mrap2tm1a/tm1a n=10 and male (B) Mrap2+/+ n=6; Mrap2tm1a/+ n=7; Mrap2tm1a/tm1a n=6 mutant mice and their food intake (C and D) in response to a 24h fast (129/Sv background). P<0.05.
Neither female nor male Mrap2tm1a/tm1a mice exhibited changes in food intake when compared with wild-type controls over the entire period. Further, Mrap2tm1a/tm1a mice did not show any difference in the rate of weight loss upon fasting compared with the wild-type mice. However, interestingly, during re-feeding after a fast, Mrap2tm1a/tm1a male mice did not re-gain weight as fast as the wild-type males (Fig. 3C and D).
Total energy expenditure measurements versus lean mass or total body weight did not show significant changes between the genotypes/sex (Supplementary Fig. 1). Analysis of respiratory quotient over a period of 48-h demonstrated that it did not differ between Mrap2tm1a/tm1a and their Mrap2+/+ littermates (Supplementary Fig. 2). In keeping with a lack of change in energy expenditure, there was no difference between Mrap2tm1a/tm1a and Mrap2+/+ mice in the expression level of Ucp1 mRNA and UCP1 protein levels in brown adipose tissue of age-matched animals, despite differences in morphology (Supplementary Fig. 3).
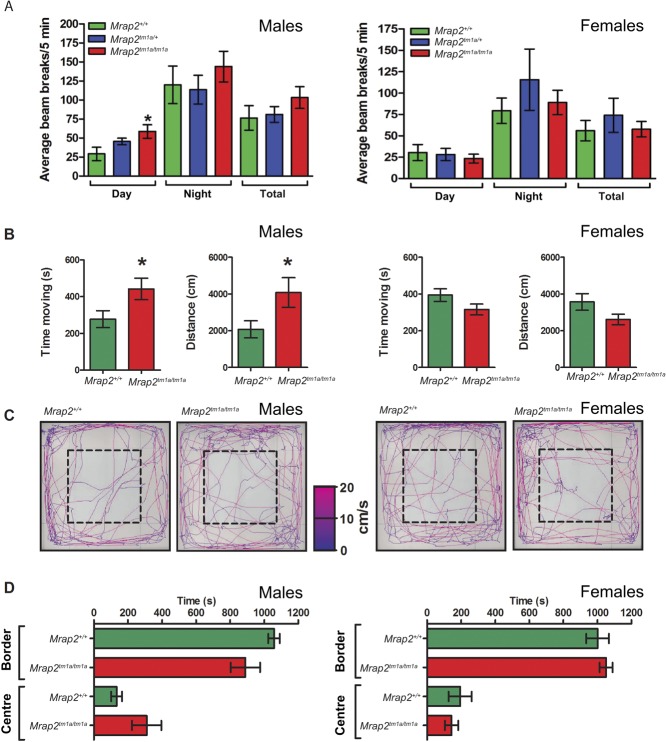
Locomotor activity measurements (average beam breaks in a 5min time period) demonstrated that male 129/Sv Mrap2tm1a/tm1a mice, compared with wild-type, moved significantly more during the daytime (Fig. 4A). No difference was observed in females.
Figure 4.
Male Mrap2tm1a/tm1a mice locomotor activity analysis. (A) Beam breaks/5 min in male (left, Mrap2+/+ n=9; Mrap2tm1a/+ n=7; Mrap2tm1a/tm1a n=11) and female (right, Mrap2+/+ n=6; Mrap2tm1a/+ n=4; Mrap2tm1a/tm1a n=10) mice are shown, with male Mrap2tm1a/tm1a mice demonstrating significantly increased locomotor activity in their home cages during the daytime. (B) Open field assessment of Mrap2tm1a/tm1a mice also indicate a significant increase in total time moving and distance travelled over 20 min in males (left), when compared with Mrap2+/+ littermates, but not females (right). (C) Representative activity traces of the centre-point of individual male mice (left) and female mice (right) in the open field. The colour of the trace indicates the velocity of the mouse from 0cm/s (blue) to 20cm/s (pink). The centre of the open field is indicated by a dashed box. (D) Neither male (left) nor female Mrap2tm1a/tm1a mice (right) displayed a significant difference in time spent in areas of the open field, compared with wild-type controls. N number for B–D is eight per group/gender, *P<0.05.
Mrap2tm1a/tm1a mice display behavioural changes when presented with a novel environment
To further examine the locomotor activity as well as novel environment exploration and anxiety-related behaviour, 20-week-old 129/Sv Mrap2tm1a/tm1a mice were subjected to an open field exploration test during the light phase. This recapitulated the sex-specific difference in locomotor activity between female and male Mrap2tm1a/tm1a mice (Fig. 4B and C). Mrap2tm1a/tm1a male mice spent more time moving and covered a greater distance compared with Mrap2+/+ mice. Although Mrap2tm1a/tm1a male mice appeared to spend more time traversing the centre of the open field than controls, the difference was not significant, P=0.075 (Fig. 4C and D). There was no difference in thigmotactic behaviour in the females, and Mrap2tm1a/tm1a mice of both sexes displayed no differences in gait, circling and rearing behaviour (data not shown).
Mrap2-deficient mice are Sim1 deficient
To further explore what might be the driving changes in body composition in Mrap2-deficient mice, we undertook transcriptomic analysis of laser microdissection PVN from 9-week-old 129/Sv Mrap2tm1a/tm1a mice and wild-type littermates (Fig. 5A). Mice of a 129/Sv genetic background, less prone to developing obesity-related co-morbidity, were used to reveal the effect of Mrap2 deficiency without secondary changes caused by hyperinsulinaemia and/or elevated glucose. We confirmed the changes observed in laser-capture material by undertaking Q-RT-PCR on whole hypothalamus extracted from a separate, second population of 129/Sv Mrap2tm1a/tm1a mice and wild-type littermates. Expression of genes that did not show any changes by microarray, such as Sf-1 and Pomc, were also confirmed by Q-RT-PCR as additional controls (Supplementary Fig. 4).
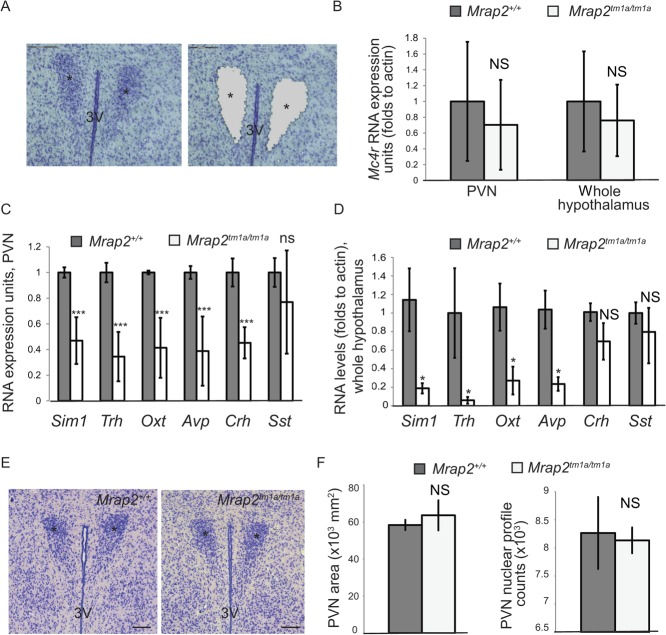
Figure 5.
Mrap2 is involved in Mc4r regulation in the hypothalamus. (A) An example of the hypothalamic section stained with cresyl violet before the microdissection (left panel) and after (right panel). Asterisks show the position of the PVN, 3 V-third ventricle, the scale bars are 150 μm. (B) Mc4r expression level in the PVN (Mrap2+/+ n=3, Mrap2tm1a/tm1a n=3) and in the whole hypothalamus (Mrap2+/+ n=4, Mrap2tm1a/tm1a n=4) as determined by the qPCR. (C) Expression of Sim1, Trh, Oxt, Avp, Crh and Sst in the PVN of 129/Sv wild type (n=4) and Mrap2tm1a/tm1a (n=3) mice. The data are represented as the mean of the microarray fluorescence values (±s.e.m.), normalized to the wild type for each gene. *P<0.05; **P<0.005; ***P<0.0005. (D) Expression of Sim1, Trh, Oxt, Avp, Crh and Sst in the whole hypothalamus of the wild-type and Mrap2tm1a/tm1a mice as determined by the qPCR. Data from male mice n=4 per genotype is shown. (E) Morphology of the PVN of 129/Sv Mrap2tm1a/tm1a mice (right panel) compared with the wild type as shown by representative images of coronal brain sections (approx. bregma –0.8 mm) stained by Nissl. (F) Average PVN area size (left graph) and stereotaxic counts of Nissl positive cells (right graph) in the PVN of the mutant 129/Sv mice (n=3) and their wild-type littermates (n=3).
We could not detect significant changes in Mc4r mRNA expression in the PVN of Mrap2tm1a/tm1a mice due to the high variability between mice within each group (Fig. 5B). However, we found that Sim1 mRNA level in the PVN of Mrap2tm1a/tm1a mice was less than 50% of that observed in wild-type littermates (Fig. 5C). Sim1 is responsible for the late stages of the differentiation of oxytocin (Oxt), arginine vasopressin (Avp), corticotrophin-releasing hormone (Crh), thyrotropin-releasing hormone (Trh) and somatostatin neurons (Sst) (Michaud et al. 1998). In keeping with this, in Mrap2tm1a/tm1a mice, PVN expression levels of Oxt, Avp, Trh and Crh were significantly decreased compared with the wild-type. Sst expression in Mrap2tm1a/tm1a was unchanged compared with Mrap2+/+ mice, although results were variable within the cohort (n=3 per group). Analysis of RNA from whole hypothalami (Fig. 5D) recapitulated these findings, except for Crh expression levels, which did not reach statistical significance. All changes were confirmed in both sexes (data not shown).
It is known that SIM1 is implicated in the development of the PVN and Sim1+/– mice exhibit a smaller PVN with reduced neuron number compared with their wild-type littermates (Michaud et al. 2001). We could not find morphological changes or a reduction in the number of neurons in the PVN of Mrap2tm1a/tm1a mice compared with wild-type (Fig. 5E and F), suggesting that unlike Sim1+/–, a lack of Mrap2 does not cause underdevelopment of the PVN.
Mrap2 deficiency increases circulating HDL and LDL cholesterol
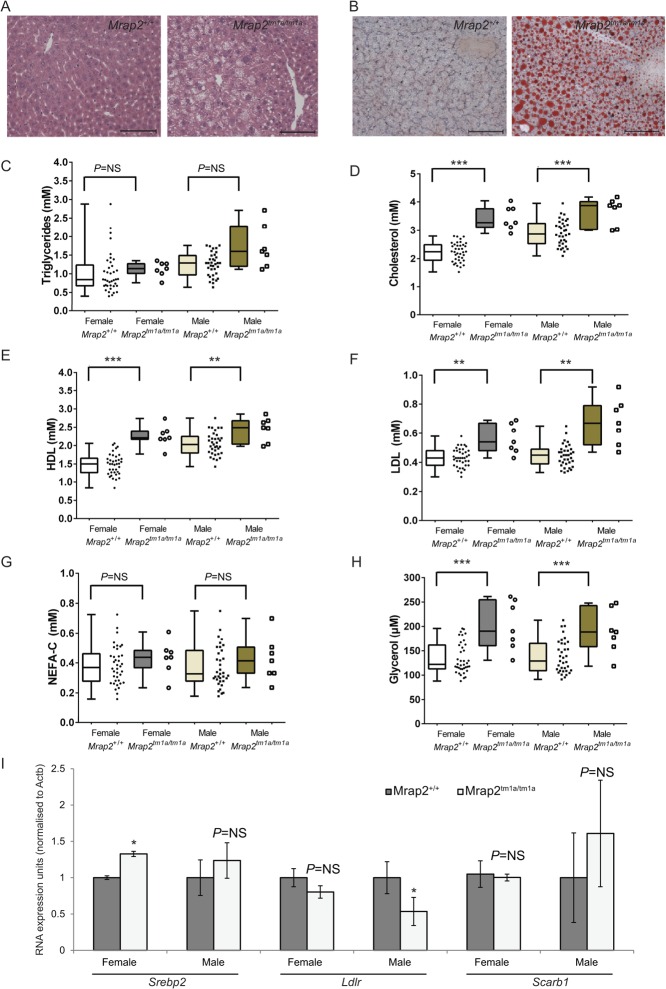
Macroscopically, the livers of Mrap2tm1a/tm1a mice were visibly pale in both sexes in both 129/Sv and C57BL/6N backgrounds, and the histological analysis showed lipid accumulation (Fig. 6A and B). There is recent evidence that the central melanocortin system directly controls peripheral lipid metabolism and circulating cholesterol (Nogueiras et al. 2007, Perez-Tilve et al. 2010). We therefore studied the cholesterol and lipid profile in C57BL/6N Mrap2tm1a/tm1a mice. The blood triacylglyceride levels (TAG) were not significantly different in Mrap2tm1a/tm1a mice compared with wild-type (Fig. 6C). However, total circulating cholesterol in Mrap2tm1a/tm1a mice was significantly higher than in wild-type controls of both sexes (Fig. 6D). HDL was elevated in both sexes with a greater percentage increase in females (Fig. 6E). LDL was significantly increased in male and female Mrap2tm1a/tm1a mice (Fig. 6F). NEFA-C levels were not significantly different between Mrap2tm1a/tm1a and Mrap2+/+ mice of either sex (Fig. 6G), whilst glycerol concentration was increased to a similar degree in mutant mice of both sexes (Fig. 6H). To investigate whether high cholesterol levels were due to a decrease in cholesterol re-uptake in the liver or an increase in cholesterol synthesis, we analysed the expression of the HDL scavenger receptor Scarb1, LDL receptor (Ldlr) and the key transcription factor of cholesterol biosynthesis Srebp2 (Shimomura et al. 1998) in the livers of 129/Sv Mrap2tm1a/tm1a mice. Interestingly, Srebp2 mRNA levels were increased in the livers of female Mrap2tm1a/tm1a mice, whereas Scarb1 and Ldlr levels were similar to the wild type (Fig. 6I). Expression analysis of livers from male Mrap2tm1a/tm1a mice showed lower levels of Ldlr mRNA whereas Scarb1 and Srebp2 transcript were similar to the wild-type male littermates (Fig. 6I).
Figure 6.
Mrap2tm1a/tm1a mice exhibit fatty liver and changes in cholesterol metabolism. Morphological changes in the liver of 129/Sv Mrap2tm1a/tm1a mutant mice as shown by H&E staining (A) and Oil Red O (B), suggesting lipid accumulation in Mrap2tm1a/tm1a. Scale bars=200μm. (C, D, E, F, G and H) Circulating TAG, total cholesterol, HDL, LDL, NEFA-C and Glycerol in 16-week C57BL/6N Mrap2tm1a/tm1a mice are shown, n=7 for each Mrap2tm1a/tm1a group, 38 for female Mrap2+/+ and 35 for Mrap2+/+ controls gender/genotype. The P-values presented on graphs are either global P-values for genotype adjusted for multiple correction testing, or (in the cases of sexual dimorphism) the P-value is the impact of genotype for that sex. NS, not significant. *P<0.05; **P<0.005; ***P<0.0005; NS, not significant. (I) Elevated expression levels of Srebp2 in Mrap2tm1a/tm1a female mice, reduced Ldlr in male Mrap2tm1a/tm1a mice and expression levels of Scarb1 in female mice and male mice (n=4 for each genotype/gender, *P<0.05, NS, not significant).
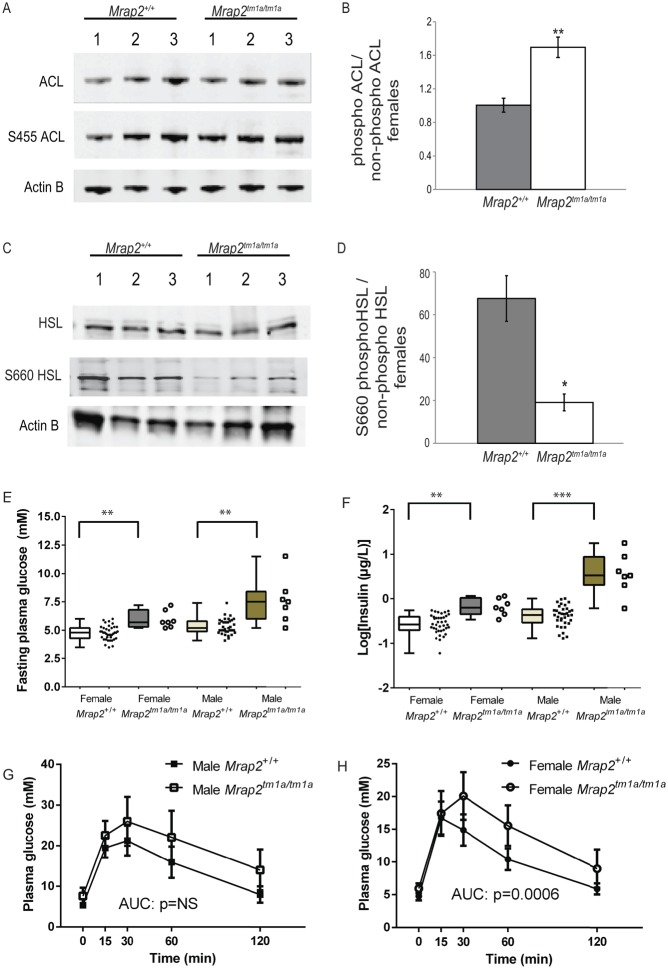
To study white fat function, we tested the protein levels and phosphorylarion state of enzymes involved in lipogenesis and fatty acid synthesis as well as phosphorylation of the rate-limiting enzyme for lipolysis of hormone sensitive lipase (HSL). Phosphorylation of ATP-citrate lyase (ACL), an enzyme responsible for the synthesis of cytosolic acetyl-CoA that serves lipogenesis and cholesterolgenesis pathways (reviewed in Chypre et al. 2012), was increased in white fat of female Mrap2tm1a/tm1a mice but not in Mrap2tm1a/tm1a male mice (Fig. 7A and B, Supplementary Fig. 5A and B).
Figure 7.
Phosphorylation of ACL and HSL are changed in female Mrap2tm1a/tm1a mice and insulin insensitivity in both genders observed at 13 weeks of age. (A) Analysis of ACL phosphorylation in white fat of the female mutant mice compared with the wild type by immunoblotting. (B) A significant increase is demonstrated in mutant mice by using band densitometry analysis of the ratio of phosphorylated to non-phosphorylated ACL normalized to β-actin. (C) Phosphorylation of HSL on S660 in white fat of the female mutant mice is decreased as assessed by western blotting. (D) Densitometry analysis of phosphorylated to non-phosphorylated HSL normalized to β-actin was calculated (n=3 per genotype; 129/Sv). (E) Fasting plasma glucose and (F) fed-state plasma insulin are higher in C57BL/6N Mrap2tm1a/tm1a mutant mice of both genders associated with significantly elevated insulin levels (log transformed due to the range of values observed in Mrap2tm1a/tm1a male mice. (G&H) IPGTT performed on 13-week-old mice of both genders, delayed glucose clearance demonstrated in Mrap2tm1a/tm1a female mice but not statistically significant in male Mrap2tm1a/tm1a mice. n=7 for each Mrap2tm1a/tm1a group, 39 for female Mrap2+/+ and 35 for Mrap2+/+ controls gender/genotype. P-values presented on graphs are either global P-values for genotype adjusted for multiple correction testing, or (in the cases of sexual dimorphism) the P-value is the impact of genotype for that sex. AUC, area under the curve; *P<0.05; **P<0.005; ***P<0.0005; NS, not significant.
HSL phosphorylation is known to be important for the enzyme activation and therefore lipolysis (Egan et al. 1992). The proportion of HSL phosphorylated on S563, S565 and S660 was analysed and it was found that the proportion of S660 phosphorylated HSL was three times lower in white fat of female Mrap2tm1a/tm1a mice compared with the wild type (Fig. 7C and D). Phosphorylation on other residues was not changed and male Mrap2tm1a/tm1a mice did not have changes in HSL phosphorylation on any residues tested (Supplementary Fig. 5C, D, E and F).
Analysis of glucose homeostasis in Mrap2-deficient mice
Mrap2tm1a/tm1a mice of both sexes on a C57BL/6N background fed on a chow diet from weaning display elevated fasting plasma insulin concentration with higher fasting blood glucose at 13 weeks of age compared with Mrap2+/+ mice (Fig. 7E and F). Glucose clearance in response to an IP glucose bolus (Fig. 7G and H) appeared delayed, although statistical analysis of the area under the curve was only significant in female mice.
Discussion
In this study, we report the generation and characterization of a murine model with a targeted Mrap2 allele (knockout-first Mrap2tm1a(EUCOMM)Wtsi). The construct used here is a Tm1a allele, which theoretically can still allow transcriptional read through (White et al. 2013). We detected low Mrap2 expression within Mrap2tm1a/tm1a homozygous mice and in the absence of a suitable antibody we cannot rule out the possibility that the animals studied were indeed ‘strong hypomorphs’ rather than of complete knockouts.
Mrap2tm1a/tm1a mutant mice on both C57/BL6N and 129/Sv background display severe early-onset obesity with a significantly increased fat mass, consistent with a recent report of Mrap2-knockout mice (Asai et al. 2013). Unlike Mrap2–/– mice on a 129/Sv genetic background (Asai et al. 2013), our C57BL/6N Mrap2tm1a/tm1a display elevated fasting insulin and blood glucose concentrations. We believe that this is an evidence of an interesting interaction between the genetic backgrounds with Mrap2, which will form the basis of a future study.
Mrap2–/– mice have been reported previously to show no increased food intake or reduction in energy expenditure and thermogenesis to account for their increased body weight. In our assessment of 8-week-old Mrap2tm1a/tm1a mice, we recapitulated and confirmed these findings. We calculate that female Mrap2tm1a/tm1a mice gained more weight compared with the wild-type mice (females: 0.117±0.041g/day; males: 0.096±0.023g/day). To achieve this, a female mutant mouse would need to deposit 0.701±0.246kcal/day and a male mouse 0.578±0.139kcal/day (Flatt 1991). This would equate to an increase in food intake of 0.232±0.081g of standard chow (for females) and 0.191±0.046g (for males), which is within the measurable limits of food intake variation. Therefore, it is possible that the causative difference is below the threshold of the detection (Tschop et al. 2012, Speakman 2013). Indeed, older animals that are significantly more obese than their wild-type counterparts, and as a result would be expected to consume larger quantities, demonstrate a subtle increase in cumulative food intake when monitored over 50 days (Asai et al. 2013). Additionally, our behavioural tests on separate cohorts of animals, independently analysed on two separate platforms, demonstrated sex-specific increased daytime locomotor and exploratory activity in Mrap2tm1a/tm1a male mice, which may indicate food-seeking behaviour. These lines of evidence would point to hyperphagia being the key driver in the development of obesity. However, importantly, obesity in Mrap2-deficient animals clearly precede any change in food intake and in paired feeding studies Mrap2-deficient animals continue to gain more weight than their wild-type counterparts (Asai et al. 2013). It is only when further food restriction was undertaken did weight gain in mutant mice become equivalent to that of wild-type mice (Asai et al. 2013). Intriguingly, this suggests a far more complex mechanism at play in Mrap2 null mice.
Our transcriptomic analysis of the PVN of the mutant mice also favoured increased energy intake as being a more likely promoter of increased body weight. It was found that Sim1 expression levels were low in the PVN of the Mrap2tm1a/tm1a mice resembling Sim1 deficiency. The reduced expression of Sim1 and its associated neuropeptides cannot be secondary to obesity alone, as this was not observed in reported hypothalamic microarray data from obese mice fed in a high fat diet (Lee et al. 2010). SIM1 is a transcription factor that regulates development of the PVN, and Sim1–/– mice die due to the abnormal hypothalamic architecture (Michaud et al. 2001). Heterozygous Sim1+/– mice exhibit a small PVN with reduced neuronal number and develop severe early-onset obesity due to hyperphagia and increased linear growth. They have an impaired response to MTII, an MC3R/MC4R agonist, indicative of a disrupted central melanocortin pathway (Holder et al. 2004, Kublaoui et al. 2006a, b, Tolson et al. 2010). Expression analysis of the PVN from Sim1+/– mice has shown an 80% decrease in Oxt expression and 20–40% decrease in Trh, Crh, Avp and Sst expression (Kublaoui et al. 2008). Compared with Sim1+/– mice, we did not detect morphological changes within the PVN of Mrap2tm1a/tm1a mice. However, the levels of Oxt, Avp and Trh in Mrap2tm1a/tm1a mice PVN were reduced, consistent with low Sim1 expression levels. Interestingly, despite these changes and also the high expression of Mrap2 in the pituitary, we found no evidence of pituitary dysfunction in Mrap2tm1a/tm1a mice. Progression through puberty and fertility appear unchanged in mutant mice, and thyroid hormone levels, T3 and T4, were normal (data not shown). Corticosterone concentrations were previously reported to be normal (Asai et al. 2013). This would suggest that the level of neuropeptide expression is sufficient for peptide production and physiological stimulation of downstream hormones, as exemplified by normal levels of T4 in the case of Trh.
In contrast to the downstream effects, the change in neuropeptide expression is likely to play a direct role in maintaining energy homeostasis as it is known that Oxt, Avp and Trh in the PVN have anorexigenic effects (reviewed in Valassi et al. 2008), and Oxt is thought to be key to the mechanism for the hyperphagia of Sim1+/− mice (Kublaoui et al. 2008). Overall, the changes in Sim1 provide further evidence that a central melanocortin pathway deficiency exists in Mrap2tm1a/tm1a mice as SIM1 has been considered to be downstream of MC4R signalling (Holder et al. 2004, Kublaoui et al. 2006a, b, Tolson et al. 2010).
Modulation of MC4R has been shown to directly affect peripheral lipid metabolism. Mc4r–/– mice had elevated plasma cholesterol and HDL levels (Nogueiras et al. 2007, Perez-Tilve et al. 2010). Both sexes of Mrap2tm1a/tm1a mice displayed elevated circulating cholesterol although there is a suggestion that males and females partition cholesterol into HDL differently, consistent with reports of sex differences in the hepatic control of cholesterol metabolism (De Marinis et al. 2008). Mrap2tm1a/tm1a female mice showed increased de novo hepatic lipogenesis; however, unlike female Mrap2tm1a/tm1a mice, male Mrap2tm1a/tm1a mice had low expression of liver LDL receptor, possibly reflecting elevated circulating LDL levels.
Mrap2tm1a/tm1a female mice had increased ACL phosphorylation in white fat, a key modification that activates ACL catalytic activity (Berwick et al. 2002) and leads to an increase in de novo lipogenesis. Along with this change we found that phosphorylation of HSL on S660, which is phosphorylated by protein kinase A upon sympathetic nervous system activation (Anthonsen et al. 1998), was decreased. It is possible that both changes in the liver and white adipocytes are due to the low sympathetic tone which is in part regulated via the central melanocortin system (Nogueiras et al. 2007, Perez-Tilve et al. 2010). Mrap2 is not expressed in white fat or liver and therefore is unlikely to influence de novo lipogenesis directly in these tissues, suggesting that MRAP2 may contribute to the melanocortin regulation of sympathetic outflow. The changes observed in white fat were only found in females and are thus unlikely to be the primary cause of MRAP2-associated obesity, although this might explain the greater severity of obesity in females.
Our study corroborates the role of MRAP2 in metabolism. The changes in cholesterol metabolism and transcriptomic profile in the PVN of Mrap2tm1a/tm1a mice support the notion that MRAP2 is involved in the MC4R signalling pathway in vivo. However, our data further highlights phenotypic differences between Mrap2-deficient and Mc4r-deficient mice. Despite both mice developing severe early-onset obesity, Mc4r–/– mice are clearly hyperphagic with decreased energy expenditure (Huszar et al. 1997, Balthasar et al. 2005), whilst Mrap2-deficient mice display no demonstrable hyperphagia or reduction in energy expenditure. We describe other additional phenotypic differences such as normal bone mineral content and density in Mrap2tm1a/tm1a mice in contrast to increased bone density in Mc4r–/– mice (Braun et al. 2012). Importantly, Asai and coworkers demonstrated that Mrap2–/– mice remain responsive to treatment with MTII, a MC3R/MC4R agonist, whilst the anorexic response to MTII is abolished in Mc4r–/– mice, suggesting at least some preservation of MCR function centrally (Marsh et al. 1999, Asai et al. 2013). We also show sex-specific differences in glucose handling as well as an exploratory activity phenotype in Mrap2tm1a/tm1a mice. Taken together, our study points towards the likelihood of MC4R-independent mechanisms and possibly MCR-independent pathways in the pathogenesis of MRAP2-associated obesity.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/JOE-16-0057.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
Funding was received from The Medical Research Council UK (MRC/Academy of Medical Sciences Clinician Scientist Fellowship Grant G0802796, to L F C, supporting T V N), Society for Endocrinology Early Career award to T V N, the Wellcome Trust (Grant No. 098051) to D W L, C L, E H W and for the M G P generation and phenotyping of the C57BL/6N background mice. R J G is supported by a Wellcome Clinical Research Training Fellowship (Grant No. Wbib92024MA). R L, D R, S O R and A P C are funded by the Medical Research Council (MRC) Metabolic Disease Unit (MRC_MC_UU_12012/1). L G was supported by Biotechnology and Biological Sciences Research Council (BBSRC), award BB/L00267/1 and Rosetrees Trust.
Acknowledgements
The authors thank the support from the funding bodies. The authors also wish to thank Sanger Mouse Genetics Project and Research Support Facility staff for generating and providing mouse phenotyping information; Natasha Karp for statistical input on the MGP mouse data; Keith Burling for insulin analysis and David Jackson for help with manuscript preparation.
References
- Anthonsen MW, Ronnstrand L, Wernstedt C, Degerman E, Holm C. 1998. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. Journal of Biological Chemistry 273 215–221. ( 10.1074/jbc.273.1.215) [DOI] [PubMed] [Google Scholar]
- Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N, Ramanathan V, Strochlic DE, Ferket P, Linhart K, et al. 2013. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science 341 275–278. ( 10.1126/science.1233000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, Ferreira M, Tang V, McGovern RA, Kenny CD, et al. 2005. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 123 493–505. ( 10.1016/j.cell.2005.08.035) [DOI] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. 2002. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. Journal of Biological Chemistry 277 33895–33900. ( 10.1074/jbc.M204681200) [DOI] [PubMed] [Google Scholar]
- Braun TP, Orwoll B, Zhu X, Levasseur PR, Szumowski M, Nguyen ML, Bouxsein ML, Klein RF, Marks DL. 2012. Regulation of lean mass, bone mass, and exercise tolerance by the central melanocortin system. PLoS ONE 7 e42183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LF, Webb TR, Chung TT, Meimaridou E, Cooray SN, Guasti L, Chapple JP, Egertova M, Elphick MR, Cheetham ME, et al. 2009. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. PNAS 106 6146–6151. ( 10.1073/pnas.0809918106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ingham N, Clare S, Raisen C, Vancollie VE, Ismail O, McIntyre RE, Tsang SH, Mahajan VB, Dougan G, et al. 2013. Mcph1-deficient mice reveal a role for MCPH1 in otitis media. PLoS ONE 8 e58156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chypre M, Zaidi N, Smans K. 2012. ATP-citrate lyase: a mini-review. Biochemical and Biophysical Research Communications 422 1–4. ( 10.1016/j.bbrc.2012.04.144) [DOI] [PubMed] [Google Scholar]
- Cone RD. 2005. Anatomy and regulation of the central melanocortin system. Nature Neuroscience 8 571–578. ( 10.1038/nn1455) [DOI] [PubMed] [Google Scholar]
- De Marinis E, Martini C, Trentalance A, Pallottini V. 2008. Sex differences in hepatic regulation of cholesterol homeostasis. Journal of Endocrinology 198 635–643. ( 10.1677/JOE-08-0242) [DOI] [PubMed] [Google Scholar]
- Egan JJ, Greenberg AS, Chang MK, Wek SA, Moos MC, Jr, Londos C. 1992. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. PNAS 89 8537–8541. ( 10.1073/pnas.89.18.8537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia M, Livesey G. 1992. Energy expenditure and fuel selection in biological systems: the theory and practice of calculations based on indirect calorimetry and tracer methods. World Review of Nutrition and Dietetics 70 68–131. ( 10.1159/000421672) [DOI] [PubMed] [Google Scholar]
- Flatt JP. 1991. Assessment of daily and cumulative carbohydrate and fat balances in mice. Journal of Nutritional Biochemistry 2 193–202. ( 10.1016/0955-2863(91)90016-X) [DOI] [Google Scholar]
- Franklin KBJ, Paxinos G. 2012. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates. Amsterdam, The Netherlands: Academic Press. [Google Scholar]
- Gorrigan RJ, Guasti L, King P, Clark AJ, Chan LF. 2011. Localisation of the melanocortin-2-receptor and its accessory proteins in the developing and adult adrenal gland. Journal of Molecular Endocrinology 46 227–232. ( 10.1530/JME-11-0011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder JL, Jr, Zhang L, Kublaoui BM, DiLeone RJ, Oz OK, Bair CH, Lee YH, Zinn AR. 2004. Sim1 gene dosage modulates the homeostatic feeding response to increased dietary fat in mice. American Journal of Physiology: Endocrinology and Metabolism 287 E105–E113. [DOI] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, et al. 1997. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88 131–141. ( 10.1016/S0092-8674(00)81865-6) [DOI] [PubMed] [Google Scholar]
- Karp NA, Melvin D, Mott RF. 2012. Robust and sensitive analysis of mouse knockout phenotypes. PLoS ONE 7 e52410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublaoui BM, Holder JL, Jr, Gemelli T, Zinn AR. 2006a. Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Molecular Endocrinology 20 2483–2492. [DOI] [PubMed]
- Kublaoui BM, Holder JL, Jr, Tolson KP, Gemelli T, Zinn AR. 2006b. SIM1 overexpression partially rescues agouti yellow and diet-induced obesity by normalizing food intake. Endocrinology 147 4542–4549. [DOI] [PubMed] [Google Scholar]
- Kublaoui BM, Gemelli T, Tolson KP, Wang Y, Zinn AR. 2008. Oxytocin deficiency mediates hyperphagic obesity of Sim1 haploinsufficient mice. Molecular Endocrinology 22 1723–1734. ( 10.1210/me.2008-0067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AK, Mojtahed-Jaberi M, Kyriakou T, Astarloa EA, Arno M, Marshall NJ, Brain SD, O’Dell SD. 2010. Effect of high-fat feeding on expression of genes controlling availability of dopamine in mouse hypothalamus. Nutrition 26 411–422. ( 10.1016/j.nut.2009.05.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Hollopeter G, Huszar D, Laufer R, Yagaloff KA, Fisher SL, Burn P, Palmiter RD. 1999. Response of melanocortin-4 receptor-deficient mice to anorectic and orexigenic peptides. Nature Genetics 21 119–122. ( 10.1038/5070) [DOI] [PubMed] [Google Scholar]
- McIntyre RE, Lakshminarasimhan Chavali P, Ismail O, Carragher DM, Sanchez-Andrade G, Forment JV, Fu B, Del Castillo Velasco-Herrera M, Edwards A, van der Weyden L, et al. 2012. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genetics 8 e1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metherell LA, Chapple JP, Cooray S, David A, Becker C, Ruschendorf F, Naville D, Begeot M, Khoo B, Nurnberg P, et al. 2005. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nature Genetics 37 166–170. [DOI] [PubMed] [Google Scholar]
- Michaud JL, Rosenquist T, May NR, Fan CM. 1998. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes & Development 12 3264–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud JL, Boucher F, Melnyk A, Gauthier F, Goshu E, Levy E, Mitchell GA, Himms-Hagen J, Fan CM. 2001. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Human Molecular Genetics 10 1465–1473. ( 10.1093/hmg/10.14.1465) [DOI] [PubMed] [Google Scholar]
- Nogueiras R, Wiedmer P, Perez-Tilve D, Veyrat-Durebex C, Keogh JM, Sutton GM, Pfluger PT, Castaneda TR, Neschen S, Hofmann SM, et al. 2007. The central melanocortin system directly controls peripheral lipid metabolism. Journal of Clinical Investigation 117 3475–3488. ( 10.1172/JCI31743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselova TV, Jackson D, Campbell DC, Clark AJ, Chan LF. 2013. Melanocortin receptor accessory proteins in adrenal gland physiology and beyond. Journal of Endocrinology 217 R1–R11. [DOI] [PubMed] [Google Scholar]
- Perez-Tilve D, Hofmann SM, Basford J, Nogueiras R, Pfluger PT, Patterson JT, Grant E, Wilson-Perez HE, Granholm NA, Arnold M, et al. 2010. Melanocortin signaling in the CNS directly regulates circulating cholesterol. Nature Neuroscience 13 877–882. ( 10.1038/nn.2569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM. 2009. Opposite effects of the melanocortin-2 (MC2) receptor accessory protein MRAP on MC2 and MC5 receptor dimerization and trafficking. Journal of Biological Chemistry 284 22641–22648. ( 10.1074/jbc.M109.022400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM. 2010. Regulation of G protein-coupled receptor signaling: specific dominant-negative effects of melanocortin 2 receptor accessory protein 2. Science Signaling 3 ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebag JA, Zhang C, Hinkle PM, Bradshaw AM, Cone RD. 2013. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 341 278–281. ( 10.1126/science.1232995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD. 1998. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. Journal of Biological Chemistry 273 35299–35306. ( 10.1074/jbc.273.52.35299) [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474 337–342. ( 10.1038/nature10163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. 2013. Measuring energy metabolism in the mouse – theoretical, practical, and analytical considerations. Frontiers in Physiology 4 34 ( 10.3389/fphys.2013.00034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolson KP, Gemelli T, Gautron L, Elmquist JK, Zinn AR, Kublaoui BM. 2010. Postnatal Sim1 deficiency causes hyperphagic obesity and reduced Mc4r and oxytocin expression. Journal of Neuroscience 30 3803–3812. ( 10.1523/JNEUROSCI.5444-09.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop MH, Speakman JR, Arch JR, Auwerx J, Bruning JC, Chan L, Eckel RH, Farese RV, Jr, Galgani JE, Hambly C, et al. 2012. A guide to analysis of mouse energy metabolism. Nature Methods 9 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valassi E, Scacchi M, Cavagnini F. 2008. Neuroendocrine control of food intake. Nutrition, Metabolism, and Cardiovascular Diseases 18 158–168. ( 10.1016/j.numecd.2007.06.004) [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154 452–464. ( 10.1016/j.cell.2013.06.022) [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a