Abstract
AML is a heterogeneous disease comprising a large number of subtypes defined by specific chromosome abnormalities. One such subtype carries the t(6;9)(p22;q34) chromosome rearrangement which leads to expression of the DEK-NUP214 chimeric gene, and has a particularly poor outcome. To provide a better understanding of the molecular etiology of these relatively rare individual AML variants, it is necessary to generate in vivo models, which can also serve as a means to evaluate targeted therapies based on their specific genetic abnormalities. Here we describe the development of a human cell AML, generated in CD34+ human hematopoietic progenitor cells xenografted into immuncompromised mice that express human myeloid cell growth factors. Within six months, these mice develop a human cell AML with phenotypic characteristics of the primary t(6;9) disease and a CD45+CD13+CD34+CD38+ immunophenotype. Gene expression studies show that members of the HOX family of genes (HOXA9, 10, B3, B4 and PBX3) are highly upregulated in the AML from this mouse model as well as from primary human t(6;9) AML. Gene expression analysis also identified several other significantly disregulated pathways involving KRAS, BRCA1 and ALK, for example. This is the first report of a humanized model of the DEK-NUP214 disease and provides a means to study the development and treatment of this particular subtype of AML.
INTRODUCTION
Acute myeloid leukemia (AML) with t(6;9)(p22;q34) is currently classified as a distinct entity by the World Health Organization, occurring in 1% of adult and 2% of pediatric AML cases. AML with t(6;9) identifies a high-risk group of AML patients, has a significantly lower complete remission rate, higher relapse rate and poor overall survival. The DEK-NUP214 (previous known as DEK-CAN) fusion gene was initially described as the consequence of a recurrent t(6;9) translocation in a subset of AML patients.1, 2 Typically, the t(6;9) is found in de novo AML, morphologically associated with type M2 in the French-American-British (FAB) classification system. Currently available chemotherapy protocols are not effective, although allogeneic stem cell transplantation was shown to be associated with better outcome.3 Although t(6;9) has been ontologically associated with the development of AML for more than 3 decades, the molecular etiology for this subgroup of AML is still unclear.
The 6;9 chromosome translocation leads to a fusion transcript which encodes a 165-kDa chimeric protein, resulting from the in-frame fusion of the DEK (at 6p22.3) and NUP214 (at 9q34.13) open reading frames. The reciprocal NUP214-DEK fusion transcript is not detectable in t(6;9)-positive patients.4 DEK-NUP214, therefore, is considered the etiological product of the t(6;9) translocation. At the time of diagnosis, the t(6;9) translocation is seen as the only cytogenetic abnormality in the majority (88%) of these AML, suggesting that the DEK-NUP214 fusion gene is the primary initiation event that drives disease development in this leukemia subgroup.3, 5 The DEK-NUP214 fusion has been primarily observed in AML, suggesting the DEK-NUP214 oncogene exclusively impacts the differentiation and maturation of myeloid lineage cells.
DEK is a nuclear protein that binds to chromatin and functions in several cellular processes including chromatin remodeling, transcriptional regulation, cellular differentiation and apoptosis.6 DEK upregulation has been clinically noted for many human tumor types, including AML.6, 7 These observations suggest that DEK may serve as a proto-oncogene.8, 9 Moreover, in the DEK-NUP214 fusion protein, all the chromatin binding domains of DEK are conserved (Figure 1a), and expression of DEK-NUP214 is consequently regulated by the DEK promoter.10 Nucleoporin 214kDa (NUP214), also known as CAN, is a gene that encodes a component of the nuclear pore complex, a structure that extends across the nuclear envelope, forming a gateway that regulates the flow of macromolecules between the nucleus and the cytoplasm.11 The protein encoded by this gene is localized to the cytoplasmic face of the nuclear pore complex, where it is required for regulated cell cycle progression and nucleocytoplasmic transport. The biological function of the NUP214 component in the fusion protein is unknown.
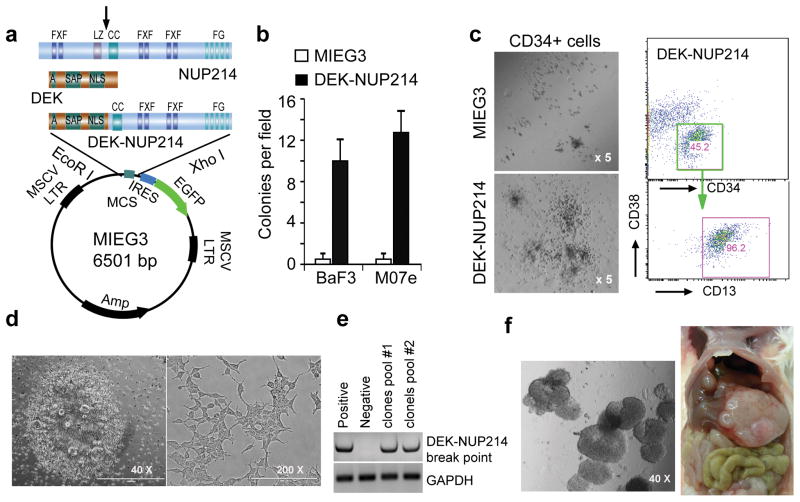
Figure 1. Transformation activity of the DEK-NUP214 fusion gene in vitro.
(a) Scheme for sub-cloning the DEK-NUP214 fusion gene. The DEK-NUP214 was excised from a pMIG vector using EcoRI/XhoI and then cloned into the MIEG3 retroviral vector. A - acidic regions; SAP - scaffold attachment factor; NLS - nuclear localization signal. NUP214: FXF - repeat motifs; LZ - leucine zipper; FG - repeat motifs. Arrows: breakpoints in leukemia. (b) Colony formation assays (CFA) in soft-agar shows the number of colonies per field calculated as an average of triplicate experiments. CFA was performed in 0.6% PCR grade agarose without the addition of cytokines and chemokines (c) Human cord blood cells were obtained from the Cord Blood Bank of GRU under an approved IRB protocol. CD34+ cells were isolated using the EasySep Cord Blood CD34 Positive Selection Kit (StemCell Technologies) following the manufacturer’s protocol. CD34+ cells were infected with either the DEK-NUP214 or the control MIEG3 vector as described previously.16 . The infected cells were then plated into methylcellulose media (MethoCult H4435, StemCell Technologies) at 1500 cell/well in 12-well plates, and cultured for 10 days. Colonies were then counted and classified under a microscope at x50 magnification (left panel). The experiment was repeated in triplicate. After counting, the colonies were pooled for flow cytometry analysis with human-specific antibodies (right panel). (d) Some adherent cells formed colonies in the DEK-NUP214 infected CD34+ plates after 2 weeks infection (left panel). These cells showed an epithelial morphology after reseeding (right panel). (e) RT-PCR analysis with primers spanning the DEK-NUP214 breakpoint shows that DEK-NUP214 is expressed in the pooled colonies. (f) A representative image of colonies in soft-agar (left panel). The DEK-NUP214 transduced adherent cells were seeded in soft-agar medium at 1000 cells/well (6-well plate) for 2 weeks. When 2 million cells were transplanted into NSG-SGM3 mice, the engrafted mice developed tumors in the kidney capsule (not shown) and liver (right panel).
The molecular pathogenesis of DEK-NUP214 is still largely unknown, due to the small number of reported patients and the lack of a representative mouse model. Evidence for an oncogenic function comes from in vitro studies that demonstrate forced expression of the DEK-NUP214 fusion gene promotes cell proliferation through upregulation of mTOR.12 Clinical data shows that STAT5 is activated in DEK-NUP214 positive AML compared with normal CD34+ cells.13 Although it was shown that the DEK-NUP214 can induce AML development in a bone marrow transduction/transplantation syngeneic mouse model,13 only ~20% of the mice in this study developed AML, and with a long latency period (~350 days), which made it difficult to perform in-depth molecular analyses. Here, we demonstrated that NSG-SGM3 immunocompromised mice, engrafted with DEK-NUP214 transduced human CD34+ cells, predominantly developed AML within an average of 6 months. We also demonstrate that this humanized mouse AML model has phenotypic and molecular features that faithfully recapitulates primary human t(6;9) AML. Thus, this novel murine model provides the opportunity to evaluate molecular etiology and therapeutic strategies to treat this devastating disease.
RESULTS AND DISCUSSION
Transformation activity of DEK-NUP214 in vitro
To investigate the oncogenic effect of DEK-NUP214 on myeloid lineage development, we examined the ability of DEK-NUP214 to transform the M07e and BaF3 cell lines, as well as human CD34+ hematopoietic progenitor cells. The M07e human megakaryoblastic leukemia and murine BaF3 pro-B cell lines are strictly dependent on either IL-3 or GM-CSF for survival, and have been frequently used to assess the oncogenic potential of individual genes by transforming them to IL3/GM-CSF independence.14, 15 We subcloned DEK-NUP214 from a pMIG vector (a kind gift from Dr. Carl Sandén from the Lund University, Lund, Sweden) into the MIEG3 retroviral vector (Figure 1a). Retroviral expression of this fusion gene in IL-3 dependent M07e and BaF3 cell lines,16 led to their transformation to cytokine independence and increased colony formation ability in soft-agar, compared with cells infected with the empty MIEG3 vector (Figure 1b). We then determined how DEK-NUP214 expression affected differentiation of human cord blood CD34+ progenitor cells. When DEK-NUP214 transduced CD34+ cells were seeded into methycellulose medium as described previously,17 significantly increased numbers of larger colonies developed compared with those seen for control vector-infected CD34+ cells (Figure 1c). Flow cytometry analysis of the immunophenotype of these cells showed that the majority expressed myeloid lineage markers (CD13+), with small subsets showing T- (CD3+) and B- (CD19+) cell linage markers (data not shown), supporting the idea that DEK-NUP214 has a fundamental role in leukemic transformation and predominantly impacts the differentiation of myeloid cells. Surprisingly, we also obtained multiple (>10) clones of adherent cells in the DEK-NUP214 transfected wells (from a 6-well plate) in the duplicate experiments. Each clone formed multiple cell layers (Figure 1d left) which, after reseeding, demonstrated an epithelial phenotype (Figure 1d right). RT-PCR analysis demonstrated that the DEK-NUP214 fusion gene was also expressed in these adherent cells (Figure 1e). When these DEK-NUP214 transduced cells were seeded in soft agar medium, they were all able to form foci (Figure 1f, left), suggesting a tumorigenic potential. Indeed, when injected into mice, these adherent cells infiltrated into multiple organs, such as liver and kidney, to form tumors (Figure 1f, right), demonstrating the strong transformation activity of the chimeric DEK-NUP214 fusion gene in human bone marrow cells.
DEK-NUP214 transduced human CD34+ cells develop AML in immunocompromized mice
With the in vitro evidence that DEK-NUP214 is related to leukemic transformation, we then used the modified NSG (NOD.Cg-Prkdcscid-Il2rgtm1WjlTg, Jackson Laboratory) transgenic mouse strain which endogenously expresses human SCF, GM-CSF, and IL-3 cytokines (NSG-SGM3)18 as the host for healthy human cord blood CD34+ progenitor cells transduced with chimeric DEK-NUP214, using previously described approaches16 (Figure 1a). Eleven primary recipient mice were engrafted with DEK-NUP214 transduced CD34+ cells that had been derived from 3 different umbilical cord blood samples (biologically independent triplicates). Two months after transplantation, ~10-20% of peripheral blood cells from all of these transplanted mice showed a human-specific CD45 immunophenotype. Of the eight recipients that died from AML (Figure 2b), >20% of the lymphocytes were human (CD45+CD13+CD34+CD38+) myeloid blast cells (Figure 2c). Peripheral blood smears also showed the typical human AML cell morphology with a larger nucleus and reduced cytoplasmic ratio (Figure 2d). One of the diseased mice also showed a large myeloid sarcoma in the abdomen (Figure 2a), with a CD45+CD13+CD38+ immunophenotype (Figure 2c, bottom panel). Myeloid sarcoma can be found in 2-8% of patients with AML19 and molecular analysis shows the AML cells from both the peripheral blood and myeloid sarcoma from this mouse harbored the DEK-NUP214 fusion gene (Figure 2e). Thus, DEK-NUP214 can transform human CD34+ progenitor cells and predominantly induces human AML in an NSG-SGM3 mouse model.
Figure 2. A mouse model engrafted with DEK-NUP214-transduced human CD34+ progenitor cells.
(a) A working flow for developing the humanized AML mouse model (left). An example of a large myeloid sarcoma (arrow) in a DEK-NUP214 AML bearing mouse is shown (right). (b) Survival curve for mice with DEK-NUP214 AML (n=11) compared with mice engrafted with empty vector infected CD34+ cells (n=4). c) Flow cytometry analysis with human-specific antibodies shows a myeloid progenitor immunophenotype (CD45+CD13+CD34+CD38+) in the bone marrow (BM), spleen (SP), peripheral blood (PB) and the myeloid sarcoma tumor. (d) A peripheral blood smear shows human AML cells with large nuclear-cytoplasmic ratios. (e) RT-PCR analysis demonstrates the presence of the transcript from the DEK-NUP214 fusion gene in the diseased mouse PB and sarcoma cells.
Upregulation of the HOXA and B gene clusters in DEK-NUP214 AML cells in the mouse model
To determine whether the molecular profiles from the humanized mouse AML model we have developed were similar to primary, t(6;9) AML, we first investigated the gene expression profiles available through three publically available microarray datasets: GSE17855,20 containing 6 DEK-NUP214 positive and 231 t(6;9)-negative AML cases, GSE1506121 which reports data from 69 normal human bone marrows (BM) and 202 t(6;9)-negative AML, as well as GSE3515922 which has 12 AML cell lines including the DEK-NUP214 positive, FKH-1 cell lime.23 The bioinformatics approaches used were described previously.24 Overall, unsupervised hierarchical cluster analysis (Figure 3a) demonstrated that the t(6;9) primary AMLs cluster independently from both the normal bone marrow cells and the t(6;9)-negative AML samples (not including the DEK-NUP214 transformed CD34+ cells generated in the current study). To define pathways that are potentially involved in DEK-NUP214 AML, we performed Gene Set Enrichment Analysis (GSEA, www.broadinstitute.org/gsea/) using these published datasets. The GSEA analysis revealed that aberrant up-regulation of HOXA and HOXB gene clusters were the most significantly affected signaling pathways associated with expression of the DEK-NUP214 fusion gene (Figure 3a and b), compared with both normal BM and t(6;9)-negative AML samples (Figure 3b). Aberrant up-regulation of the HOXA gene cluster has been associated with various AML subtypes,25 but up-regulation of both HOXA and HOXB gene clusters is only reported in those AML with either a normal karyotype or an NPMc mutation. 25 This study also showed that HOXA and HOXB gene clusters are highly expressed in CD34+ progenitor cells which is consistent with >50% of CD34+ cells seen in both the primary DEK-NUP214 leukemia cells and those from the mouse models (Figure. 2c and 4d). In addition to disregulation of the HOX pathways (Supplementary Figure S1), GSEA analysis also showed five other significantly disrupted pathways (Supplementary Table S1), some of which have been related to leukemia development (KRAS, JAK2) and others (BRCA1, ALK and PRC2/SUZ12), more typically seen in solid tumors (Supplementary figures 1-5).
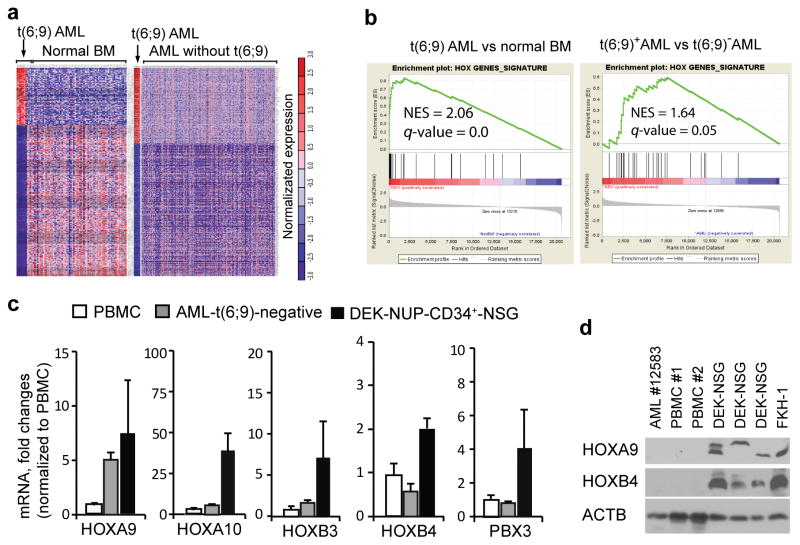
Figure 3. Upregulation of HOXA and B genes in DEK-NUP214 induced human AML in a mouse model.
(a) Gene expression profiling of AML cases with or without the t(6;9) translocation. Three publically available microarray datasets were reanalyzed: GSE17855 20 containing 6 DEK-NUP214 positive and 231 t(6;9)- negative AML cases, GSE15061 21 including 69 normal human bone marrow (BM) and 202 t(6;9)-negative AML, as well as GSE35159 22 which has 12 AML cell lines including the DEK-NUP214 positive FKH-1 cells. The bioinformatics approaches used were described previously.24 Heatmap analysis of gene expression profiling (FDR < 0.05) of t(6;9) AML (n=6) plus the FKH-1 cell line (carrying DEK-NUP214) compared with healthy human bone morrows (BM, n=69) (left panel) and t(6;9)-negative AML (image not drawn to scale). (b) Gene set enrichment analysis (GSEA) shows activation of the HOX transcription factor pathway in t(6;9) AML compared to normal BM and t(6;9)-negative AML (right panel). (c) Real-time RT-PCR with human specific primers shows upregulation of HOXA and B family genes in spleen AML cells from mice engrafted with DEK-NUP214 transduced human CD34+ progenitor cells, compared with healthy human peripheral blood mononuclear cells (PBMC) as well as primary DEK-NUP214 negative AML samples.
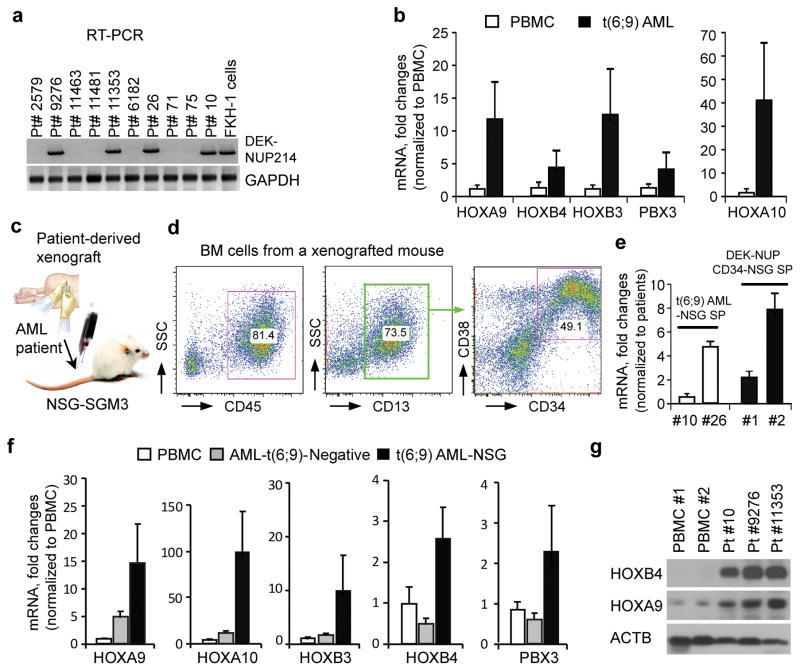
Figure 4.
Upregulation of HOXA and B cluster genes in t(6;9) AML patients as well as in patient-derived mouse xenografts. (a) RT-PCR analysis shows 4 primary AML carrying the DEK-NUP214 fusion transcript. Seventy eight primary AML samples were screened by RT-PCR using primers spanning the DEK-NUP214 fusion gene breakpoint. The FKH-1 cell line is used as a positive control. (b) Quantitative RT-PCR shows mRNA levels of HOXA and B genes in these t(6;9) AML (n=4) compared with healthy mononuclear cells (PBMC, right, n=3). (c) Scheme of the AML patient-derived xenograft model. (d) A typical immunophenotype of bone marrow (BM) cells from a t(6;9) AML engrafted mouse. Flow cytometry analysis was performed using antibodies specifically recognizing human antigens. (e) qPCR analysis of the fusion transcript in xenografts of primary AML and AML arising in the NSG model, show a range of expression levels between different leukemias from both types of AML and the expression levels were comparable. (f) Real-time RT-PCR shows upregulation of HOXA and B family genes in spleen cells from patient-derived xenografted mice (n=4), compared with either normal PBMC or primary DEK-NUP214 negative AML samples.
Among the HOX genes, we focused on HOXA9/4 and HOXB3/4, because they were the most highly upregulated. Using quantitative RT-PCR and Western blot analysis we demonstrated that, consistent with the microarray data, the transcriptional levels of HOXA9/10, HOXB3/4 and PBX3 genes were also highly increased in cells from the diseased mouse spleens compared with either peripheral blood nuclear cells (PBMC) from healthy human samples or xenografted AML cells that do not express the DEK-NUP214 fusion gene (Figure 3c). Western blot analysis, using specific antibodies for human HOXA9 and HOXB4 proteins, showed that protein levels of both genes were also remarkably increased in the DEK-NUP214 transduced mouse model (Figure 3d). However, due to the current non availability of reliable HOXA10, HOXB3 and PBX3 antibodies, we were not able to compare relative protein levels for these genes. Thus, the mouse model we have generated recapitulates the human t(6;9) disease, both in immunophenotype and molecular signature. The HOXA/B genes have been shown to play an important role in the regulation of differentiation, proliferation and self-renewal of hematopoietic stem and progenitor cells 26 as well as in leukemia transformation.27 Therefore, it seems intuitive that abnormal regulation of these genes may play a potential collaborative effect during DEK-NUP214 leukemogenesis.
HOXA and B genes are upregulated in primary t(6;9) AML patients and mouse xenografts
To extend our observations to primary human AML, we used RT-PCR to screen 78 de novo AML samples from both the GRU Cancer Center Tissue Bank and the University of Michigan. We identified four cases expressing the DEK-NUP214 fusion gene (Figure 4a). Using quantitative RT-PCR, we confirmed the high level expression of the HOXA/B genes in these t(6;9) AML, compared with normal PBMC (Figure 4b). For comparison, we then engrafted primary bone marrow cells from each of these four t(6;9) AML samples into NSG-SGM3 mice (Figure 4c), all of which developed AML within 6 months. Flow cytometry analysis of the BM, SP and PB from these mice showed a CD45+CD13+CD34+CD38+ immunophenotype (Figure 4d), which is consistent with the observations from DEK-NUP214 transduced CD34+ cells in the NSG-SGM3 mouse model (Figure 2c). We next compared the relative levels of the DEK-NUP214 mRNA in the primary AML samples and AML from the CD34+ transformation model. QPCR analysis shows a range of expression levels between different primary AML (Figure 4e), which was also seen in the AML from the transformation model, and that relative expression levels across this range were not significantly different (Figure 4e). Consistently, qRT-PCR using primers specific for human HOX genes, also demonstrated higher expression of HOXA and B genes in spleen cells from the primary t(6;9) AML mouse xenografts compared with either normal PBMC or primary DEK-NUP214 negative AML samples (Figure 4f). To determine whether the protein levels of these HOX genes were also increased, we performed Western blot analysis using human-specific antibodies for human HOXA9 and HOXB4 proteins. Consistent with mRNA expression data, Western blot analysis also showed that both HOXA9 and HOXB4 were remarkably increased in the t(6;9) AML xenografts (Figure 4g).
In summary, we have successfully established the first humanized mouse model of t(6;9) AML. Molecular analysis of these diseased mice shows that the same genetic changes were seen in both the AML arising in the mice engrafted with DEK-NUP214 transduced and transplanted CD34+ cells (Figure 2c), and xenografts of primary t(6;9) AML. Thus, the humanized mouse AML model described here, recapitulates the phenotypic and molecular genetic features of primary human t(6;9) AML. This faithful murine model can now be used to evaluate therapeutic strategies for the treatment of DEK-NUP214 disease. Furthermore, our results also suggest that, since aberrant upregulation of HOXA and B genes correlate with DEK-NUP214 expression in primary and experimentally induced AML, these genes may play an important role in the regulation of differentiation, induced proliferation and self-renewal of hematopoietic stem and progenitor cells 26 as well as in leukemia transformation.25, 27
Supplementary Material
Footnotes
CONFLICT INTEREST: The authors declare no conflict of interest.
References
- 1.Soekarman D, von Lindern M, Daenen S, de Jong B, Fonatsch C, Heinze B, et al. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood. 1992;79:2990–2997. [PubMed] [Google Scholar]
- 2.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Molecular and cellular biology. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slovak ML, Gundacker H, Bloomfield CD, Dewald G, Appelbaum FR, Larson RA, et al. A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare 'poor prognosis' myeloid malignancies. Leukemia. 2006;20:1295–1297. doi: 10.1038/sj.leu.2404233. [DOI] [PubMed] [Google Scholar]
- 4.Oyarzo MP, Lin P, Glassman A, Bueso-Ramos CE, Luthra R, Medeiros LJ. Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of flt3 gene mutations. American journal of clinical pathology. 2004;122:348–358. doi: 10.1309/5DGB-59KQ-A527-PD47. [DOI] [PubMed] [Google Scholar]
- 5.Sanden C, Gullberg U. The DEK oncoprotein and its emerging roles in gene regulation. Leukemia. 2015 doi: 10.1038/leu.2015.72. [DOI] [PubMed] [Google Scholar]
- 6.Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Privette Vinnedge LM, Kappes F, Nassar N, Wells SI. Stacking the DEK: from chromatin topology to cancer stem cells. Cell cycle. 2013;12:51–66. doi: 10.4161/cc.23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Privette Vinnedge LM, Ho SM, Wikenheiser-Brokamp KA, Wells SI. The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer. PLoS One. 2012;7:e46985. doi: 10.1371/journal.pone.0046985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ageberg M, Gullberg U, Lindmark A. The involvement of cellular proliferation status in the expression of the human proto-oncogene DEK. Haematologica. 2006;91:268–269. [PubMed] [Google Scholar]
- 11.Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanden C, Ageberg M, Petersson J, Lennartsson A, Gullberg U. Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC cancer. 2013;13:440. doi: 10.1186/1471-2407-13-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oancea C, Ruster B, Henschler R, Puccetti E, Ruthardt M. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia. 2010;24:1910–1919. doi: 10.1038/leu.2010.180. [DOI] [PubMed] [Google Scholar]
- 14.Palacios R, Henson G, Steinmetz M, McKearn JP. Interleukin-3 supports growth of mouse pre-B-cell clones in vitro. Nature. 1984;309:126–131. doi: 10.1038/309126a0. [DOI] [PubMed] [Google Scholar]
- 15.Avanzi GC, Brizzi MF, Giannotti J, Ciarletta A, Yang YC, Pegoraro L, et al. M-07e human leukemic factor-dependent cell line provides a rapid and sensitive bioassay for the human cytokines GM-CSF and IL-3. Journal of cellular physiology. 1990;145:458–464. doi: 10.1002/jcp.1041450310. [DOI] [PubMed] [Google Scholar]
- 16.Ren M, Qin H, Kitamura E, Cowell JK. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood. 2013;122:1007–1016. doi: 10.1182/blood-2013-03-489823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren M, Qin H, Ren R, Cowell JK. Ponatinib suppresses the development of myeloid and lymphoid malignancies associated with FGFR1 abnormalities. Leukemia. 2013;27:32–40. doi: 10.1038/leu.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood. 2011;117:3076–3086. doi: 10.1182/blood-2010-08-301507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamauchi K, Yasuda M. Comparison in treatments of nonleukemic granulocytic sarcoma: report of two cases and a review of 72 cases in the literature. Cancer. 2002;94:1739–1746. doi: 10.1002/cncr.10399. [DOI] [PubMed] [Google Scholar]
- 20.Balgobind BV, Van den Heuvel-Eibrink MM, De Menezes RX, Reinhardt D, Hollink IH, Arentsen-Peters ST, et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica. 2011;96:221–230. doi: 10.3324/haematol.2010.029660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills KI, Kohlmann A, Williams PM, Wieczorek L, Liu WM, Li R, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114:1063–1072. doi: 10.1182/blood-2008-10-187203. [DOI] [PubMed] [Google Scholar]
- 22.Saito Y, Nakahata S, Yamakawa N, Kaneda K, Ichihara E, Suekane A, et al. CD52 as a molecular target for immunotherapy to treat acute myeloid leukemia with high EVI1 expression. Leukemia. 2011;25:921–931. doi: 10.1038/leu.2011.36. [DOI] [PubMed] [Google Scholar]
- 23.Hamaguchi H, Nagata K, Yamamoto K, Fujikawa I, Kobayashi M, Eguchi M. Establishment of a novel human myeloid leukaemia cell line (FKH-1) with t(6;9)(p23;q34) and the expression of dek-can chimaeric transcript. British journal of haematology. 1998;102:1249–1256. doi: 10.1046/j.1365-2141.1998.00900.x. [DOI] [PubMed] [Google Scholar]
- 24.Ren M, Cowell JK. Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease. Blood. 2011;117:6837–6847. doi: 10.1182/blood-2010-07-295725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29:1279–1289. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abramovich C, Humphries RK. Hox regulation of normal and leukemic hematopoietic stem cells. Current opinion in hematology. 2005;12:210–216. doi: 10.1097/01.moh.0000160737.52349.aa. [DOI] [PubMed] [Google Scholar]
- 27.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.