Summary
Remodeling DNA methylation in mammalian genomes can be global as seen in pre-implantation embryos and primordial germ cells (PGCs), or locus-specific, which can regulate neighboring gene expression. In PGCs global and locus-specific DNA demethylation occur in sequential stages, with an initial global decrease in methylated cytosines (stage I) followed by a Tet methylcytosine dioxygenase (Tet)-dependent decrease in methylated cytosines that act at imprinting control regions (ICRs) and meiotic genes (stage II). The purpose of the two-stage mechanism is unclear. Here we show that Dnmt1 preserves DNA methylation through stage I at ICRs and meiotic gene promoters, and is required for the pericentromeric enrichment of 5hmC. We discovered that the functional consequence of abrogating two-stage DNA demethylation in PGCs was precocious germline differentiation leading to hypogonadism and infertility. Therefore, bypassing stage-specific DNA demethylation has significant consequences for progenitor germ cell differentiation and the ability to transmit DNA from parent to offspring.
eTOC Blurb
Hargan-Calvopina et al. shed light on functional significance of two-stage DNA demethylation – a global decrease followed by locus-specific loss – in primordial germ cells. Dnmt1 preserves DNA methylation in stage I at specific loci, including pericentromeric regions. Abrogating the two-stages results in precocious germline differentiation, leading to hypogonadism and infertility.
Introduction
The germline has one essential role and that is to pass a genome and an epigenome from parent to child. In model organisms such as Drosophila, Xenopus and Zebrafish the germline is considered immortal, created by a process known as preformation. Although the preformation model has some species-specific differences, the unifying event is the allocation of specific RNAs and proteins generated by the oocyte to a small number of cells in the embryo endowing them with germline fate. Embryonic cells that do not inherit these germline-specific molecules become somatic cells (Ephrussi et al., 1991; Strome and Wood, 1983; Wang and Lehmann, 1991).
In mice, and possibly all mammals including humans, germline fate is not preformed. Instead, with each generation the germline is created de novo. Using the mouse as a model it is now accepted that germline cells are specified at around embryonic (E) day E6.25 due to bone morphogenetic signaling from the extra-embryonic ectoderm (Lawson, 1999). Furthermore, elegant epiblast transplantation studies performed almost twenty-years ago revealed that distal epiblast cells transplanted to the proximal region can also yield germline cells, confirming that the specification of the mouse germline is an inductive process (Tam and Zhou, 1996).
The mouse epiblast in vivo is pluripotent and epigenetically dynamic. It is generated from the inner cell mass of the pre-implantation blastocyst between E3.5 and E4.5, and continues to exist up until the end of gastrulation. Using single-base resolution bisulfite sequencing to accurately measure DNA methylation, it is now appreciated that the mouse E3.5 inner cell mass cells are globally demethylated (Smith et al., 2012). The mechanism leading to this highly demethylated state begins at fertilization through a combination of Tet dioxygenase 3 (Tet3) driven demethylation of the paternal genome in the fertilized egg, and replication coupled DNA demethylation (Guo et al., 2014; Peat et al., 2014; Shen et al., 2014). The hypomethylated landscape of the inner cell mass cells is then rapidly reversed by de novo DNA methylation leading to the creation of the highly methylated primed epiblast in post-implantation embryos at E5.5.
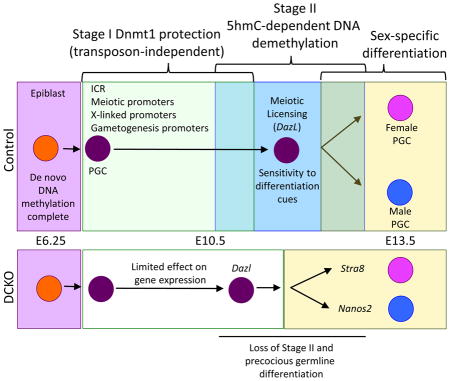
Specification of the mouse germline occurs in the epiblast at E6.25, yielding a highly methylated population of around forty pioneering germline cells at E7.25 called primordial germ cells (PGCs) (Saitou and Yamaji, 2012). Using immunofluorescence it was proposed that the germline passively loses DNA methylation between E8.0-E13.5 (Seki et al., 2005). However using single-base resolution genome-wide sequencing, a new model for PGC demethylation was proposed suggesting that PGC demethylation proceeded in two-stages (Kobayashi et al., 2013; Seisenberger et al., 2012; Vincent et al., 2013). For example, starting from E8.0 as the PGCs migrate from the base of the allantois into the hind-gut endoderm, the PGCs globally lose DNA methylation to around 50% of the levels observed in the epiblast (Seisenberger et al., 2012). This event is referred to as stage I DNA demethylation and is accompanied by repression of the protein Ubiquitin-Like with PHD and Ring Finger Domains 1 (Uhrf1) as well as repression of the de novo DNA methyltransferases (Dnmt3a and Dnmt3b) (Kagiwada et al., 2013). Stage I DNA demethylation is independent from the activity of Tet1 and Tet2 (Vincent et al., 2013). Then starting from E10.5, PGCs undergo locus-specific DNA dimethylation referred to as stage II, which involves the activity of Tet1 and Tet2 as well as DNA replication to create a hypomethylated germline epigenetic ground state at E13.5 (Dawlaty et al., 2013; Hackett et al., 2013; Hajkova et al., 2008; Kagiwada et al., 2013; Seisenberger et al., 2012; Yamaguchi et al., 2013b).
At the conclusion of stage I, the genomic sites protected from global DNA methylation include imprinting control regions (ICRs), endogenous retrovirus called Intracisternal A particles (IAPs) and the promoters of genes involved in meiosis and gamete generation (Seisenberger et al., 2012). The purpose of protecting these loci during stage I, only to be demethylated three days later is unclear. Similarly the enzyme/s responsible for protecting these sites are also unknown. It could be rationalized that the enzyme DNMT1 is responsible for protecting ICRs and meiotic gene promoters through stage I as it is the only DNA methyltransferase expressed throughout the PGC period (Kagiwada et al., 2013). Furthermore, a Dnmt1 knockout causes up-regulation of post migratory germ cell-specific genes in the somatic cells of the post-implantation embryo, (Maatouk et al., 2006), however its role specifically in regulating the two-stage PGC demethylation process is unknown.
Therefore, in order to determine the role of DNMT1 in protecting PGCs from precocious DNA demethylation in stage I, we created a conditional Dnmt1 deletion using Cre-LoxP recombination in PGCs using Blimp1-Cre. We discovered that DNMT1 maintains methylation at imprinting control regions (ICRs) and the endogenous retrovirus called Intracisternal A particle (IAP), and that loss of Dnmt1 leads to significantly fewer PGCs at the conclusion of stage II DNA demethylation due to precocious germline differentiation into oocytes and prospermatogonia in males and females respectively. Together, our data demonstrate that a small number of highly potent loci in mouse PGCs are directly regulated by Dnmt1, and removing Dnmt1 prematurely leads to precocious germline differentiation between E11.5–E13.5. The outcome of our work has important implications for gene regulation during cell lineage differentiation and illustrates the importance of commissioning stage-wise epigenetic events in order to maximize developmental potential.
Results
A conditional deletion of DNMT1 in PGCs leads to loss of 5mC and 5hmC prior to E13.5
Dnmt1 mutant embryos die before mid-gestation due to abnormal embryo development and differentiation (Li et al., 1992). Therefore, to determine whether DNMT1 has a role in PGC formation we used a mouse strain called Dnmt12lox (Jackson-Grusby et al., 2001) and crossed the strain to Blimp1-Cre (BC) to promote a Dnmt1 deletion at the time of PGC specification (Ohinata et al., 2005; Ohinata et al., 2008). In previous studies we found that recombination efficiency of BC in PGCs at E9.0 is 85%, and at E11.5 it is 100% (Li et al., 2015).
To determine whether DNMT1 is required for PGC development before E9.5, we created BC:Dnmt1fl/− conditional knockout (DCKO) mice and performed whole mount immunofluorescence at E9.5 to identify OCT4+ PGCs in the hind-gut endoderm (Figure S1A). We found that DCKO embryos are indistinguishable from controls (+:Dnmt12lox) at this stage of development. To quantify PGC number, we crossed Dnmt12lox to Oct4-GFP (Lengner et al., 2007), and bred the double homozygotes to BC:Dnmt1fl/+. PGCs were sorted from individual embryos at E10.5 by isolating the GFP+ cells with fluorescent activated cell sorting (FACS) (Figure S1B and Figure 1A). Using this approach, we found no difference in the average number of PGCs per embryo at E10.5 either.
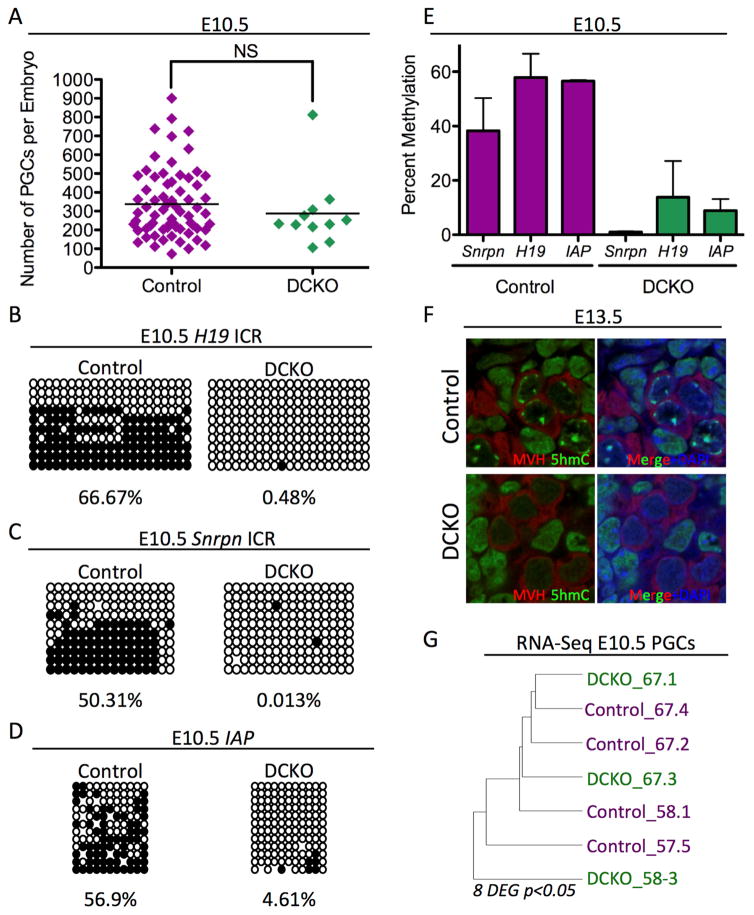
Figure 1. Dnmt1 Conditional knockout PGCs are hypomethylated.
(A) Total umber of PGCs sorted at E10.5. Error bars, ± SEM; NS: p=0.3986, two-tailed unpaired t test; n =78 biological replicates. (B–D) Bisulfite PCR of sorted PGCs from Control and DCKO embryos at E10.5 evaluating the H19 ICR (B), Snrpn ICR (C) and IAP EZ (D). For each locus at least 20 clones were sequenced. (E) Graphical representation of Bisulfite PCR methylation levels found in respective loci from cells at E10.5 n=2 biological replicates. (F) Immunofluorescence of E13.5 DCKO and Control male gonads evaluating MVH positive PGCs (red), 5hmC (green), and DAPI (blue) (n=3). (G) Unsupervised hierarchical clustering RNA-Seq analysis of PGCs at E10.5. p<0.05 and FDR of 10%. DEG = Differentially expressed genes. See also Figure S1
At E10.5, PGCs are exiting the hind-gut and initiating stage II locus-specific DNA demethylation (Hargan-Calvopina et al., 2015; Seisenberger et al., 2012). In wild type PGCs, whole-genome DNA methylation levels are quantified as being on average ~25–30% of CpGs with retained methylation particularly at ICRs, IAPs and non-CG island (non-CGI) promoters (Kobayashi et al., 2013; Seisenberger et al., 2012). To determine whether DNMT1 is required for maintaining methylation at these sites, we isolated genomic DNA from GFP+ PGCs isolated by FACS at E10.5, and performed bisulfite-conversion (BS) followed by polymerase chain reaction (PCR) and sequencing for the H19 and Snrpn ICRs, as well as IAP (Figure 1B–D). Consistent with previous reports (Hajkova et al., 2002), we discovered that control PGCs were methylated at these three genomic locations at E10.5. In contrast, all three loci were severely hypomethylated in the DCKO sorted PGCs (Figure 1B–D, and quantified in Figure 1E). Therefore, our results demonstrate that DNMT1 functions to maintain methylation at discreet regions of the PGC genome up to E10.5.
As PGCs enter the genital ridge at E10.5, their pericentromeric heterochromatin becomes uniquely enriched in 5hmC (Hargan-Calvopina et al., 2015; Yamaguchi et al., 2013a). Previous work has shown that DNMT3b is primarily responsible for DNA methylation at pericentromeric regions in mouse embryonic stem cells (Okano, 1999), yet Dnmt3b is not expressed in PGCs. To determine whether Dnmt1 functions upstream of the pericentromeric enrichment of 5hmC by TETs, we performed immunofluorescence at E13.5 using an antibody that recognizes 5hmC and a germ cell specific protein called mouse vasa homologue (MVH). As expected, control PGCs exhibit a punctate 5hmC staining pattern unique to PGCs. In contrast, DCKO PGCs are significantly depleted in 5hmC foci (Figure 1F, quantified in Figure S1C), with no 5hmC enrichment at E10.5 either (Figure S1D). To determine whether loss of 5hmC affects the deposition of histone H3 lysine 9 trimethylation (H3K9me3), a marker of pericentromeric heterochromatin, we co-stained female and male E13.5 gonads for the germline marker MVH together with H3K9me3 (Figure S1E and S1F). Our results show that H3K9me3 localization in female and male DCKO PGCs is indistinguishable from controls. Therefore deleting Dnmt1 does not affect H3K9me3 enrichment of pericentromeric heterochromatin in PGCs, despite the severe depletion of 5hmC.
To determine how loss of DNA methylation at E10.5 affects gene expression we performed RNA-Sequencing of Oct4-GFP+ PGCs isolated by FACS at E10.5 (Figure 1G). Using unsupervised hierarchical clustering we found that control and DCKO PGCs did not cluster as distinct groups, suggesting that the transcriptional program between mutant and control PGCs is almost identical. To identify differentially expressed genes (DEGs) we compared n=4 control and n=3 DCKO PGC replicates and identified eight differentially expressed genes (DEGs) with ≥2 fold change in gene expression. The only repressed gene in the DCKO PGCs was Dnmt1. The seven up-regulated genes (Zfp598, Vps37c, Slc25a27, Prnp, Cyp51, Chtf18, 4930470H14Rik) were not specific to any particular biological process, and notably do not correspond to known imprinted genes. Taken together, deleting Dnmt1 in PGCs prior to E10.5 causes almost no impact on gene expression, but instead has a significant effect on DNA methylation including the deposition of 5hmC.
Loss of Dnmt1 leads to precocious activation of the meiotic program in female germ cells
In order to determine whether Dnmt1 has any role in PGC development, we used FACS to isolate Oct4-GFP+ PGCs from female embryos at E13.5 (Figure 2A). Unlike at E10.5 where PGC number in DCKO e, brups was similar to controls, we discovered that the DCKO female gonads had significantly fewer PGCs at E13.5 (Figure 2A). One hypothesis for this decrease in PGC number is failed repression of retrotransposons such as IAP, similar to what was previously reported for the somatic cells of E9.5 Dnmt1 knockout mice (Walsh et al., 1998). To address this, we performed immunofluorescence for IAP as well as a second class of retrotransposons called long interspersed nuclear elements (L1) using an antibody that recognizes open reading frame 1p (ORF1p) (Figure S2A and S2B) (Pastor et al., 2014). We found that both IAP and L1ORF1p are repressed in DCKO PGCs similar to controls. Therefore, the reduction in PGC number at E13.5 is most likely not caused by the de-repression of retrotransposons.
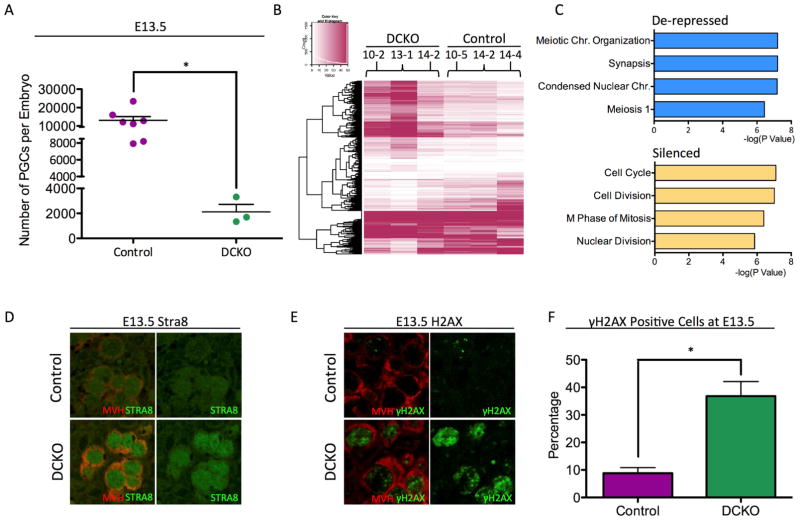
Figure 2. Dnmt1 conditional knockout female PGCs precociously turn on the meiotic program at E13.5.
(A) Total number of Control and DCKO female PGCs sorted at E13.5. Error bars, ± SEM; p=0.0092, two-tailed unpaired t test; n =10 biological replicates. (B) Heat map showing differentially expressed genes (DEGs) between DCKO and control PGCs (C) Gene ontology analysis of de-repressed and silenced genes in E13.5 DCKO female PGCs using DAVID Bioinformatics Resources 6.7. (D) Immunofluorescence of Control and DCKO E13.5 female gonads for MVH (red), and STRA8 (green) (n=3). (E) Immunofluorescence of Control and DCKO E13.5 female gonads for MVH (red), and gamma H2aX (green) (n=3). (F) Quantification of γH2AX foci in Control and DCKO PGCs (MVH) at E13.5. Error bars, ± SEM; p=0.0079, two-tailed Unpaired t test; n =3 biological replicates. FPKM stands for Fragments Per Kilobase of transcript per Million mapped reads. See also Figure S2, and Table S3
To identify the cause of fewer PGCs in DCKO embryonic ovaries at E13.5 we performed RNA-Seq of Oct4-GFP+ PGCs at E13.5 (n=3 biological replicates of each) and identified statistically significant DEG with ≥2 fold change in gene expression. In total we discovered 1,307 DEGs, with 863 genes de-repressed (expressed at higher levels) in the DCKO PGCs, and 442 genes that were silenced (Figure 2B, Table S3). Using gene ontology analysis we discovered genes involved in meiotic prophase I were significantly enriched in the DCKO PGCs (Figure 2C). In contrast, genes involved in cell division, and mitosis were silenced (Figure 2C). This result suggests that DCKO PGCs are exiting mitosis and entering meiosis earlier than controls.
One of the de-repressed genes in the DCKO PGCs is called Stimulated by retinoic acid 8 (Stra8), which induces female germ cells to enter meiosis at E14.5 (Anderson et al., 2008; Baltus et al., 2006; Bowles et al., 2006). To confirm that female germline cells are precociously entering meiosis, we performed immunofluorescence for STRA8 and discovered that STRA8 protein was more often detected in DCKO MVH+ PGCs at E13.5 compared to controls (Figure 1D). As an alternate approach, we evaluated the percentage of MVH+ PGCs that were also positive for gamma H2AX, which marks meiotic double strand breaks (Anderson et al., 2008). Our results show that almost 40% of PGCs in the DCKO mutant ovaries are positive for gamma H2AX, whereas less than 10% of control PGCs are positive for this marker (Figure 2E and quantified in Figure 2F). Taken together, our studies suggest that DNMT1 functions in female PGCs to desensitize precocious entrance into the meiotic program and the formation of double strand breaks.
The promoters of meiotic genes are methylated at E9.5 in wild type PGCs
To determine whether any of the de-repressed genes at E13.5 are methylated at an earlier time point in PGCs, we utilized the whole genome bisulfite sequencing (WGBS) data-set of PGCs sorted from wild-type embryos at E9.5, E10.5, E11.5 and E13.5 by FACS (Seisenberger et al., 2012). Of the 863 de-repressed genes covered in the WGBS data set (Seisenberger et al., 2012), we identified 96 gene promoters with ≥ 20% average methylation at E9.5 (Figure 3A, Table S3), suggesting that these promoters may be direct targets of DNMT1. Heatmap analysis of these 96 promoters shows that they mostly retain methylation in PGCs through to E11.5, before becoming almost completely demethylated by E13.5 (Figure 3B).
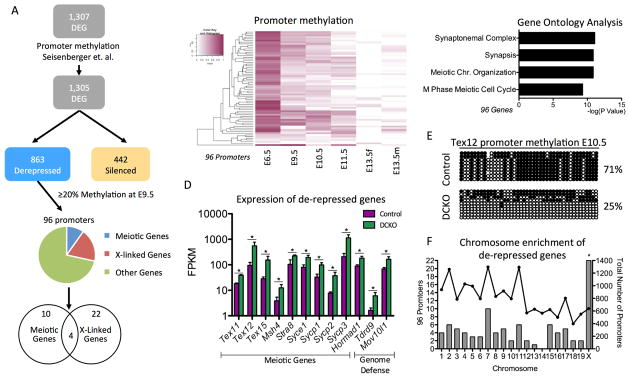
Figure 3. Meiotic genes regulated by DNMT1 have promoter methylation at E9.5.
(A) Analysis pipeline of DEGs in female PGCs at E13.5 to determine whether any de-repressed genes had promoter methylation of ≥ 20% in wild-type E9.5 PGCs (B) Heat map showing promoter methylation of the 96 DEG at E13.5 with promoter methylation of ≥ 20% at E9.5. (C) Gene ontology analysis of the 96 genes de-repressed genes with promoter methylation ≥ 20% at E9.5. (D) Examples of de-repressed genes at E13.5 with promoter methylation of ≥ 20% at E9.5. The genes with ≥ 2-fold difference in expression and FDR < 5% were considered differentially expressed. (E) Bisulfite PCR of the Tex12 promoter comparing Control and DCKO PGCs at E10.5 (F). Hypergeometric test to determine the significance of the number of methylated promoters per chromosome. Left axis: Number of de-repressed genes in the chromosome with promoter methylation of ≥ 20% at E9.5 (Bars). Right axis: Number of genes in the chromosome with E9.5 promoter methylation available (Lines). Hypergeometric test to evaluate the enrichment of the methylated and de-repressed genes in each chromosome. * Refers to statistical significance of hypergeometric test. FPKM stands for Fragments Per Kilobase of transcript per Million mapped reads. See also Table S1.
Similar to the RNA-Seq data set, gene ontology analysis of the 96 de-repressed genes with promoter methylation in wild-type PGCs at E9.5 revealed a significant enrichment in functional categories of meiosis (Figure 3C), including Stra8, testis expressed (Tex) genes, and genes that encode for critical components of the synaptonemal protein complex (Sycp) (Figure 3D). We also identified two genome-defense genes, Mov10l1 and Tdrd9 that exhibit promoter methylation in wild-type PGCs at E9.5, and are also de-repressed in DCKO female PGCs at E13.5 (Figure 3D). To confirm that one of the meiotic promoters was methylated at E10.5, we performed BS-PCR for the Tex12 promoter in GFP+ PGCs isolated by FACS at E10.5. This experiment revealed that the Tex12 promoter is methylated in control PGCs, whereas the DCKO PGCs are demethylated (Figure 3E).
A second category of genes with ≥20% promoter methylation that were de-repressed at E13.5 in DCKO PGCs, were X-linked genes (Figure 3A). To probe this further, we performed a hypergeometric significance test taking into account all promoters at E9.5 from the Seisenberger data set and discovered that the 96 DEG that are methylated in wild type PGCs at E9.5, yet de-repressed in DCKO PGCs at E13.5 are significantly enriched in X-linked genes (Figure 3F). Taken together, we discovered that DNMT1 safeguards precocious expression of meiotic and X-linked genes in female PGCs between E11.5–E13.5 (Supplemental Table 1).
Loss of Dnmt1 leads to precocious differentiation in male germ cells
Initially, it was hypothesized the XY and XX germ cells are intrinsically programmed to enter meiosis (McLaren and Southee, 1997). However, more recent studies revealed that retinoic acid secreted from the mesonephros as well as retinoic acid produced within the ovary induces the meiotic program in female PGCs. In contrast, male PGCs are protected from retinoic acid-induced meiosis due to sertoli cell expression of Cyp26b1 (Bowles et al., 2006; Mu et al., 2013). To determine whether male PGCs precociously activate the meiotic program in the absence of DNMT1, we used FACS to isolate Oct4-GFP+ PGCs from male embryos at E13.5 (Figure 4A). Similar to female DCKO PGCs, we discovered significantly fewer male DCKO PGCs in the testis relative to controls, and IAP and L1ORF1p were not expressed (Figure S3D and S3E).
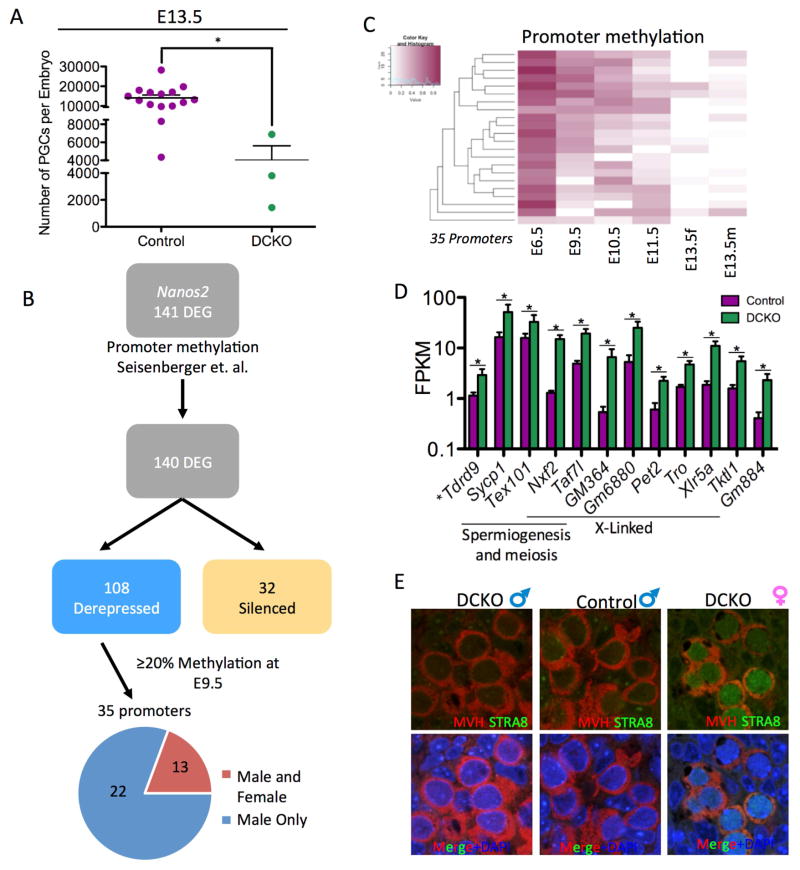
Figure 4. Dnmt1 conditional knockout male PGCs precociously activate the male differentiation pathway.
(A) Total number of male PGCs sorted at E13.5. Error bars, ± SEM; p=0.0274, two-tailed Unpaired t test; n =18 biological replicates. (B) Analysis pipeline to determine which of the de-repressed RNAs at E13.5 had ≥ 20% DNA methylation in wild-type PGCs at E9.5. (C) Heat map showing the methylation status of the 35 de-repressed DEG at E13.5 with promoter methylation at E9.5. (D) Expression levels of de-repressed in common to males and female DCKO PGCs that also had promoter methylation of ≥ 20% at E9.5. FPKM stands for Fragments Per Kilobase of transcript per Million mapped reads. The genes with ≥ 2-fold difference in expression and FDR < 5% were considered differentially expressed. (E) Immunofluorescence of E13.5 male gonads for MVH (red), and STRA8 (green). Included on the right is a positive control for STRA8 staining using an E13.5 DCKO female. See also Figure S3, Table S2, and Table S4.
To identify DEGs in DCKO male PGCs, we performed RNA-Seq of the Oct4-GFP+ DCKO and control PGC population, and identified 141 statistically significant DEGs in male PGCs including the gene Nanos2, an RNA binding protein responsible for initiating prospermatogonia differentiation after E13.5 (Suzuki and Saga, 2008). Specifically, we found that similar to females the majority of DEGs (109) were de-repressed, whereas only 32 genes were silenced (Figure 4B, Table S4). Notably, only the meiotic gene Sycp1 was de-repressed whereas all other meiotic prophase I genes identified in female PGCs were unaffected suggesting that the male PGCs are most likely not entering meiosis in the absence of DNMT1.
To determine whether any of the de-repressed genes identified at E13.5 in male PGCs were methylated at E9.5 in wild type germ cells, we again examined the Seisenberger WGBS data set (Seisenberger et al., 2012). We discovered that of the 108 de-repressed genes with available promoter methylation, thirty-five exhibited ≥ 20% promoter methylation at E9.5 (Figure 4C). When comparing male and female PGCs, we discovered a small number of DEG with methylated promoters in common including meiotic genes such as Tex101, Sycp1, Taf7l, the genome defense gene Tdrd9, as well as several X-linked genes (Figure 4D, Supplemental Table 2). Genes that were unique to male DCKO PGCs included the genome-defense gene Piwil4, and a male meiotic gene Gtsf1 (Figure S3A, Supplemental Table 2). Similar to female PGCs, the 35 genes that were de-repressed at E13.5 in male DCKO PGCs and were methylated at E9.5 in wild type cells, are also enriched in X-linked genes (Figure S3B). Notably, we also discovered that DNMT1 is responsible for repressing XIST expression in male PGCs given that it was de-repressed in our RNA-Seq data set. In order to verify that E13.5 male DCKO PGCs are not entering meiosis, immunofluorescence was used to assay for STRA8 expression in male control and DCKO PGCs. Female DCKO PGCs were included as a positive control (Figure 4E). Our results show that male control and DCKO PGCs lack detectable levels of STRA8 at E13.5, and therefore are not entering meiosis.
Loss of Dnmt1 leads to the de-repression of Dazl at E11.5
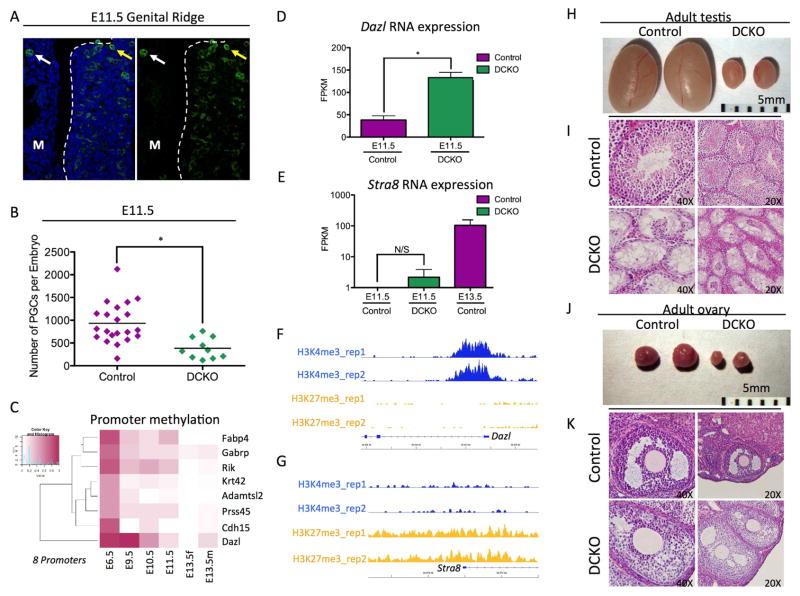
Due to the loss of PGCs and the de-repression of meiotic genes at E13.5, we next sought to determine whether the meiotic program was initiated in female and male PGCs prior to sex-determination at E11.5. At this stage the majority of PGCs have colonized the genital ridge (Figure 5A). Unlike at E10.5 where PGC number is equivalent in control and DCKO embryos, we found that DCKO embryos had around half the number of PGCs compared to controls. To determine whether there were any transcriptional differences between DCKO and control PGCs at this stage in development we performed RNA-Seq of GFP+ PGCs isolated by FACS from individual DCKO and control embryos (Table S5). In total we discovered 43 DEGs with ≥ 2-fold change in gene expression with 24 genes expressed at higher levels in the DCKO PGCs, and 19 genes that were silenced. Of the 24 de-repressed genes we identified 8 with ≥ 20% average promoter methylation at E9.5 in wild type PGCs (Figure 5C). Of the eight genes identified in this analysis, only one was germ cell specific, a gene called Deleted in azoospermia like (Dazl) (Figure 5D). Dazl is critical for both male and female PGC formation where it functions as a licensing factor for meiosis, and represses pluripotent gene expression enabling male and female germ cell differentiation (Haston et al., 2009; Lin et al., 2008; Ruggiu et al., 1997; Schrans-Stassen et al., 2001).
Figure 5. Dazl is de-repressed at E11.5 in mutant PGCs.
(A) Immunofluorescence of an E11.5 Genital ridge for SSEA1+ PGCs (green) and DAPI (blue). M refers to the mesonephros. White arrows point to a PGC in the mesonephors, the yellow arrows point to a PGCs that has colonized the genital ridge. (B) Total number of germ cells sorted at E11.5. Error bars, ± SEM; p=0.0009, two-tailed Unpaired t test; n =31 biological replicates. (C) Heat map showing promoter methylation of 8 DEG at E11.5 with promoter methylation of ≥ 20% at E9.5. (D) Dazl expression levels in E11.5 Control and DCKO PGCs. Genes with ≥ 2-fold difference in expression and FDR < 10% were considered differentially expressed. (E) Stra8 expression levels in E11.5 Control and DCKO PGCs. E13.5 PGCs are included as a control. Genes with ≥ 2-fold difference in expression and FDR < 10% were considered differentially expressed. (F, G) H3K27me3 and H3K4me3 peaks at the Dazl locus (F) and Stra8 locus (G) at E11.5. (H) Control (left) and DCKO (right) testes from 6 month male mice (n=1). (I) Histology of Adult testis comparing control (top) and DCKO (bottom) (n=1). (J) Control (left) and DCKO (right) ovaries from 6 month female mice (n=1) (K) Histology of adult ovary comparing control (top) and DCKO (bottom) (n=1). See also Table S5.
Given that Dazl was significantly up-regulated in DCKO PGCs at E11.5, yet Stra8, the critical master regulator of meiosis was not (Figure 5D and Figure 5E), we next evaluated the chromatin state at the 5’ end of the Dazl and Stra8 genes using the previously published ChIP-Seq reference map of PGCs at E11.5 (Sachs et al., 2013) (Figure 5F and Figure 5G). Analysis of the ChIP-Seq data revealed that the first exon and intron of Dazl gene is highly enriched in the active histone H3K4me3 mark, where as H3K27me3 is absent. In contrast the Stra8 promoter is enriched in the repressive histone mark H3K27me3 with very little enrichment of H3K4me3. Therefore, we propose that loss of DNA methylation at E11.5 does not have an immediate impact on Stra8 expression due to the presence of H3K27me3 at the promoter and within the gene body, whereas Dazl is sensitized to changes in DNA methylation on account of being depleted of H3K27me3 and instead enriched in H3K4me3.
Given the reduced number of PGCs in male and female gonads at E13.5, we finally determined the outcome on adult mice at six months of age. We discovered that deleting Dnmt1 from the time of PGC specification leads to hypogonadism in DCKO mutant mice compared to litter mate controls (Figure 5H and 5J). Using histology we show that the adult testis of DCKO mice are devoid of germ cells (Figure 1I and Figure S3F). However adult ovaries contain oocytes that are capable of being recruited into folliculogenesis, although the meiotic potential of these oocytes is unknown (Figure 5K).
Discussion
In this study we discovered that DNMT1 regulates expression of the meiotic licensing and germ cell-specific differentiation gene DazL in PGCs. We show that DNMT1 functions to prevent precocious meiosis in female PGCs, and prospermatogonia differentiation in male PGCs. We also show that DNMT1 is essential for maintaining DNA methylation at ICRs and meiotic promoters prior to E10.5, and is responsible for maintaining the 5mC substrate for conversion to 5hmC between E9.5 and E13.5. Finally, we show that DNMT1 is not required for transposon repression during PGC development. Results from this work have important implications for our understanding of the role of DNA methylation during the earliest stages of germline differentiation, as well as a potential role for DNMT1 in maintaining methylation in the absence of UHRF1. Future work should be aimed at determining whether DNMT1 functions independently from UHRF1 in PGCs to safeguard PGCs from precocious differentiation.
Ever since the discovery of the two-stage DNA demethylation process in PGCs it has been unclear why mammals don’t simply deploy DNA demethylation in a single passive step as was originally hypothesized twenty years ago. Our study indicates that the two-stage mechanism functions to desensitize the progenitor pool of PGCs from sex-specific differentiation cues in the gonad and to prevent precocious differentiation. We propose that mechanistically, the stage I of DNA demethylation and therefore locus-specific protection from DNA demethylation may involve mechanisms analogous to the ones that function in the pre-implantation embryo upstream of DNMT1 and the oocyte (o)-specific version called DNMT1o (Hirasawa et al., 2008; Howell et al., 2001). Notably, protection of ICRs in pre-implantation embryos involves the activities of TRIM28, SETDB1, ZFP57, DPPA3 and H3K9me2/3 (Li et al., 1998; Messerschmidt et al., 2012; Nakamura et al., 2012). A conditional deletion of Setdb1 in PGCs using Tissue non-specific alkaline phosphatase (Tnap)-Cre demonstrates an important role for H3K9me3 in repressing endogenous retroviruses (ERVs) and consequently the maintenance of DNA methylation at these sites, however Setdb1 does not appear to regulate meiotic gene expression (Liu et al., 2015). In contrast, indirect evidence in ESCs reveals that the late demethylating CGI promoters discovered in PGCs are enriched in Zfp57 binding sites and therefore may be regulated by H3K9me2 (Seisenberger 2012). Finally, evaluating methylation levels at ICRs in Dppa3 mutant PGCs identifies a modest role for DPPA3 in protecting ICRs, but this effect is very mild compared to DPPA3’s activity in pre-implantation embryos (Nakashima et al. 2013). Taken together, although the global levels of DNA methylation in pre-implantation embryos and PGCs at the end of stage I are similar, we propose that the underlying chromatin landscape in hypomethylated PGCs is different from pre-implantation embryos with the emergence of new protected sites such as meiotic and germ cell differentiation genes including Dazl and Stra8. In future studies it will be important to better understand the chromatin landscape of PGCs prior to E10.5, particularly as gene regulation in PGCs up to E10.5 is mostly independent of promoter DNA methylation status, similar to what has been proposed in human PGCs (Gkountela et al., 2015).
In female mice, Stra8 RNA is expressed from E12.5 due to retinoic acid signaling from the neighboring mesonephros and coordinated loss of the Polycomb Repressive Complex 1 enzyme Rnf2 and H3K27me3 from the Stra8 promoter (Yokobayashi et al., 2013). In males, Stra8 is not expressed at E12.5 due to the creation of testis cords (tubules), which express Cyp26b1 to protect male PGCs from the retinoic acid inducing signal (Bowles et al., 2006; Brennan et al., 2002; Hacker et al., 1995; Jeske et al., 1995; MacLean et al., 2007). Here, we show that repression of Stra8 in male PGCs up to E11.5 is not the role of Dnmt1, and instead loss of Dnmt1 sensitizes female PGCs to meiotic entry in the ovarian niche, possibly through the up regulation of Dazl, which licenses germ cells to undergo meiosis. In future studies, it will be interesting to determine whether over riding the meiotic switch in DCKO female PGCs by modulating cell cycle genes is sufficient to rescue PGC number.
Another critical finding in our study was the de-repression of X-linked genes in both male and female PGCs with a conditional deletion in Dnmt1. X chromosome reactivation in female PGCs begins soon after PGC specification, with loss of Xist and loss of H3K27me3 enrichment from the inactive X chromosome (Chuva de Sousa Lopes et al., 2008). This aligns chromosome-wide remodeling of the X-chromosome with the timing of stage I global DNA demethylation. In male germline cells, Xist is expressed after birth in pro-spermatogonia and during the earliest stages of meiosis, however it is not involved in male meiotic sex-chromosome inactivation (McCarrey et al., 2002; McCarrey and Dilworth, 1992). Given that a deletion in Dnmt1 caused increased X-linked gene expression in both male and female PGCs, this phenomenon is most likely not a consequence of X-reactivation in males, although it may accelerate X-reactivation in females on account of DNA methylation being retained at many X-linked genes until E11.5 (Sugimoto and Abe, 2007). Instead, our results are consistent with studies in embryonic stem cells demonstrating that repression of X-linked genes in males can be attributed to locus-specific effects on X-linked gene expression which are independent of X chromosome inactivation (Oda et al., 2006).
In summary, assigning Dnmt1 the task of simultaneously protecting and marking the meiotic program in PGCs provides a relatively simple and reversible approach for preventing PGCs from activating the meiotic program until the gonadal niche can instruct male or female fate from a gender-neutral position. In females, demethylation of the meiotic program by Tet1 and Tet2 is required for meiotic entrance through a 5hmC intermediate (Dawlaty et al., 2013; Yamaguchi et al., 2012). However, our results show that 5hmC is no longer enriched in DCKO PGCs indicating that 5hmC itself is not required for meiosis. Instead, our data indicate that the role of Dnmt1, 5mC and 5hmC in the mammalian germline is to facilitate DNA demethylation and meiosis at the appropriate time. This stage-specific requirement for DNA demethylation in the germline, and the critical role for Dnmt1 in preventing precocious differentiation have critical implications for ensuring the appropriate niche-specific interactions of germline cells that are differentiated in vitro.
Experimental Procedures
Mice
All the animal experiments performed for this study were approved by The UCLA Institutional Animal Care and Use Committee, also known as the Chancellor’s Animal Research Committee (ARC). Embryos were obtained from crosses between Oct4-IRES-Gfp (OG) (Lengner et al., 2007); Dnmt12lox (Jackson-Grusby et al., 2001) homozygous females and Blimp1-Cre (BC) males. Embryos were staged by the detection of a vaginal plug on the morning after time-mating pairs were established (0.5 dpc).
Immunofluorescence
Gonads from various time points were fixed in 4% paraformaldehyde overnight and placed in 70% ethanol. Following this they were embedded in paraffin and sectioned for staining. Staining was performed using conventional methods with some exceptions in the case of 5hmC (Hargan) Permeabilization was performed using 0.5% TX-100 in Phosphate buffered saline (PBS), and washes were performed using 0.2% tween in PBS. The following antibodies were used: Goat anti human Mvh 1:100 (abcam), Rabbit anti mouse 5hmC 1:100 (Active Motif), Goat anti mouse Oct4 1:100(Santa Cruz), Rabbit anti mouse H2AX (Millipore), Rabbit anti mouse Stra8 1:100 (a kind gift from Dr. Cathryn Hogarth and Dr Michael Griswold at Washington State University), rabbit anti-LINE Orf1p 1:300 (provided by Alex Bortvin at the Carnegie Institution for Science), and rabbit anti-IAP Gaga antibody 1:300 (provided by Bryan Cullen at Duke). Visualization was performed by using species-specific secondary antibodies with a conjugated fluorescent tag. Images were captured using a Zeiss LSM 780 confocal microscope.
Sorting
Gonads were harvested from embryos at E10.5, E11.5 and E13.5. The gonads were washed once in calcium and magnesium free PBS (Gibco) before being transferred to a 15ml conical tube containing 3ml of 0.25% Trypsin (Gibco). This was placed in 37°C for 5 minutes followed by gentle flicking and placing it back in 37°C for 5 more minutes. Trypsin was neutralized with 3ml of mouse embryonic flibroblast media and the Gonads were further dissociated by pipetting several times. The resulting solution containing the dissociated gonads was then centrifuged for 5 minutes at 1.3K RPM and resuspended in 1% BSA prior to sorting. 7AAD (BD) was added at 1:50, and only 7AAD negative, GFP positive cells were sorted and used for further analysis.
Genotyping
The head of the mouse embryo was removed and placed in a separate 1.5ml microcentrifuge tube. Tissue was degraded using proteinase K. This was incubated at 55C for 2 hours. Subsequently the temperature was raised to a boil for 10 minutes. The samples were then centrifuged at 13.2K RPM for 5 minutes. 100ul of the supernatant was then transferred to a clean 1.5ml microcentrifuge tube and 1ul was used for the PCR reaction. PCR products were run on a 2% gel at 90 volts for 1 hour. For sexing the embryos the result is two bands around 300bp in males, whereas females exhibit only one band equivalent in size to the smaller fragment in males. The Blimp-Cre transgene results in a band at 200bp. For Dnmt1 fl/fl wildtype band at 334bp and a mutant band at 368bp. The Dnmt1 delta PCR results in a band at 1.1kb. See supplemental experimental procedures, supplemental experimental procedures table 2.
Bisulfite-PCR
GFP positive PGCs at E10.5 and E13.5 were sorted and collected in an empty 1.5ml microcentrifuge tube. DNA was extracted using the zymo quick gDNA mini prep kit (Zymo). The DNA was subsequently subjected to bisulfite treatment using the EZ DNA methylation kit (Zymo). Gene specific PCR amplifications were performed by using primers against the Snrpn DMR1 as described in (El-Maarri et al., 2001), IAP as described in (Hajkova et al., 2002) and the H19 DMR, as described in (Kagiwada et al., 2013). PCR products were run on a 1.2% agarose gel and purified using the QIAquick gel extraction kit (Qiagen) and ligated to a pCR-Topo2.1 cloning vector (TOPO cloning kit, Invitrogen). At least ten clones were picked for analysis by sequencing. See supplementary experimental procedures, supplemental experimental procedures table 2.
RNA Libraries
GFPpositive PGCs at E10.5, E11.5, and E13.5 were sorted and collected in a 1.5ml microcentrifuge tube containing 350ul of RLT buffer. RNA was extracted using the RNeasy Micro Kit (Qiagen). The RNA was subsequently amplified through the use of the Ovation RNA-Seq System V2 kit (Nugen), and bar codes were added using the Encore Rapid Library Systems kit (Nugen). Libraries were sequenced on an Illumina 2500. For the E10.5 libraries 4 biological replicates were used for control and 3 biological replicates were used for the DCKO samples. For the E11.5 libraries 8 biological replicates were used for the control and 10 biological replicates were used for the DCKO samples. For the E13.5 samples 3 biological replicates were used for each set of control and DCKO libraries for both the female and male samples See supplemental experimental procedures, supplementary methods table 1
Statistics
Statistical analysis between two groups was performed using a non-parametric T-Test. In all cases p<0.05 was considered significant.
RNA-Seq Analysis
The RNA-seq reads were aligned to the mouse reference genome mm9 using TopHat (Trapnell et al., 2009). Differential gene expression analysis was performed using Cuffdiff (Trapnell et al., 2012). The genes with RPKM=0 were removed from the analysis. The multiple testing errors were corrected by the false discovery rate (FDR). The genes with ≥ 2-fold difference in expression and FDR < 5% were considered differentially expressed. For the E10.5 and E11.5 libraries genes with ≥ 2-fold difference in expression and FDR < 10% were considered differentially expressed. The raw data are deposited to GEO (accession number GSE74938)
Identifying de-repressed genes with promoter methylation
The DNA methylation data (BS-seq) of mouse PGC from E6.5 to E13.5 were downloaded from Seisenberiger (2012) (Seisenberger et al., 2012). The BS-seq reads were mapped against the mm9 mouse genome using BS Seeker 2 (Guo et al., 2013). Genome-wide DNA methylation profiles were generated by determining methylation levels for each cytosine in the genome. Because bisulfite treatment converts unmethylated cytosines (Cs) to thymines (Ts) after PCR amplification, the methylation level at each cytosine was estimated as #C/(#C+#T), where #C is the number of methylated reads and #T is the number of unmethylated reads. The methylation level per cytosine serves as an estimate of the percentage of cells that have a methylated cytosine at a specific locus. We only included cytosines that are covered by at least three reads. The promoter region is defined as the region between 2,500 bp upstream and 500 bp downstream of the transcription start site (TSS). The de-repressed genes with the promoter methylation level ≥ 20% at E9.5 were selected for further analysis.
Enrichment of methylated and de-repressed genes in chromosome X
We used hypergeometric test to evaluate the enrichment of the methylated and de-repressed genes in each chromosome. The test uses the hypergeometric distribution to calculate the statistical significance of the enrichment of the genes among all genes with both data of methylation and expression in the chromosome. As a result, we found Chromosome X is significantly enriched with these genes, in both male data and female data.
Highlights.
Dnmt1 is responsible for maintaining DNA methylation in PGCs
Dnmt1 has no role in transposon repression in PGCs
Precocious germline differentiation is restrained by Dnmt1
The 5hmC epigenetic mark is not required for meiotic entry
Acknowledgments
The authors would like to thank the UCLA BSCRC flow cytometry core for flow and FACS assistance. We would also like to thank Cathryn Hogarth and Michael Griswold for their generous gift of the STRA8 antibody. This work was supported by an R01 grant from the NIH (NIH/NICHD R01 HD058047) awarded to ATC. As well as the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, and by grants from Academia Sinica and National Health Research Institutes, Taiwan (NHRI-EXI03-10324SC) to P.-Y.C. We would also like to acknowledge the support of the California Institute for Regenerative Medicine (CIRM) Predoctoral training grant (TG2-01169) for funding JHC.
Footnotes
Author Contributions
J.H.-C designed and performed the experiments and wrote the manuscript. S.T., H.C., Z.H., and S.A.L. designed and performed the experiments. M.R.Y, Y.S.C., and P.Y.C performed the RNA-seq and WGBS analyses. A.T.C. designed and oversaw all experiments, maintained all University compliance and reporting requirements, wrote the manuscript, and obtained the research funding.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet. 2006;38:1430–1434. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Brennan J, Karl J, Capel B. Divergent vascular mechanisms downstream of Sry establish the arterial system in the XY gonad. Dev Biol. 2002;244:418–428. doi: 10.1006/dbio.2002.0578. [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined Deficiency of Tet1 and Tet2 Causes Epigenetic Abnormalities but Is Compatible with Postnatal Development. Dev Cell. 2013 doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed]
- El-Maarri O, Buiting K, Peery E, Kroisel P, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan C, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genetics. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X-L, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Bu T-P, Hu B, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008 doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hargan-Calvopina J, Cook H, Vincent J, Nee K, Clark A. The Aorta Gonad Mesonephros organ culture recapitulates 5hmC reorganization and replication-dependent and independent loss of DNA methylation in the germline. Stem Cells Dev. 2015 doi: 10.1089/scd.2014.0410. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haston K, Tung J, Reijo Pera R. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One. 2009;4:e5654. doi: 10.1371/journal.pone.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C, Bestor T, Ding F. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jeske YW, Bowles J, Greenfield A, Koopman P. Expression of a linear Sry transcript in the mouse genital ridge. Nat Genet. 1995;10:480–482. doi: 10.1038/ng0895-480. [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. Embo J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KD, Roelen NR, Zeinstra BA, Davis LM, Wright AM, Korving CV, Hogan JP, BL Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:373–376. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C, Camargo F, Hochedlinger K, Welstead G, Zaidi S, Gokhale S, Scholer H, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith A. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 1998;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yu J, Hosohama L, Nee K, Gkountela S, Chaudhari S, Cass AA, Xiao X, Clark AT. The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. Embo J. 2015;34:748–758. doi: 10.15252/embj.201489319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gill M, Koubova J, Page D. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;12:1685–1687. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- Liu S, Brind’Amour J, Karimi M, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz M. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2015;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MR, Lei H, Li E, Bartolomei MS, Resnick JL. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- MacLean G, Li H, Metzger D, Chambon P, Petkovich M. Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology. 2007;148:4560–4567. doi: 10.1210/en.2007-0492. [DOI] [PubMed] [Google Scholar]
- McCarrey J, Watson C, Atencio J, Ostermeier G, Marahrens Y, Jaenisch R, Krawetz S. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis. 2002;34 doi: 10.1002/gene.10163. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Dilworth DD. Expression of Xist in mouse germ cells correlates with X-chromosome inactivation. Nat Genet. 1992;2:200–203. doi: 10.1038/ng1192-200. [DOI] [PubMed] [Google Scholar]
- McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol. 1997;187:107–113. doi: 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- Messerschmidt D, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 Is Required for Epigenetic Stability During Mouse Oocyte to Embryo Transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Mu X, Wen J, Guo M, Wang J, Li G, Wang Z, Wang Y, Teng Z, Cui Y, Xia G. Retinoic acid derived from the fetal ovary initiates meiosis in mouse germ cells. J Cell Physiol. 2013;228:627–639. doi: 10.1002/jcp.24172. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, NK, Li E, Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton S, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005:1–7. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M. A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Blimp1-mVenus and stella-ECFP double transgenic reporter. Reproduction. 2008 doi: 10.1530/REP-08-0053. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Okano MB, Harber DW, Li DAE. DNA methyltransferases Dnmt3a and Dnmt3b are essential for denovo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc’his D, et al. MORC1 represses transposable elements in the mouse male germline. Nat Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat J, Dean W, Clark S, Krueger F, Smallwood S, Ficz G, Kim J, Marioni J, Hore T, Reik W. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 2014;9:1990–2000. doi: 10.1016/j.celrep.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, Mc Kay S, Kilanowski F, Saunders P, Dorin J, Cooke H. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- Sachs M, Onodera C, Blaschke K, Ebata K, Song J, Ramalho-Santos M. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrans-Stassen BHGJ, Saunders PTK, Cooke HJ, Rooij DGd. Nature of the spermatogenic arrest in Dazl −/− mice. Biol Repro. 2001;65:771–776. doi: 10.1095/biolreprod65.3.771. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:1309–1317. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev. 2008;22:430–435. doi: 10.1101/gad.1612708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PPL, Zhou SX. The allocation of epiblast cells to ectoderman and germline lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. Stage-Specific Roles for Tet1 and Tet2 in DNA Demethylation in Primordial Germ Cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;68:1177. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, Zhang Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013a;23:329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Science. 2013b;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, Lohizen M, Roloff T, Peters A. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 2013;495:236–240. doi: 10.1038/nature11918. [DOI] [PubMed] [Google Scholar]