Abstract
We characterized 549 new human leukocyte antigen (HLA) class I and class II alleles found in newly registered stem cell donors as a result of high‐throughput HLA typing. New alleles include 101 HLA‐A, 132 HLA‐B, 105 HLA‐C, 2 HLA‐DRB1, 89 HLA‐DQB1 and 120 HLA‐DPB1 alleles. Mainly, new alleles comprised single nucleotide variations when compared with homologous sequences. We identified nonsynonymous nucleotide mutations in 70.7% of all new alleles, synonymous variations in 26.4% and nonsense substitutions in 2.9% (null alleles). Some new alleles (55, 10.0%) were found multiple times, HLA‐DPB1 alleles being the most frequent among these. Furthermore, as several new alleles were identified in individuals from ethnic minority groups, the relevance of recruiting donors belonging to such groups and the importance of ethnicity data collection in donor centers and registries is highlighted.
Keywords: genetic diversity, hematopoietic stem cell transplantation, human leukocyte antigens, new alleles, sequencing‐based typing, next‐generation sequencing
High‐throughput human leukocyte antigen (HLA) typing of newly registered hematopoietic stem cell donors at high resolution continuously generates a large number of HLA allele sequences which often lead to the identification of new alleles. Recently, the development and use of new sequencing techniques [next‐generation sequencing (NGS)] in the field of HLA typing have also cost‐efficiently facilitated the identification and characterization of these new alleles.
More than 13,000 HLA alleles have been identified so far 1, 2 and many new HLA alleles are likely to be identified. In this work, we describe 549 new HLA class I and class II alleles that were found in potential stem cell donors registered with DKMS donor centers in the United States, Poland and Germany, a total of 101 HLA‐A, 132 HLA‐B, 105 HLA‐C, 2 HLA‐DRB1, 89 HLA‐DQB1 and 120 HLA‐DPB1 alleles (Table S1, Supporting information). These new alleles have been reported to the IMGT/HLA Database and have been included in the monthly nomenclature updates by the World Health Organization (WHO) Nomenclature Committee for factors of the HLA system between June 2009 3 and March 2015 4.
All new HLA alleles were genotyped at the ASHI‐accredited laboratory HistoGenetics (Ossining, NY) using sequencing‐based typing (SBT) 5, 6 and NGS 7 methods. The most homologous equivalents for all new alleles were determined by locus alignment of the DNA sequences from all known HLA alleles cataloged in Release 3.21.0 of the IMGT/HLA Database 2. The definition of most homologous alleles used in this analysis was previously described elsewhere 5, 6.
The identification of DNA sequence variations was performed after comparison of each new HLA allele to its most homologous equivalent (Table 1 and Table S1). In all HLA loci, most new alleles (505, 92.0%) comprised single nucleotide variations. Only 15 new alleles differed in at least three nucleotides from their respective most homologous alleles: HLA‐A*29:48, HLA‐B*40:298 and HLA‐C*15:65 with five nucleotide variations, HLA‐A*02:527, HLA‐B*13:71, HLA‐B*37:40 and HLA‐DQB1*06:168 with four nucleotide variations, and HLA‐A*24:290, HLA‐A*26:68, HLA‐B*13:72, HLA‐B*13:79, HLA‐B*37:55, HLA‐C*05:107, HLA‐DPB1*198:01 and HLA‐DPB1*299:01 with three nucleotide variations (Table S1).
Table 1.
Description of new HLA alleles that were found in at least three potential HSC donors
| HLA locus | New allele | Most homologous allele | NVa | CAb | Codon changec | AA changed | Type of mutation | IRe | Accession no.f |
|---|---|---|---|---|---|---|---|---|---|
| A | A*01:01:10 | A*01:01:01:01 | 1 | 21 | GCG≫GCA | A135A | Synonymous | 3 | FJ898483 |
| C | C*03:221 | C*03:02:01 | 1 | 10 | GAG≫AAG | E55K | Nonsynonymous | 3 | KF220330 |
| C | C*06:02:34 | C*06:02:01:01 | 1 | 14 | CCC≫CCG | P20P | Synonymous | 4 | KC875926 |
| DQB1 | DQB1*02:41 | DQB1*02:01:01 | 1 | 10 | ATG≫ATA | M14I | Nonsynonymous | 3 | KJ190369 |
| DQB1 | DQB1*03:109 | DQB1*03:01:01:01 | 1 | 31 | CTC≫GTC | L87V | Nonsynonymous | 5 | KF220337 |
| DQB1 | DQB1*05:11:02 | DQB1*05:11:01 | 1 | 1 | GCA≫GCG | A38A | Synonymous | 3 | KC959337 |
| DQB1 | DQB1*05:52 | DQB1*05:04 | 1 | 1 | GGG≫AGG | G20R | Nonsynonymous | 5 | KF220336 |
| DQB1 | DQB1*05:69 | DQB1*05:01:01:01 | 2 | 9 | GGG≫AGG | G70R | Nonsynonymous | 3 | KF695009 |
| GCC≫ACC | A71T | Nonsynonymous | |||||||
| DQB1 | DQB1*06:90 | DQB1*06:03:01 | 1 | 4 | GTG≫ATG | V24M | Nonsynonymous | 4 | KC592353 |
| DPB1 | DPB1*173:01 | DPB1*304:01 | 1 | 1 | TTC≫TAC | F35Y | Nonsynonymous | 3 | KF015578 |
| DPB1 | DPB1*177:01 | DPB1*04:01:01:01 | 1 | 8 | GGC≫AGC | G84S | Nonsynonymous | 3 | KF015574 |
| DPB1 | DPB1*178:01 | DPB1*04:01:01:01 | 1 | 8 | ATG≫GTG | M87V | Nonsynonymous | 6 | KC904495 |
| DPB1 | DPB1*182:01 | DPB1*16:01:01 | 1 | 2 | GAG≫GAC | E57D | Nonsynonymous | 7 | KC904489 |
| DPB1 | DPB1*189:01 | DPB1*02:01:02 | 1 | 9 | ATC≫CTC | I65L | Nonsynonymous | 4 | KF015568 |
| DPB1 | DPB1*190:01 | DPB1*04:02:01:01 | 1 | 6 | GAT≫GCT | D55A | Nonsynonymous | 19 | KC603589 |
| DPB1 | DPB1*191:01 | DPB1*02:01:02 | 1 | 7 | CGG≫TGG | R32W | Nonsynonymous | 4 | KC904487 |
| DPB1 | DPB1*201:01 | DPB1*90:01 | 1 | 1 | GCG≫GTG | A36V | Nonsynonymous | 8 | KC875999 |
| DPB1 | DPB1*206:01 | DPB1*13:01:01 | 1 | 4 | ATA≫ATG | I76M | Nonsynonymous | 3 | KC875996 |
| DPB1 | DPB1*208:01 | DPB1*06:01 | 1 | 1 | GTG≫GGG | V42G | Nonsynonymous | 4 | KC603588 |
| DPB1 | DPB1*215:01 | DPB1*04:01:01:01 | 1 | 9 | GAG≫GAC | E57D | Nonsynonymous | 3 | KF036208 |
| DPB1 | DPB1*221:01 | DPB1*03:01:01 | 1 | 5 | AGG≫GGG | R75G | Nonsynonymous | 3 | KF152906 |
| DPB1 | DPB1*222:01 | DPB1*03:01:01 | 1 | 5 | GAG≫GTG | E26V | Nonsynonymous | 4 | KF128972 |
| DPB1 | DPB1*301:01 | DPB1*05:01:01 | 1 | 3 | CGC≫CAC | R91H | Nonsynonymous | 3 | KF882494 |
| DPB1 | DPB1*393:01 | DPB1*13:01:01 | 1 | 3 | GCG≫ACG | A17T | Nonsynonymous | 3 | KF712329 |
HLA, human leukocyte antigen; HSC, hematopoietic stem cell.
NV, number of nucleotide variations between new and homologous allele.
CA, number of complementary alleles. These alleles were defined as those whose DNA sequences showed highest similarity to the new allele's DNA sequence and that showed a maximum number of synonymous substitutions 5.
The codon sequence of the most homologous allele (listed first) is compared with the codon sequence of the respective new allele (listed second). Nucleotide changes are given in bold.
AA change, amino acid change. Numbering starts from the first codon of the mature protein. Amino acid from the most homologous allele (listed first), altered codon number and the compared amino acid of the respective new allele (listed second) are displayed.
IR, number of individuals reported carrying the new allele within the current sample.
GenBank accession number of new allele (Exon 2 and 3 for class I alleles and Exon 2 for class II alleles).
Furthermore, 388 new alleles presented at least one nonsynonymous nucleotide variation followed by 145 new alleles with only synonymous nucleotide variations and 16 new alleles which comprised nonsense mutations (null alleles). Detailed information regarding the number of new alleles per locus and type of mutation is shown in Figure 1. Note that since only two new HLA‐DRB1 alleles were identified, this locus was omitted in Figure 1. The new alleles HLA‐DRB1*07:01:17 and HLA‐DRB1*11:173 showed a synonymous mutation at codon position 77 and a nonsynonymous mutation at codon position 67, respectively (Table S1).
Figure 1.
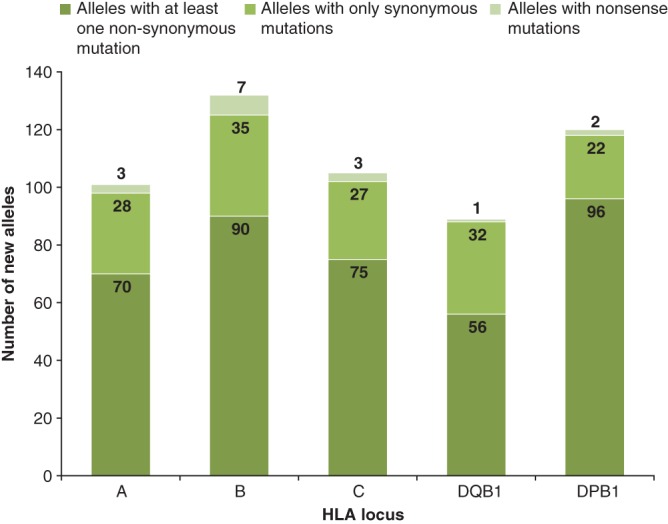
Number of new human leukocyte antigen (HLA) alleles according to type of mutation found after comparison with their respective most homologous alleles. Number of alleles per locus is indicated. Type of mutation (i.e. nonsynonymous, synonymous and nonsense mutations) is color coded.
In new HLA class I alleles, most nucleotide variations were observed in codon positions at the beginning of Exon 3 (positions 91 to 136), notably for HLA‐A alleles most of the nucleotide variations were found between Exon 3 positions 131 to 155. In new HLA class II alleles (HLA‐DQB1 and HLA‐DPB1), the nucleotide variations distributed evenly along Exon 2.
Some new alleles (49, 8.9%) comprised codon alterations that are unique among HLA alleles (Table 2), thus underlining the polymorphic nature of the HLA system. A total of 34 novel alleles presented nonsynonymous mutations introducing new amino acids in the respective codon position, 14 alleles presented synonymous mutations with new DNA codon changes, and one new allele presented a nonsense mutation that leads to a premature stop codon (null allele). Of these variations, 47 were found in 44 DNA sequence positions (11 along Exons 2 and 3 of HLA class I alleles and 33 along Exon 2 of HLA class II alleles) that have not yet been reported as polymorphic.
Table 2.
Newly identified HLA alleles with new nucleotide variations
| Alleles with novel changes | Codon numbera | Regular amino acid | Regular codonb | New amino acid | New codonb | Type of mutation |
|---|---|---|---|---|---|---|
| A*01:01:64 | 83 | Glycine | GGC | Glycine | GGT | Synonymous |
| A*01:165 | 133 | Tryptophan | TGG | Cysteine | TGC | Nonsynonymous |
| A*02:01:112 | 83 | Glycine | GGC | Glycine | GGT | Synonymous |
| B*41:34 | 106 | Aspartic Acid | GAC | Alanine | GCC | Nonsynonymous |
| B*44:02:33 | 92 | Serine | TCT | Serine | TCC | Synonymous |
| B*46:59 | 157 | Arginine | AGA | Lysine | AAA | Nonsynonymous |
| C*04:179 | 16 | Glycine | GGC | Aspartic Acid | GAC | Nonsynonymous |
| C*04:192 | 73 | Threonine | ACT | Aspartic Acid | GAT | Nonsynonymous |
| C*06:02:34 | 20 | Proline | CCC | Proline | CCG | Synonymous |
| C*06:151 | 132 | Serine | TCC | Proline | CCC | Nonsynonymous |
| C*07:334 | 89 | Glutamic Acid | GAG | Glycine | GGG | Nonsynonymous |
| C*07:376 | 23 | Isoleucine | ATC | Threonine | ACC | Nonsynonymous |
| DQB1*02:44 | 64 | Glutamine | CAG | Lysine | AAG | Nonsynonymous |
| DQB1*03:102 | 52 | Proline | CCG | Alanine | GCG | Nonsynonymous |
| DQB1*05:01:15 | 67 | Valine | GTC | Valine | GTT | Synonymous |
| DQB1*05:68 | 43 | Aspartic Acid | GAC | Asparagine | AAC | Nonsynonymous |
| DQB1*06:04:10 | 67 | Valine | GTC | Valine | GTA | Synonymous |
| DPB1*02:01:17 | 52 | Glycine | GGG | Glycine | GGA | Synonymous |
| DPB1*03:01:04 | 49 | Threonine | ACG | Threonine | ACA | Synonymous |
| DPB1*04:01:11 | 84 | Glycine | GGC | Glycine | GGA | Synonymous |
| DPB1*04:01:13 | 25 | Leucine | CTG | Leucine | TTG | Synonymous |
| DPB1*04:01:14 | 15 | Cysteine | TGC | Cysteine | TGT | Synonymous |
| DPB1*04:01:27 | 82 | Glutamic Acid | GAG | Glutamic Acid | GAA | Synonymous |
| DPB1*04:02:05 | 42 | Valine | GTG | Valine | GTT | Synonymous |
| DPB1*05:01:05 | 31 | Asparagine | AAC | Asparagine | AAT | Synonymous |
| DPB1*14:01:02 | 34 | Glutamic Acid | GAG | Glutamic Acid | GAA | Synonymous |
| DPB1*169:01 | 18 | Phenylalanine | TTT | Valine | GTT | Nonsynonymous |
| DPB1*180:01 | 63 | Lysine | AAG | Threonine | ACG | Nonsynonymous |
| DPB1*186:01 | 80 | Asparagine | AAC | Serine | AGC | Nonsynonymous |
| DPB1*187:01 | 77 | Cysteine | TGC | Phenylalanine | TTC | Nonsynonymous |
| DPB1*193:01 | 25 | Leucine | CTG | Valine | GTG | Nonsynonymous |
| DPB1*194:01 | 24 | Phenylalanine | TTC | Leucine | TTG | Nonsynonymous |
| DPB1*195:01 | 22 | Glutamine | CAG | Arginine | CGG | Nonsynonymous |
| DPB1*212:01 | 20 | Glycine | GGG | Arginine | AGG | Nonsynonymous |
| DPB1*216:01N | 78 | Arginine | AGA | Stop | TGA | Nonsense |
| DPB1*222:01 | 26 | Glutamic Acid | GAG | Valine | GTG | Nonsynonymous |
| DPB1*298:01 | 23 | Arginine | CGC | Serine | AGC | Nonsynonymous |
| DPB1*323:01 | 19 | Asparagine | AAT | Serine | AGT | Nonsynonymous |
| DPB1*325:01 | 53 | Arginine | CGG | Tryptophan | TGG | Nonsynonymous |
| DPB1*329:01 | 21 | Threonine | ACA | Isoleucine | ATA | Nonsynonymous |
| DPB1*336:01 | 50 | Glutamic Acid | GAG | Glutamine | CAG | Nonsynonymous |
| DPB1*360:01 | 22 | Glutamine | CAG | Leucine | CTG | Nonsynonymous |
| DPB1*376:01 | 52 | Glycine | GGG | Glutamic Acid | GAG | Nonsynonymous |
| DPB1*404:01 | 25 | Leucine | CTG | Glutamine | CAG | Nonsynonymous |
| DPB1*407:01 | 38 | Phenylalanine | TTC | Leucine | TTA | Nonsynonymous |
| DPB1*420:01 | 81 | Tyrosine | TAC | Cysteine | TGC | Nonsynonymous |
| DPB1*425:01 | 38 | Phenylalanine | TTC | Leucine | TTA | Nonsynonymous |
| DPB1*426:01 | 48 | Valine | GTG | Methionine | ATG | Nonsynonymous |
| DPB1*435:01 | 14 | Glutamic Acid | GAA | Glycine | GGA | Nonsynonymous |
Numbering starts from the first codon of the mature protein.
Codon alterations are printed in bold.
New alleles were found predominately only once (494, 90.0%), yet 55 (10.0%) new alleles were found more often, 12 of which were found more than three times. The most frequently identified new alleles belonged to the HLA‐DPB1 locus: DPB1*190:01 was reported 19 times, DPB1*201:01 8 times, DPB1*182:01 7 times and DPB1*178:01 6 times (Table 1). These alleles are thus likely to be common.
In order to trace the origins of the new HLA alleles, self‐assessed parentage records of the carriers of these new alleles were analyzed (Table S2). As carriers of new alleles are registered with different DKMS donor centers (in the United States, Poland and Germany) that record parentage information differently, the corresponding data were processed separately (Figure 2A, B). In the United States, parentage information is documented along ethnic groups (such as Mediterranean or North American) while in Germany these data are based on nationalities. In Poland no parentage information is recorded.
Figure 2.
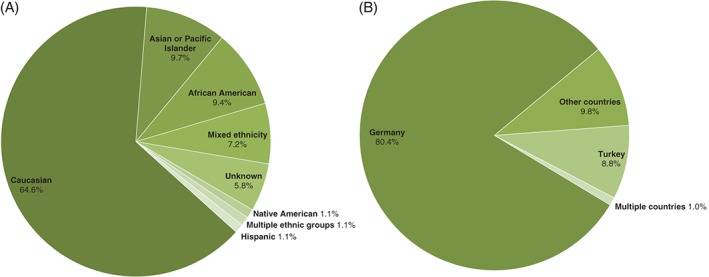
Ethnic origins of registered stem cell donors carrying new alleles based on self‐assessed parentage records. A) Ethnic groups of donors registered in the United States who carry new alleles. The category ‘mixed ethnicity’ refers to individuals who specified at least two ethnic groups. B) Nationalities of donors registered in Germany who carry new alleles. Origins that appeared only once were summarized as ‘other countries’, this includes Austria, Croatia, Colombia, Greece, Iraq, Korea, Luxembourg, Poland, Russia and Spain. The categories ‘multiple ethnic groups’ and ‘multiple countries’ refer to cases when a new allele was observed in individuals from at least two different ethnic groups or countries.
Most new alleles were found in potential donors from the United States (277 alleles, 50.5%) followed by donors from Poland (155 alleles, 28.2%) and Germany (102 alleles, 18.6%) (Table 3). Lastly, 15 (2.7%) new alleles were found in more than one donor center (indicated as ‘≥2 countries’ in Table 3) and hence likely from individuals with different origins. These alleles were excluded from further parentage analysis.
Table 3.
Number of new HLA alleles found in potential HSC donors registered with DKMS donor centers located in the United States, Poland and Germany
| HLA locus | United Statesa | Polanda | Germanya | ≥2 countriesb | Total |
|---|---|---|---|---|---|
| A | 66 | 26 | 9 | — | 101 |
| B | 73 | 41 | 18 | — | 132 |
| C | 56 | 29 | 20 | — | 105 |
| DRB1 | 1 | — | 1 | — | 2 |
| DQB1 | 32 | 26 | 28 | 3 | 89 |
| DPB1 | 49 | 33 | 26 | 12 | 120 |
| Total | 277 | 155 | 102 | 15 | 549 |
HLA, human leukocyte antigen; HSC, hematopoietic stem cell.
Location of DKMS donor centers.
New alleles found in more than one DKMS donor center.
Figure 2 illustrates the self‐assessed parentage information of potential donors from the United States (Figure 2A) and Germany (Figure 2B). To facilitate the comparison between donor centers, the ethnicity data from potential donors registered with DKMS in the United States were combined into broader ethnic groups according to Table S3.
In the United States, more than half of the new alleles (172 alleles, 62.1%) were identified in Caucasians (self‐described as Eastern Europeans, Mediterranean, Middle Eastern, North Americans, Northern Europeans, Other Whites and Western Europeans), followed by 27 new alleles (9.8%) found in individuals with mixed ethnicity. Furthermore, we observed a high percentage of new alleles (63 alleles, 22.7%) in underrepresented ethnic groups: 27 (9.8%) new alleles were identified in individuals of African descent (i.e. African American, Black Caribbean), 25 (9.0%) in Asians or Pacific Islanders (i.e. Chinese, South Asian), 9 (3.2%) in individuals with Native American parentage (i.e. Alaska Native or Aleut, Native American Indian) and 2 (0.7%) in individuals with Hispanic ancestry (i.e. White Caribbean). The remaining new alleles were found in individuals with unknown origin (12 alleles, 4.3%) and in individuals from different ethnic groups (‘multiple ethnic groups’ in Figure 2A; 3 alleles, 1.1%).
In Germany, most new alleles (82, 80.4%) were found in individuals with German parentage. Ten new alleles (9.8%) were found in individuals with nationalities listed only once (‘other countries’ in Figure 2B, including, for example, Greece and Russia), nine (8.8%) in individuals with Turkish parentage and one allele (1.0%) was found in two individuals with different nationalities (‘multiple countries’ in Figure 2B), one donor of German descent and another donor of Turkish descent.
Overall, we observed a larger ethnic diversity in the origin of new alleles in the United States when compared with those new alleles identified in potential donors registered with DKMS in Germany. However, as self‐assessment of ethnicity data is documented differently in both centers, some of the distinct nationalities reported by potential donors in Germany could also include different ethnic groups. The collection of parentage data in any form (i.e. ethnicity, race, geographic ancestry) is important, as this information not only assesses the origin of potential donors but also provides an insight into the new alleles' derivation which might be relevant for optimal HLA matching in hematopoietic stem cell transplantation (HSCT).
Additionally, as previously reported for other new HLA class I 5 and class II 6 alleles, many of the new alleles described here were also often identified in individuals from underrepresented ethnic groups, highlighting once again the importance of global donor recruitment efforts 8 with particular focus on country‐specific ethnic minority groups 9, 10.
To summarize, we characterized 549 new HLA class I and II alleles identified in newly registered DKMS stem cell donors in the United States, Poland, and Germany due to high‐resolution HLA typing at donor recruitment. When compared with their most homologous alleles, these new alleles exhibited mostly single nucleotide variations that lead to nonsynonymous mutations. Specifically, we identified DNA sequence positions in Exons 2 and 3 with novel codon changes for HLA class I and class II alleles which emphasize the high polymorphism of the HLA system. The fact that new alleles are continuously found after years of HLA typing research demonstrates the importance of high‐resolution HLA typing methods. The recent rise of NGS in the HLA field and the use of new approaches such as full‐length gene typing 7, 11, 12, 13 will not only improve the identification of new HLA alleles but also help to discover new features in previously described HLA alleles.
Conflict of interest
The authors have declared no conflicting interests.
Supporting information
Table S1. Description of new HLA alleles. Nucleotide substitutions found after comparison of each new allele's sequence with their most homologous allele
Table S2. Origin and HLA phenotypes of potential stem cell donors carrying new HLA alleles
Table S3. Broad ethnic categories used to classify ethnicity data from potential donors registered with DKMS in the United States
Acknowledgment
The authors would like to thank Rolando Silva‐González (DKMS German Bone Marrow Donor Center) for his contribution to this work.
References
- 1. Marsh SGE, Albert ED, Bodmer WF et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 2010: 75: 291–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015: 43: D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marsh SG. Nomenclature for factors of the HLA system, update June 2009. Tissue Antigens 2009: 74: 364–6. [DOI] [PubMed] [Google Scholar]
- 4. Marsh SG. Nomenclature for factors of the HLA system, update March 2015. Tissue Antigens 2015: 86: 48–52. [DOI] [PubMed] [Google Scholar]
- 5. Hernandez‐Frederick CJ, Giani AS, Cereb N et al. Identification of 2127 new HLA class I alleles in potential stem cell donors from Germany, the United States and Poland. Tissue Antigens 2014: 83: 184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hernandez‐Frederick CJ, Cereb N, Giani AS et al. Three hundred and seventy‐two novel HLA class II alleles identified in potential hematopoietic stem cell donors from Germany, the United States, and Poland. Tissue Antigens 2014: 84: 497–502. [DOI] [PubMed] [Google Scholar]
- 7. Cereb N, Kim HR, Ryu J, Yang SY. Advances in DNA sequencing technologies for high resolution HLA typing. Hum Immunol 2015: 76: 923–927. [DOI] [PubMed] [Google Scholar]
- 8. Schmidt AH, Sauter J, Pingel J, Ehninger G. Toward an optimal global stem cell donor recruitment strategy. PLoS One 2014: 9: e86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt AH, Solloch UV, Baier D et al. Criteria for initiation and evaluation of minority donor programs and application to the example of donors of Turkish descent in Germany. Bone Marrow Transplant 2009: 44: 405–12. [DOI] [PubMed] [Google Scholar]
- 10. Pingel J, Solloch UV, Hofmann JA, Lange V, Ehninger G, Schmidt AH. High‐resolution HLA haplotype frequencies of stem cell donors in Germany with foreign parentage: how can they be used to improve unrelated donor searches? Hum Immunol 2013: 74: 330–40. [DOI] [PubMed] [Google Scholar]
- 11. Lange V, Bohme I, Hofmann J et al. Cost‐efficient high‐throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics 2014: 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erlich HA. HLA typing using next generation sequencing: an overview. Hum Immunol 2015: 76: 887–890. [DOI] [PubMed] [Google Scholar]
- 13. Cereb N, Yang SY. OR14 Characterization of HLA class I new alleles with insertions and deletions in exons and introns. Human immunology 2015: 76: 24, (Abstract). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Description of new HLA alleles. Nucleotide substitutions found after comparison of each new allele's sequence with their most homologous allele
Table S2. Origin and HLA phenotypes of potential stem cell donors carrying new HLA alleles
Table S3. Broad ethnic categories used to classify ethnicity data from potential donors registered with DKMS in the United States


