Abstract
Recent advances in understanding different RNAs and unique features of their biology have revealed a wealth of information. However, approaches to identify small molecules that target these newly discovered regulatory elements have been lacking. The application of new biochemical screening and design-based technologies, coupled with a resurgence of interest in phenotypic screening, has resulted in several compelling successes in targeting RNA. A number of recent advances suggest that achieving the long-standing goal of developing drug-like, biologically active small molecules that target RNA is possible. This review highlights advances and successes in approaches to targeting RNA with diverse small molecules, and the potential for these technologies to pave the way to new types of RNA-targeted therapeutics.
Functional RNAs come in many different shapes and sizes, and represent some of the key regulators of biological processes. Yet, finding small molecules that target RNA continues to be a challenge. Connelly et al. review the emerging field of RNA-targeted small-molecule probes and therapeutics, and offer a broad view of available approaches used to discover new RNA-binding chemical entities.
Main Text
Introduction
RNA transcripts have emerged as key regulators of diverse biological phenomena. Classically RNA was viewed as a carrier of genetic information that exists solely to transmit a message for protein coding and guide the process of protein biosynthesis. Modern views encompass an expanded role for RNA, with a diverse range of RNA molecules now understood to have broad and far-reaching roles in modulating gene expression and other biological processes by various mechanisms (Breaker and Joyce, 2014). One recent report estimated that while ∼85% of the human genome is transcribed into RNA, only ∼3% of those transcripts code for protein, indicating that vast majority of RNAs are noncoding (Hangauer et al., 2013). Furthermore, a large number of newly discovered noncoding RNAs are disease associated, both in cancer and nontumorigenic diseases (Cheetham et al., 2013, Esteller, 2011). Thus, the realization that RNAs contribute to disease states apart from coding for pathogenic proteins is likely to provide a wealth of previously unrecognized therapeutic targets (Cooper et al., 2009). Given that many of these RNAs adopt discrete secondary and tertiary structures and have pivotal roles in biology, they are attractive targets for small molecules.
In mammals, the biological functions of RNAs are highly diverse and act at many levels of regulation. RNAs vary greatly in sequence length, ranging from small hairpins consisting of a few nucleotides to long noncoding RNAs (lncRNAs) consisting of several thousand nucleotides. One example of small RNA molecules is microRNAs (miRNAs), which are approximately 22 nucleotide sequences produced by DICER processing of RNA hairpins, and function to suppress gene expression by inhibiting or degrading mRNAs. Another example of a noncoding RNA is telomeric repeat RNA (TERRA), which consists of oligomeric sequences of polymorphic RNA G-quadruplexes and is believed to be required for proper telomere function (Rippe and Luke, 2015). Oligonucleotide repeat expansion diseases are a series of pathologies that occur when oligomeric expansions of RNA result in a disease state, such as (CAG)n in Huntington's disease (Galka-Marciniak et al., 2012) or (GGGCCC)n in amyotrophic lateral sclerosis (ALS) (DeJesus-Hernandez et al., 2011). Larger lncRNAs such as HOTAIR, and many others, contribute to oncogenesis through direct binding and sequestration of various tumor suppressors and other proteins, although this mechanism is not always clear (Cheetham et al., 2013). New families of functional, noncoding RNAs continue to be discovered, including PIWI-interacting RNAs, small nuclear RNAs, small nucleolar RNAs, and others (Esteller, 2011). In addition to canonical, noncoding RNA sequences, alternative splicing of an mRNA can provide multiple gene products from the same transcribed RNA sequence; the misregulation of this process is often disease associated (Scotti and Swanson, 2015). Posttranscriptional modifications such as methylation have also been discovered to modulate RNA structure and function (Chen et al., 2016, Helm and Alfonzo, 2014, Li and Mason, 2014).
Apart from mammalian systems, RNA plays a pivotal role in both viruses and bacteria. The orthogonality of homologous functional elements between viral or bacterial transcriptomes/genomes and mammalian transcriptomes makes these RNA structures excellent targets for anti-infective drugs, a concept that has gained significant attention. Historically, genomes of retroviruses such as HIV have been shown to contain noncoding, cis-acting functional elements such as the transactivation response (TAR) hairpin, the frameshift signal (FSS), or the Rev responsive element (RRE) (Le Grice, 2015). These RNAs have multifunctional roles that regulate polymerase activity, viral infectivity, and progression at multiple levels and have been studied as drug targets. Similarly, viral RNAs such as the internal ribosomal entry site (IRES) enable cap-independent translation and have been studied as therapeutically relevant targets in both poliovirus and hepatitis C virus (HCV) (Davis and Seth, 2011). Bacteria also contain naturally occurring RNA aptamers called riboswitches that bind to secondary metabolites and regulate gene expression in bacteria. More recent work demonstrated that riboswitches are not unique to bacteria, and these RNAs have emerged as a major area of study in RNA biology, ranging from serving as model systems to study RNA structure to potential drug targets (Breaker and Joyce, 2014).
In addition to new functions, the development of modern analytical techniques has ushered in an explosion in our knowledge of RNA structure. Early views tended to consider RNAs as highly flexible molecules largely devoid of discrete structure. However, current views consider RNA to exist as a dynamic ensemble of structural conformations, including well-defined secondary and tertiary folds (Salmon et al., 2014). Traditionally, X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy have been the two techniques used to characterize atomic-resolution RNA structures. Although not without technical challenges, both of these methods yielded profound insights and high-resolution structures of RNA molecules with a range of sizes and structural features. Cryoelectron microscopy (cryo-EM) has recently undergone a revolution in the resolution it can achieve and now competes with X-ray crystallography in terms of the structural analysis of increasingly large RNA structures (Garmann et al., 2015). On the other hand, advances in NMR spectroscopy and computational modeling have been critical in developing the view of RNAs as dynamic systems (Stelzer et al., 2011). Another complementary biophysical technique, small-angle X-ray scattering (SAXS), is also emerging as a powerful way to analyze the fold adopted by a particular RNA in solution, as it offers a view of a global shape (Fang et al., 2015). Chemical probing techniques such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) have further proved highly useful in evaluating RNA structure at single-nucleotide resolution, and an entire viral genome may now be structurally analyzed in a single experiment (Watts et al., 2009). A frontier in this area is the application of such probing techniques to RNA structures in vivo, with a goal of confirming the biologically relevant conformations of RNA (Spitale et al., 2013).
Despite the development of such powerful techniques for the analysis of RNA structure and discovery of new function, our ability to identify or design inhibitors that bind to and perturb the function of RNA lags far behind. To date, the most commonly employed method to target disease-associated RNAs is based on the use of antisense technologies such as small interfering RNAs. Antisense technology is highly effective, and a small number of drugs using this approach have been approved (McClorey and Wood, 2015). However, this approach primarily relies on derivatized oligonucleotide structures, which suffer from poor cell permeability and distribution due to their requisite anionic character, limiting their application as therapeutics. Small molecules, however, often offer the advantage of having desirable properties such as good absorption, distribution, and oral bioavailability. In addition, small molecules have the potential to recognize RNA by virtue of secondary or tertiary structure, as opposed to sequence. Thus, small molecules provide an orthogonal means to target different RNA structural elements such as bulges, loops, junctions, pseudoknots, or higher-order structure by virtue of recognizing and binding unique folds, rather than complementary sequence.
Early approaches to identify RNA-binding molecules led to the discovery of aminoglycosides, potent, clinically used molecules that bind to RNA through electrostatic interactions with, at best, modest selectivity and systemic toxicity. However, more recent approaches have favored the development of more drug-like RNA-binding compounds that display improved physiochemical properties and more well-defined patterns of selectivity. For the purposes of this review, we use the term “drug-like” to refer to compounds predicted or demonstrated to have good medicinal chemistry properties such as potency, solubility, selectivity, distribution, and in some rare cases oral bioavailability. These compounds tend to have molecular weight below ∼500, fewer than five hydrogen-bond donors/acceptors, low polar surface area, and no reactive/promiscuous scaffolds. A desirable feature of such compounds is that they interact with RNA not through intercalation, or sequence or electrostatic complementarity, but by means of specific molecular recognition events unique to the particular RNA target. New techniques are increasingly required to define the rules for identifying, designing, and studying RNA-binding small molecules, and advancing these molecules as biological probes and potential therapeutics. However, overcoming the challenges of the highly anionic nature of RNA, compounded by its flexible, dynamic structure, remains the main roadblock for the development of small-molecule inhibitors. Moreover, many putative small-molecule binding sites in RNA are much more polar and solvent exposed than binding sites on proteins, complicating ligand design efforts. Despite these challenges, there have been a number of promising results reported recently suggesting that the long sought-after goal of developing small-molecule drugs that target RNA may indeed be achievable. This review discusses recent developments in technologies and approaches used to identify such inhibitors. We focus mostly on advances reported since several other comprehensive reviews (Disney et al., 2014, Guan and Disney, 2012, Le Grice, 2015, Thomas and Hergenrother, 2008) and exclude ribosome-binding molecules, as these have been well reviewed elsewhere (Arenz and Wilson, 2016, Hermann, 2005, Poehlsgaard and Douthwaite, 2005).
Design- and Structure-Based Approaches
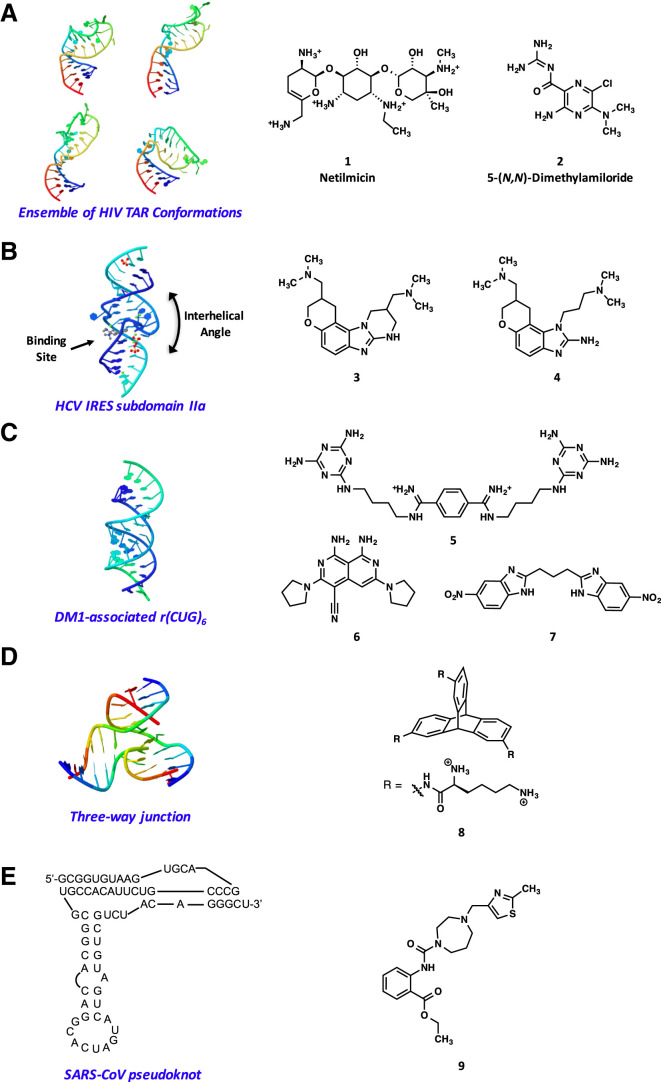
The flexible and dynamic nature of RNA structures represents one of the challenges associated with targeting RNAs with small molecules. However, the ability to identify small molecules that bind to specific RNA targets through structure-based approaches that take dynamics into account provides a viable avenue to identify potent binders (Shortridge and Varani, 2015). In a recent example, the Al-Hashimi laboratory has successfully used NMR residual dipolar coupling data to calculate a dynamic ensemble of the HIV-1 TAR element RNA, a prominent target for the inhibition of HIV replication (Le Grice, 2015). The dynamic ensemble was then used as a starting point for a virtual screen to identify small molecules that target the entire structural landscape of TAR (Stelzer et al., 2011). A 51,000-member small-molecule library was screened against 20 conformers of the TAR ensemble to identify six small molecules, including netilmicin (1) and 5-(N,N)-dimethylamiloride (2) (Figure 1A), which bind to TAR with high affinity (KD = 55 nM to 122 μM) and inhibit its interaction with the Tat peptide in vitro with Ki values ranging from 710 nM to 169 μM. Several of the identified compounds showed significant deterioration in the binding affinities when measured in the presence of excess tRNA, indicating nonspecific tRNA binding; however, no change in the KD values for 1 and 2 was observed. Furthermore, netilmicin was found to selectively bind TAR over other RNAs that resemble the TAR hairpin, including an HIV-2 TAR variant (negligible affinity), the prokaryotic ribosomal A-site hairpin (35-fold weaker affinity), and the HIV-1 RRE hairpin (86-fold weaker affinity). In addition, netilmicin bound to a TAR mutant with a deletion of a single cytosine bulge residue with 16-fold weaker affinity than wild-type TAR. Netilmicin also showed unique interactions involving the bulge, upper stem, and apical loop of TAR as determined by NMR chemical shift mapping experiments. In addition, netilmicin specifically inhibited Tat-mediated activation of the HIV-1 promoter by 81% in T cell lines and inhibited HIV-1 replication in an HIV-1 indicator cell line, TZM-bl, and the HIV-1 NL4-3 isolate, with an IC50 value of ∼23 μM. Taken together, the discovery of netilmicin as a potent binder of TAR demonstrates the ability to use NMR-informed computational dynamic ensembles as a suitable foundation for identifying RNA-targeting small molecules.
Figure 1.
Structure- and Design-Based Approaches for Identifying RNA-Binding Molecules
(A) Representative conformers of wild-type HIV-1 TAR RNA (left) used to identify TAR-binding small molecules (right) by virtual screening (Stelzer et al., 2011).
(B) Crystal structure of the HCV IRES subdomain IIa RNA in complex with the benzimidazole inhibitor 4 showing the small-molecule-induced change in the interhelical angle of the RNA (left), and structures of the identified HCV translation inhibitors that bind to subdomain IIa (right) (Parsons et al., 2009).
(C) Structure of the r(CUG)6 duplex (left) used to rationally design a highly selective CUG binding ligand 5 based on the triaminotriazine scaffold (right) (Nguyen et al., 2015, Wong et al., 2014). Structures of r(CUG)exp binding compounds 6 and 7 identified by high-throughput screening efforts are also shown (right) (Childs-Disney et al., 2013, Rzuczek et al., 2015).
(D) Representative structure of a three-way junction (left) for which RNA-binding triptycene-based scaffolds such as compound 8 (right) were developed (Barros et al., 2016a).
(E) Structure of the SARS-CoV RNA pseudoknot involved in an essential −1 ribosomal frameshift (left) used in a virtual screen to identify a SARS-pseudoknot-binding ligand (right) (Park et al., 2011).
In another RNA virus, the IRES of HCV binds to the host cell's ribosomal 40S subunit, initiates viral translation in a cap-independent fashion, and adopts an ordered structure dominated by independently folded RNA domains (Parsons et al., 2009). The Hermann laboratory reported the three-dimensional structure of the IRES subdomain IIa determined by X-ray crystallography and showed that it adopts an overall bent architecture with an L-shaped conformation that is stabilized by three divalent metal ions (Dibrov et al., 2007). Importantly, the determined L-shaped conformation is in agreement with previously reported NMR studies on the full domain II (Lukavsky et al., 2003) and cryo-EM investigations of IRES-40S complexes (Boehringer et al., 2005, Spahn et al., 2001). It is likely that maintaining the architecture of subdomain IIa is critical for correct binding of the viral mRNA at the ribosome. A high-throughput screen of a 180,000-member library against a 29-mer oligonucleotide representing the IRES subdomain IIa using a mass spectrometry-based screening method identified a benzimidazole-containing compound with a KD of ∼100 μM to the IRES IIa model (Seth et al., 2005). Subsequently, a new class of benzimidazole-containing compounds was developed after extensive structure-activity relationship studies, and several analogs were identified with submicromolar affinity (Figure 1B). Using information gathered from crystallography, Hermann and coworkers developed several fluorescence-based assays to assess the binding of the benzimidazole-based ligands to the RNA (Parsons et al., 2009). A fluorescence resonance energy transfer (FRET)-based assay monitoring the interhelical angle between the stems flanking the internal loop in IIa via a measurement of the distance between the stem termini was developed and used to determine the ability of the benzimidazole (3) to bind to an oligonucleotide mimic of IRES IIa. Addition of the benzimidazole (3) resulted in a dose-dependent quenching of FRET with an EC50 value of 0.6 μM in 2 mM Mg2+ and was unchanged in the presence of excess competitor tRNA. These data suggest that the benzimidazole acts as an HCV translation inhibitor by inducing a conformational widening of the interhelical angle in the IRES subdomain IIa RNA. In addition, the impact of 3 on HCV translation was assessed in human cells expressing a reporter under control of the HCV IRES, and the compound successfully inhibited IRES-driven translation at low micromolar concentrations. X-ray crystallographic analysis of subdomain IIa in complex with a benzimidazole ligand (4, Figure 1B) verified the widening of the interhelical angle (Dibrov et al., 2012). The initial benzimidazole ligands were further explored through subsequent rounds of synthesis of rationally designed ligands based on the cocrystal structure of the IIa RNA in complex with the ligand (Ding et al., 2014, Rynearson et al., 2014). Recent cryo-EM studies indicate that domain IIa adopts a bent conformation in complex with the 80S- and 40S-ribosomal subunits (Quade et al., 2015, Yamamoto et al., 2015). Therefore, the more linear ligand-bound IIa domain structure identified by crystallography may prevent the conformation necessary for this interaction (Yamamoto et al., 2015). Additional screening of subdomain IIa RNA with modular ligands that contain the 3,5-diaminopiperidine heterocycle revealed N-amido substituted α-amino acid conjugates that bind subdomain IIa with micromolar affinities and arrest the RNA in a bent state with a 90° interhelical angle in solution (Carnevali et al., 2010). Further adaptation of the previously mentioned structure-based FRET assay monitoring the interhelical angle of the IIa RNA to a high-throughput screening format has also led to the identification of additional IIa RNA-binding compounds (Zhou et al., 2013).
A rational design approach was similarly extended to the development of small molecules that bind to the r(CUG)exp (expansion of r(CUG) repeats) RNA associated with myotonic dystrophy type 1 (DM1). DM1 is a multisystem disorder that affects skeletal and smooth muscle as well as the eye, heart, endocrine system, and CNS, causing symptoms including myotonia, wasting of the muscle, and cardiac defects. r(CUG)exp is located in the 3′ UTR of the dystrophia myotonica protein kinase mRNA and causes disease through a gain-of-function mechanism, whereby the RNA binds to and sequesters proteins involved in RNA biogenesis, such as the MBNL (muscleblind-like) family of splicing regulators. Based in part on a previously developed ligand for the HIV-1 frameshift site RNA stem loop and on the X-ray crystal structure of r(CUG)6, the Zimmerman group designed an RNA-groove-binding inhibitor (5) for r(CUG)exp comprising two triaminotriazine units connected by a bisamidinium linker (Figure 1C) (Wong et al., 2014). The triaminotriazine units were selected for their propensity to form base triplets with U-U mismatches (and are known as Janus-wedge units), while the bisamidinium moiety was suggested as a groove-binding scaffold for CUG recognition. Compound 5 exhibited low micromolar affinity (KD = 8 ± 2 μM) for r(CUG)12 by ITC, showed selective binding over other targets including tRNA, glutathione S-transferase-tagged MBNL1, HIV-1 frameshift site RNA, and r(CCUG)8, and was capable of disrupting the MBNL1-r(CUG)12 interaction in vitro with an apparent Ki of 8 ± 2 μM as determined by electrophoretic mobility shift assay. The compound showed good cell permeability in vitro, reduced MBNL1-r(CUG)exp ribonuclear foci formation in a cell-culture model for DM1, and partially restored the misregulated splicing of two pre-mRNAs, cardiac troponin T (cTNT) and insulin receptor. In vivo, the compound suppressed the phenotype associated with r(CUG)exp RNA-induced toxicity in a DM1 transgenic Drosophila model. The ligand has been further used as the foundation for the rational design of multitarget agents for DM1 that bind to CTGexp DNA and inhibit the formation of the r(CUG)exp transcript, bind to r(CUG)exp and inhibit sequestration of MBNL1, and cleave r(CUG)exp in an RNase-like manner (Nguyen et al., 2015). This work stands as a particularly notable and relatively rare use of rational design of RNA-binding small molecules with validated activity in an animal model. In addition to this work, other groups have reported several other r(CUG)exp-binding compounds. For example, the Disney group reported a substituted naphthyridine (6, Figure 1C) that was identified by high-throughput screening, and was shown to inhibit the r(CUG)exp-MBNL1 interaction with an IC50 of 2 ± 0.4 μM using a FRET-based assay (Chen et al., 2012, Childs-Disney et al., 2013). The naphthyridine binds to r(CUG)12 with a KD of 125 nM and interacts with the UU loops in r(CUG)exp to displace MBNL1. In cell models, 6 improved DM1-associated pre-mRNA splicing defects with specificity for MBNL1-regulated splicing events and caused reductions in nuclear foci formation. Compound 6 has also been subsequently identified by the Disney laboratory to be an inhibitor of miR-544 (see below) (Haga et al., 2015). The Disney laboratory also reported benzimidazoles (7, Figure 1C) identified by screening of an RNA-focused small-molecule library, and oligomeric Hoechst dye-like compounds that bind r(CUG)exp in cells and had promising effects on DM1-associated defects in cell models (Childs-Disney et al., 2012, Pushechnikov et al., 2009, Rzuczek et al., 2015).
Another design-based approach for the recognition of specific RNA structures comes from the Chenoweth group, which has recently reported several papers describing the use of triptycene-based molecular scaffolds to recognize RNA junctions. Three-way junctions are ubiquitous throughout the transcriptome, although few small molecules that recognize them are known. The Chenoweth group initially reported a simple triptycene functionalized with cationic groups that bound to both DNA and RNA three-way junctions, and demonstrated that these compounds exhibit cell permeability as well as cytotoxicity (Barros and Chenoweth, 2014, Barros and Chenoweth, 2015). Further work showed that diverse triptycene molecules can be synthesized in a modular and efficient fashion, including solid-phase methods that are likely to have use in the preparation of libraries of triptycenes going forward, with the potential for specific junction recognition (Barros et al., 2016b, Yoon et al., 2016). Finally, a recent report demonstrated that functionalized triptycenes such as 8 (Figure 1D) can also be used to modulate the Escherichia coli rpoH RNA temperature sensor, indicating utility in recognizing RNA structures in bacteria as well (Barros et al., 2016a). To date, most of the work in this area has focused on molecular recognition and the development of synthetic methods to access new triptycene scaffolds; however, it is clear that this approach has potential for broad applications in RNA recognition and targeting in future efforts.
In another structure-based approach, Park et al. (2011) performed an in silico screen to discover compounds that target the frameshifting signal pseudoknot in the severe acute respiratory syndrome coronavirus (SARS-CoV). SARS-CoV utilizes an essential programmed −1 ribosomal frameshift (−1 RF) to synthesize key replication components. The stability and structure of the RNA pseudoknot present in the −1 RF site is essential for efficient frameshifting and viral replication, indicating that it may be a valuable target for small molecules. The authors used a 3D structural model of the SARS pseudoknot to perform a virtual ligand screen. Docking of approximately 80,000 compounds from the commercially available chemical database LeadQuest produced a set of high-ranking compounds. Hit validation was performed using a pseudoknot-driven dual luciferase reporter assay in both biochemical and cell-based assays, and a novel ligand that inhibits the −1 RF of SARS-CoV was identified that dramatically decreased the −1 RF in vitro and inhibited −1 RF efficiency in HEK293 cells with an IC50 of approximately 0.45 μM (9, Figure 1E). Selectivity was evaluated using similar reporter assays on the pseudoknot from the pea enation mosaic virus, where the compound had no activity. In a later report by Woodside, binding studies by surface plasmon resonance revealed an apparent KD of 210 ± 20 μM (Ritchie et al., 2014). It remains unclear how a compound with this KD could have such potent cellular effects. Finally, analysis using optical tweezers revealed that ligand binding appears to influence the ensemble of conformations populated by the RNA, rather than influencing its mechanical stability. Based on these studies, the authors propose a mechanism whereby the ligand reduces −1 RF efficiency by decreasing the conformational plasticity of the SARS pseudoknot (Ritchie et al., 2014).
In addition to designing ligands de novo on the basis of RNA structure, a related strategy is to redesign cognate ligands for a particular RNA. This is primarily applicable to riboswitches, naturally occurring aptamers that regulate gene expression by changing conformation in the presence of a metabolite or small-molecule ligand. Over the last decade, several groups have used structure-guided rational design to identify and optimize small molecules that bind to selected riboswitch aptamers based on the structures of their cognate ligands, including small molecules targeting cyclic diguanylate monophosphate (Furukawa et al., 2012), glmS (Blount et al., 2006, Lunse et al., 2011), purine (Gilbert et al., 2009), and lysine (Blount et al., 2007) riboswitches. In some cases, synthetic or natural analogs of cognate ligands have also demonstrated potent antibacterial activity, suggesting the potential for developing riboswitch-targeting antibiotics (Blount et al., 2007, Kim et al., 2009, Lee et al., 2009, Mansjo and Johansson, 2011, Ott et al., 2009). Recently, an analog of riboflavin, 5FDQD, was designed via medicinal chemistry optimization to bind to riboswitches that recognize the natural coenzyme flavin mononucleotide (FMN) and regulate the homeostasis of FMN and riboflavin, as reported by the Breaker laboratory in collaboration with several industrial groups (Blount et al., 2015). In vitro, 5FDQD binds to and triggers function of an FMN riboswitch, shows antibacterial activity against Clostridium difficile, and prevents antibiotic-induced C. difficile infection (CDI) in mice nearly as effectively as fidaxomicin, a current CDI treatment. It is clear that in many cases where substantial information about the structure and dynamics of a given RNA is available, this information may be leveraged in the design of small-molecule binders with good affinity and biological activity.
Informatics-Based Approaches
Another powerful tool that has successfully been used to design small molecules that bind to RNA targets is the informatics-driven, rational design approach termed Inforna, developed by the Disney laboratory. Inforna utilizes a database of experimentally determined RNA-motif-small-molecule interactions identified through a selection-based strategy called two-dimensional combinatorial screening (2DCS) and structure-activity relationships through sequencing (StARTS) (Disney et al., 2008). In brief, a small-molecule library is conjugated to an agarose microarray surface, which is then probed for binding to a library of small RNA motifs, including hairpins, loops, or bulges, that are likely to be found as components of larger cellular RNAs. Quantification of specific bound RNAs is accomplished by excision of each individual spot of the array and sequencing of the corresponding bound oligonucleotides. Next, StARTS is used to score each RNA-motif-small-molecule interaction (Velagapudi et al., 2010). Inforna can be used to identify lead compounds for an RNA of interest by comparing the structural motifs present in a target RNA with the motifs in the database of annotated RNA-motif-small-molecule interactions derived from 2DCS. For a given RNA of interest, the Inforna software provides the targetable structural motifs for the input RNA, the corresponding lead small-molecule hits, and the fitness of the predicted RNA-small-molecule interactions as determined by StARTS analysis (Velagapudi et al., 2014). A powerful demonstration of the abilities of Inforna is the discovery of a compound that binds to the miRNA miR-96 and inhibits miR-96 biogenesis with selectivity similar to that of an miR-96 antagomir (Velagapudi et al., 2014). The discovered small molecule inhibits Drosha processing of pri-miR-96, leading to the upregulation of its target FOXO1 and the induction of apoptosis in MCF7 breast cancer cells.
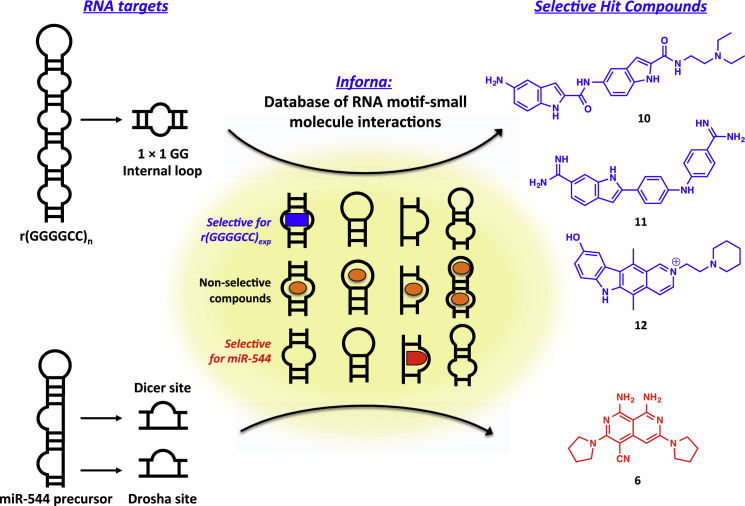
Recently, the Disney laboratory has successfully employed Inforna to identify small-molecule ligands for several other disease-associated RNA structures. One prominent example is the repeat expansion r(GGGGCC)exp, the most common genetic cause of frontotemporal dementia and ALS (c9FTD/ALS) (Su et al., 2014). This repeat expansion RNA forms nuclear foci that sequester various RNA-binding proteins, causing toxicity and undergoing repeat-associated non-ATG (RAN) translation to produce c9RAN proteins that form neuronal inclusions throughout the CNS. Based on the determined mixed hairpin/G-quadruplex structure of r(GGGGCC)exp, lead small molecules that bind to the RNA were identified using Inforna (Figure 2 ). The hit compounds were experimentally screened using a biochemical TO-PRO-1 displacement assay, and three lead compounds were identified and validated. Kinetic binding studies with r(GGGGCC)8 revealed KDs of 16, 10, and 9.7 μM for compounds 10, 11, and 12, respectively (Figure 2). The compounds showed similar binding affinities to r(CGG)12 and r(GGCC)4, both of which possess 1 × 1 GG internal loops structurally similar to that of r(GGGGCC)8; however, the compounds had significantly decreased binding affinities for a hairpin with a fully paired stem, demonstrating modest selectivity for 1 × 1 GG internal loops. Furthermore, the compounds successfully engaged r(GGGGCC)66 RNA in COS7 cells as demonstrated by competitive Chem-CLIP, validating binding in cellulo. In HEK293 cells, expression of r(GGGGCC)66 resulted in the synthesis of poly(GP) and poly(GA) proteins through RAN translation, and treatment with two of the lead compounds (11 and 12) significantly decreased both poly(GP) and poly(GA) protein levels, demonstrating the ability of the small molecules to inhibit RAN translation. These compounds were also capable of inhibiting nuclear foci formation as determined by RNA fluorescence in situ hybridization. In addition, compound 12 inhibited RAN translation and foci formation in iNeurons with the r(GGGGCC) expansion, suggesting the potential of a small-molecule inhibitor of r(GGGGCC)exp as a lead structure for therapeutic development for treatment of c9FTD/ALS.
Figure 2.
Application of Inforna to the Identification of Small Molecules that Bind to Disease-Associated RNAs
A database of RNA motif-small molecule interactions was used to predict compounds that bind to the 1 × 1 nucleotide GG internal loops present in the c9FTD/ALS-associated r(GGGGCC)exp (top, blue), or to the 1 × 1 nucleotide UU internal loops present in the Dicer and Drosha processing sites of the cancer associated miR-544 hairpin precursor (bottom, red). Structures of validated lead compounds that bind to r(GGGGCC)8 and the miR-544 hairpin precursor are shown in blue and red, respectively (Haga et al., 2015, Velagapudi et al., 2014).
Similarly, the Disney and Finney laboratories used Inforna to identify small molecules that bind to the miRNA miR-544, which silences mammalian target of rapamycin (mTOR), as a means to interrogate the role of miR-544 in tumor cell growth under hypoxic conditions (Haga et al., 2015). The precursor hairpin structure of miR-544 was parsed into its composite motifs (the 1 × 1 nucleotide UU internal loops present in the Dicer and Drosha processing sites), which were then screened in Inforna (Figure 2). Identified compounds were screened and tested for their ability to modulate miR-544 biogenesis, and five potential lead compounds were identified. The most potent compound (6, Figure 2) successfully disrupted miR-544-mediated inhibition of its validated target BMI1 as demonstrated by a fluorescence-based reporter assay in cellulo, and caused accumulation of pre-miR-544 and a decrease in mature miR-544 levels by qRT-PCR. The compound selectively bound RNAs containing the 1 × 1 nucleotide UU internal loop of the Dicer and Drosha sites over control RNAs containing a UA pair with binding constants in the midnanomolar range, and showed no binding to a DNA control by gel-shift assay. Impressively, microarray analysis of cells treated with 6 revealed changes in mRNA and miRNA expression similar to that of cells transfected with the miR-544 antagomir, indicating that the compound is as selective as an oligonucleotide-based antisense inhibitor. However, the Disney laboratory also reported that 6 binds to r(CUG)exp RNA as well (see discussion above) (Chen et al., 2012, Childs-Disney et al., 2013). In immunodeficient mice implanted with MDA-MB-231-GFP-luc cells, pretreatment with 6 as well as postimplantation intraperitoneal injection impeded tumor growth compared with untreated cells, and qRT-PCR analysis of developed tumors revealed decreased levels of miR-544, HIF-1α, and ATM and increased levels of pre-miR-544 and mTOR. Taken together, this work stands as a powerful example of the Inforna platform's ability to identify highly selective RNA-binding small molecules that modulate miRNA biogenesis in vivo. Combined with the r(GGGGCC)exp and other examples, primarily focused on precursor miRNA inhibitors (Haga et al., 2015, Luo and Disney, 2014, Velagapudi et al., 2014), it is clear that Inforna has matured as a highly useful tool for the discovery of RNA-binding molecules.
Fragment-Based Approaches
Fragment-based drug discovery (FBDD) is a modern method for the development of highly potent inhibitors with desirable, drug-like properties. In brief, this method employs sensitive biophysical techniques to identify low molecular weight compounds with weak but specific interactions to the biomolecule of interest. Once identified, compounds may be elaborated via multiple strategies into potent, selective inhibitors. While this method has enjoyed much success in developing enzyme inhibitors, it has only recently begun to be used broadly in the arena of RNA-binding small molecules (Davidson et al., 2011).
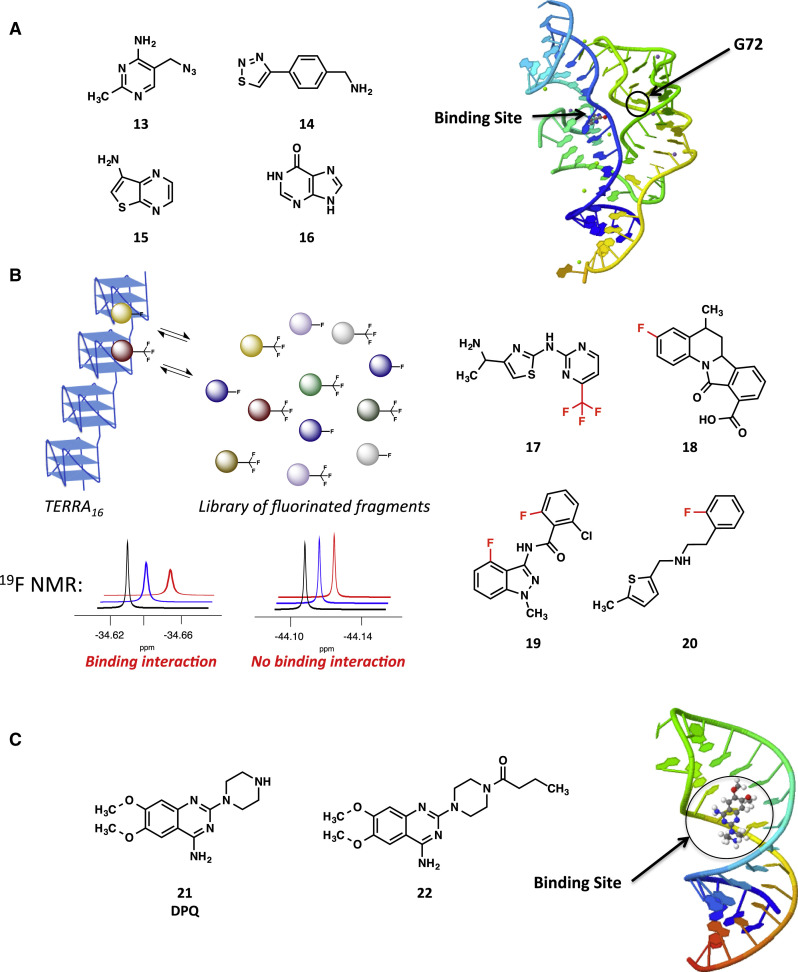
For example, Cressina et al. (2011) reported a fragment-based approach for identifying novel thiamine pyrophosphate (TPP) riboswitch ligands. TPP riboswitches are located in the 5′ UTR of various bacterial mRNAs and regulate the expression of the thiMD operon by changing conformation in the presence of TPP. In this study, a library of 1,300 structurally and chemically diverse fragments that were “rule of three” compliant (molecular weight ≤300 Da; c log p ≤ 3; no more than three hydrogen-bond donors and acceptors) and had ≥95% purity and ≥1 mM aqueous solubility was screened by equilibrium dialysis, whereby [3H]thiamine and fragments were placed in a separate chamber from the riboswitch and allowed to equilibrate. The concentration of the radiolabeled ligand was then measured to identify fragments that disrupt the thiamine-RNA interaction. From this screen, 20 hits were initially identified and further validated by NMR. Selectivity of the hits was also assessed using equilibrium dialysis against a structurally different lysine riboswitch present in Bacillus subtilis lysC. Several of the compounds (13–16, Figure 3A) were later investigated in the Weeks and Ferré-D’Amaré laboratories through in-depth biophysical studies to provide insight into the mechanism of binding of the fragments (Warner et al., 2014). Here, cocrystal structures of the E. coli thiM TPP riboswitch with the fragments revealed that the fragment occupies the same binding site as the aminopyridine of TPP. However, binding of the fragments resulted in a rearrangement of an unoccupied site, G72 (Figure 3A). Additional studies via SAXS and SHAPE showed that complete folding is achieved when TPP is bound and only partial folding when fragment 16 is bound. Taken together, these results suggest that the small-molecule fragments are competitive with TPP itself, but may stabilize different folding pathways.
Figure 3.
Fragment-Based Discovery of RNA-Binding Molecules
(A) Chemical structures of four TPP riboswitch-binding fragments (left) and the cocrystal structure of TPP riboswitch bound to 16, depicting a rearrangement of G72 (right) (Cressina et al., 2011, Warner et al., 2014).
(B) 19F-NMR fragment-based screening strategy to identify chemically diverse compounds that bind to TERRA (left), in which 19F-NMR signals are perturbed or broadened by compounds binding. Chemical structures of four representative hits that bind to TERRA2 are shown on the right. Fluorines are indicated in red (Garavis et al., 2014).
(C) Fragment-based screening hit (DPQ) and improved analog that bind to the influenza A promoter RNA. Also shown is a structure of the promoter RNA in complex with DPQ (Bottini et al., 2015, Lee et al., 2014).
The FBDD strategy has also been implemented to identify novel and chemically diverse fragments that bind to TERRA in the Campos-Olivas and González laboratories (Garavis et al., 2014). The r(UUAGGG)n sequence folds into G-quadruplexes that are required for telomere heterochromatin formation in cancer cells, and is therefore an attractive anticancer target. A library of 355 fluorinated fragment compounds was screened against RNA containing 16 r(UUAGGG) repeats (TERRA16) using 19F-NMR spectroscopy (Figure 3B). Spectra of the fragments were collected in the presence and absence of TERRA16 and compared to identify perturbations in the peak intensity and width upon addition of the RNA target. Of the 20 molecules identified by the initial screening, seven were further validated (representative hits, 17–20, are shown in Figure 3B). Six of these compounds were shown to interact with a shorter TERRA construct of two repeats (TERRA2) and four compounds demonstrated selectivity for TERRA over a duplex DNA and phenylalanine tRNA by 1H- and 19F-NMR, respectively. While all of the compounds interacted with the DNA analog of TERRA2, the compounds were shown to favor the parallel conformation, which is the predominant conformation in the RNA G-quadruplexes present in TERRA.
In the context of viral RNA, Göbel and coworkers reported a successful screen for fragments that inhibit the HIV Tat/TAR interaction using an FRET displacement assay (Zeiger et al., 2014). They validated fragment binding to TAR RNA using 1H-NMR and 1H-1H nuclear Overhauser effect spectroscopy (NOESY) experiments and identified several hits with affinities ranging from 40 to 20,000 μM. While Tat/TAR is a well-studied target, new viral RNAs have also been evaluated by fragment-based approaches. A powerful example of this has been the study of genomic elements of the influenza A virus. The influenza A viral genome contains a promoter region, which is a highly conserved sequence located on both the 5′ and 3′ termini that folds into a partial duplex and binds to the RNA-dependent RNA polymerase (RdRp). This promoter region has been considered an attractive drug target, and previous studies have demonstrated that neomycin binds with a submicromolar affinity (Kim et al., 2012). Subsequently, the Varani and Choi groups reported a fragment-based screen for small molecules that bind to the influenza A virus promoter (Lee et al., 2014). A library of 4,279 fragments was evaluated by RNA-observed 1H-NMR. Through this screen, 6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine (DPQ, 21) was identified and studied as an inhibitor (Figure 3C). DPQ has an affinity of 61 μM for the hairpin and an IC50 of 549 μM against influenza A. The binding mode of DPQ was investigated by NMR, and the structure revealed that the compound binds to the promoter in the major groove of its internal loop. Inhibition may occur due to a structural change that prevents the bending of the helix and the association of RdRp subunits. Subsequent work focused on SAR studies with DPQ to improve its cellular activity against influenza (Bottini et al., 2015). Since the piperazinyl secondary amine was not involved in any significant contacts (Figure 3C), 15 analogs with modifications to this group were prepared. Of these compounds, 22 displayed an improved antiviral IC50 of 44 μM while maintaining a KD value comparable with that of DPQ. Although fragment-based approaches have yet to yield a selective, high-affinity RNA binder, going forward it is probable that these approaches will play a large role in assessing the druggability of newly discovered RNA structures. Thus, the value of fragment-based approaches may be not only in developing leads for specific RNA structures, but also in helping to decide which RNA structures are druggable at all.
Small-Molecule Microarray Screening
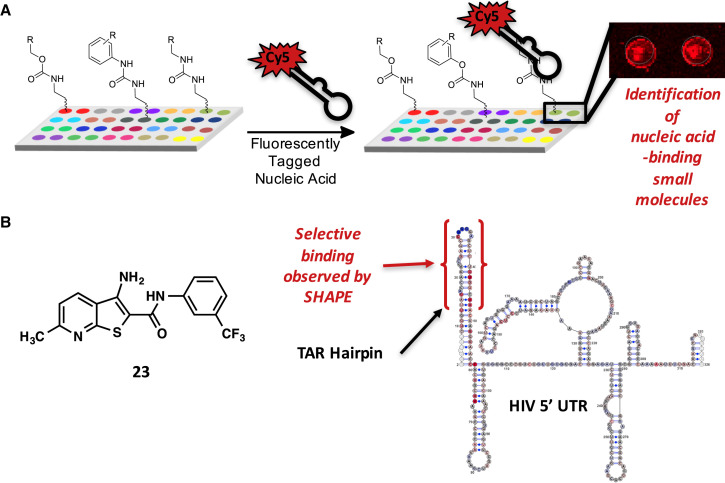
The small-molecule microarray (SMM) approach was originally developed by Schreiber for the discovery of ligands for “undruggable” proteins such as transcription factors and has been widely applied in that field (Bradner et al., 2006a, Bradner et al., 2006b, Hergenrother et al., 2000, Hong et al., 2014, Kawasumi et al., 2005, Koehler et al., 2003). In this technique, small molecules are spatially arrayed with a robotic microarrayer and covalently linked to a glass surface. Next, the array is incubated with a fluorescently labeled biomolecule. Arrays are imaged using a fluorescence scanner, and fluorescence intensity is quantified for each spot on the array. A statistical analysis reveals spots with large increases in fluorescence upon incubation, corresponding with discrete molecular interactions between the oligonucleotide and associated small molecule.
Our laboratory has advanced the use of SMMs as a platform for the identification of small molecules that bind to structured nucleic acids, building on the original work that employed aminoglycosides (Figure 4A) (Bryan and Wong, 2004, Disney et al., 2004). First, we assembled a library of 20,000 drug-like compounds, each of which contains an amine or alcohol group to covalently react with the array surface. This library can be screened in less than a day with little optimization. In one example, we were able to identify a small molecule that binds to the HIV TAR hairpin (Sztuba-Solinska et al., 2014). A fluorescently labeled TAR hairpin was screened against our library on the SMM platform, and three compounds were identified that bound to TAR but not other oligonucleotides, including an miR-21 hairpin and three distinct DNA sequences. A derivative of one of these compounds (23) bound reversibly to the TAR hairpin with a KD of 2 μM (Figure 4B). SHAPE profiling was used to probe compound binding in the context of the entire HIV 5′ UTR, and ligand binding could be mapped specifically to the TAR hairpin and not other structural elements (Figure 4B). Thus, microarrays are useful to identify potent, noncationic compounds that bind selectively to RNA structures even as simple as hairpins in the context of complex structures such as UTRs. In a more recent study, we used the SMM platform to identify molecules that selectively bind to the DNA G-quadruplex responsible for controlling the expression of MYC, an oncogene that is dysregulated in many cancers (Felsenstein et al., 2015). Given the prevalence of quadruplexes in the promoter regions of oncogenes (Ohnmacht and Neidle, 2014) and in mRNA, it is highly likely that SMMs will be a useful technology for identifying selective inhibitors of RNA quadruplexes as well.
Figure 4.
Use of Small-Molecule Microarrays to Identify RNA-Binding Molecules
(A) Small-molecule microarray screening approach.
(B) Structure of a molecule discovered by SMM screening to bind specifically to the HIV TAR hairpin, but not other structural elements of the HIV 5′ UTR (Sztuba-Solinska et al., 2014).
Dynamic Combinatorial Screening
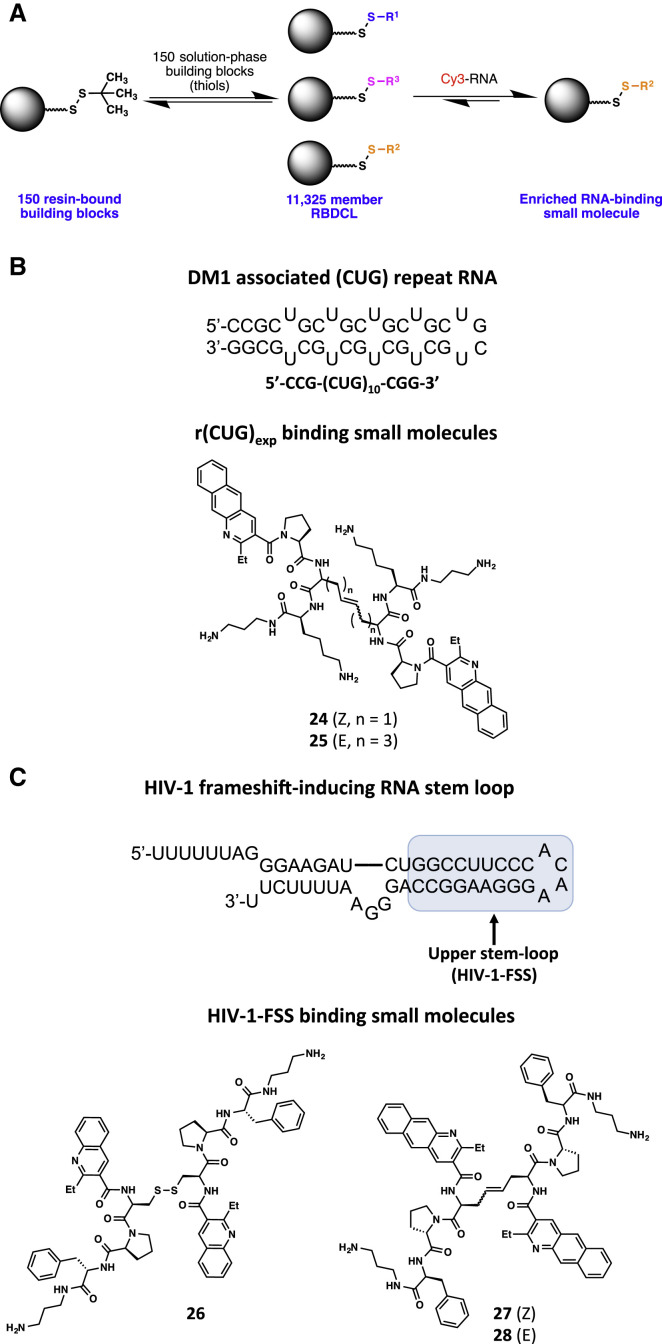
Dynamic combinatorial chemistry (DCC) is a powerful method of evolution-driven small-molecule discovery. Using DCC, small-molecule fragments can undergo recombination in the presence of a target of interest, producing substantial chemical diversity, thereby allowing selection and amplification of the highest-affinity binders through an equilibrium shift. A resin-bound DCC (RBDCC) approach was used by the Miller laboratory to screen a library of 11,325 compounds for small molecules that bind to r(CUG)exp involved in DM1 (Gareiss et al., 2008). The library was created from 150 resin-bound, cysteine-containing peptides and an identical set of solution-phase peptides, where formation of a disulfide bond between the resin-bound and solution-phase cysteines provides the combinatorial library (Figure 5A). The library was screened with a fluorescently labeled r(CUG)10, and four compounds were identified with low micromolar binding affinities for r(CUG)10. These compounds inhibited the (CUG)109-MBNL1 binding interaction with Kis ranging from 2.8 to 3.8 μM in an enzyme fragment complementation assay. Second-generation binders (24 and 25, Figure 5B) showed enhanced selectivity by surface plasmon resonance with 25 demonstrating a significant preference for (CUG)10 over (CCUG)10 RNA, a 38-fold preference versus (CAG)10, no measurable binding to a duplex CUG-CAG sequence, and an 89-fold reduction in binding to HIV-1 FSS RNA. These improved compounds blocked DM1-associated translational defects in cell-culture models, and, importantly, two compounds (24 and 25) successfully improved alternative pre-mRNA splicing defects in a DM1 mouse model, suggesting the therapeutic application of a small molecule that targets r(CUG)exp (Ofori et al., 2012).
Figure 5.
Dynamic Combinatorial Approaches to Identify RNA-Binding Molecules
Generic depiction of a dynamic combinatorial chemistry approach (A). Targets for dynamic combinatorial chemistry include both (B) DM1-associated (CUG) repeat RNA and (C) the HIV-1 frameshift-inducing RNA stem loop (Gareiss et al., 2008, Hilimire et al., 2015, Ofori et al., 2014). RBDCL, resin-bound dynamic combinatorial library.
RBDCC was further applied by the Miller laboratory to the discovery of compounds that bind to an RNA target that regulates frameshifting in HIV, the precise control of which is crucial for viral proliferation (McNaughton et al., 2007). The production of the essential Gag-Pol polyprotein in HIV requires a −1 nucleotide ribosomal frameshift that is in part directed by a highly conserved downstream RNA stem loop. RBDCC with a library of 11,325 members was used to identify a set of disulfide-containing peptides that bind to the HIV-1 FSS RNA (Figure 5C). The lead compound (26) had an affinity of 4.1 ± 2.4 μM for HIV-1 FSS as determined by surface plasmon resonance and a solution-phase KD of 0.35 ± 1.1 μM by fluorescence titration (McNaughton et al., 2007). Furthermore, the presence of either excess competitor yeast tRNA or total yeast RNA had no effect on the binding affinity, indicating good selectivity (Palde et al., 2010). No saturable binding was observed to a DNA homolog of the HIV-1 FSS or to unrelated RNA hairpins, and single or multiple mutations in the loop region of the HIV-1 FSS caused 2- and 4-fold reductions in the binding, respectively. Optimization of 26 produced an olefin bioisostere that had enhanced biostability and comparable affinity (Palde et al., 2010). Additional SAR efforts produced high-affinity binders (27 and 28) that altered frameshifting in HEK293FT cells and strongly inhibited viral infectivity in a pseudotyped HIV assay (Ofori et al., 2014). A series of N-methyl derivatives also bind the HIV-1 FSS RNA with low nanomolar affinity and high selectivity. These compounds readily penetrated cell membranes and inhibited infectivity in a pseudotyped HIV assay, which correlated with compound-induced changes in the expression of the Gag-Pol polyprotein (Hilimire et al., 2015). These two targets demonstrate that RBDCC is a powerful approach to identifying potent, selective, biologically active RNA-binding small molecules by leveraging the power of selection approaches.
Phenotypic Assays
In addition to the biochemical and biophysical approaches discussed above, phenotypic assays have also shown great promise for the discovery of biologically active, RNA-binding small molecules. These approaches use a phenotypic readout, such as a reporter gene assay, to report on the activity of a particular RNA or RNA-associated pathway in intact cells. For example, the Deiters laboratory has demonstrated the use of reporter gene assays to identify compounds that act against specific miRNAs, including miR-21 (Gumireddy et al., 2008, Naro et al., 2015) and miR-122 (Young et al., 2010). However, it is not clear whether these compounds bind directly to the miRNA itself or to another intracellular target. Another prominent example of phenotypic screening comes from the study of spinal muscular atrophy (SMA), a life-threatening motor neuron disease that is caused by the deficiency of the survival of motor neuron (SMN) protein (Palacino et al., 2015). In SMA cells, the lack of fully functional SMN protein is a consequence of homozygous deletion of the SMN1 gene. Both the SMN1 and SMN2 genes can produce the SMN protein. Although the SMN2 gene is able to partially compensate for low levels of the SMN protein, a single-nucleotide transition in SMN2 results in the exclusion of exon 7 and ultimately in a further decrease in full-length (FL) SMN mRNA and protein production. Researchers at the Novartis Institutes for Biomedical Research utilized a phenotypic assay to screen for small molecules with the ability to reduce exclusion of exon 7 and increase FL-SMN protein levels. Two SMN2 reporter genes, FL and Δ7, were designed within the NSC34 motor neuron cell line to indicate either exon 7 inclusion or exclusion, respectively (Figure 6A). Compounds from the Novartis compound library (∼1.4 × 106 compounds) were screened against the two reporter genes, and those that gave complementary results reporting a simultaneous increase in FL-SMN and a decrease in Δ7 were reported as hits, leading to the identification of NVS-SM2 (29, Figure 6A). Gene-expression analysis by RNA sequencing (RNA-seq) revealed that the compound regulates a discrete set of splicing variants, indicating fairly selective splicing modulation. Furthermore, 29 demonstrated promising in vivo activity, while NVS-SM3 (30), the structure of which closely resembles that of NVS-SM2, was inactive (Figure 6A). Total correlation spectroscopy NMR and surface plasmon resonance confirmed that 30 binds directly to the U1 small nuclear ribonucleoprotein (snRNP) 5′ splice site (ss), while computational modeling was used to suggest a binding mode of the compound to the U1 snRNP-SMN pre-mRNA (Figure 6A). Taken together, these results suggest a model in which NVS-SM2 functions by binding near the nGA site, stabilizing the U1 snRNP-SMN exon 7 5′ ss complex, and therefore enabling exon 7 inclusion. It is also of note that this work is not the first example of SMN2 splicing inhibitors. In an earlier example, a team from PTC Therapeutics and Hoffmann-La Roche discovered a series of compounds that display excellent in vivo activity in modulating the splicing of SMN2 and potential for the treatment of SMA (Naryshkin et al., 2014). These molecules display remarkably high specificity as measured by RNA-seq and oral bioavailability in a mouse model for SMA. It is speculated that these molecules interact with specific RNA structures or RNA-protein complexes within the SMN2 pre-mRNA, although the specific binding targets of these molecules have yet to be identified.
Figure 6.
Use of Phenotypic Assays to Identify RNA-Binding Molecules
(A) Schematic of a reporter gene assay that utilizes luciferase expression to measure exon 7 exclusion/inclusion in SM2 splicing. Chemical structures of NVS-SM2 and its inactive analog NVS-SM3. Putative binding site of NVS-SM2 within the crystal structure of the U1 snRNP-SMN pre-mRNA with rainbow coloring indicating 5′ (blue) to 3′ (red) (bottom) (Palacino et al., 2015).
(B) Depiction of a whole-cell phenotypic screening assay (top left) to identify inhibitors of riboflavin biosynthesis (right). In the absence of riboflavin, ribocil produces a clear zone as a result of growth inhibition. Cocrystal structure of the FMN riboswitch bound to ribocil B (bottom left) (Howe et al., 2015).
In another example, researchers at the Merck Research Laboratories used a phenotypic assay to discover a riboswitch-binding small molecule (Howe et al., 2015). The concept of targeting riboswitches has been a long-standing goal since their initial discovery in 2002 (Blount and Breaker, 2006, Winkler et al., 2002), and most approaches have focused on redesigning cognate ligands to develop improved inhibitors, as described above. In this work, a small molecule deemed ribocil was reported as an inhibitor of the FMN riboswitch, a structured, metabolite-responsive element in the promoter region of the ribB gene. Due to its role in riboflavin biosynthesis, the FMN riboswitch is essential for bacterial growth, and is therefore an attractive target for antibacterial activity, as previously demonstrated (Blount et al., 2015). Deletion strains missing two genes that play a role in earlier steps of riboflavin biosynthesis, ribA and ribB, were shown to have a significant reduction in bacterial burden compared with the wild-type control. An internal library of ∼57,000 small molecules was screened in a phenotypic assay to identify lead compounds that exhibit such antibacterial effects and produce a clear zone of growth inhibition (Figure 6B). One hit compound, racemic ribocil, caused not only a reduction in bacterial burden but also a reduction in the levels of FAD and FMN, effects that had also been observed in deletion strains. Further investigation showed only one of the enantiomers of ribocil, ribocil B (31) was active, whereas ribocil A (32) was inactive. The specific target interactions were determined by identifying 19 ribocil-resistant mutants, all of which contained mutations in the FMN riboswitch within the ribB gene, pointing to this structured RNA as the small-molecule target. Furthermore, ribocil displayed selective microbiological activity against E. coli MB5746 possessing either the native FMN riboswitch or orthologous FMN aptamers (which was suppressed by the addition of exogenous riboflavin), and was inactive against yeast and human cells that lack the cognate target. The Fusobacterium nucleatum FMN aptamer was cocrystallized with ribocil to identify the key interactions between the riboswitch and the ligand (Figure 6B). Remarkably, ribocil appears to make many of the same interactions with the riboswitch as the cognate ligand, and despite having no apparent structural similarity to FMN itself, it has a KD of 16 nM. The discovery of ribocil is a robust demonstration that phenotypic screens are an excellent way to identify drug-like chemical structures that bind to RNA.
Conclusion
In summary, the increased understanding of RNA structure and function is likely to lead to a broad variety of potential therapeutic targets for small molecules. The continued development of technologies for druggability assessment and inhibitor discovery against new RNA targets will be invaluable. For example, targeting a riboswitch, which has evolved to bind to a small-molecule metabolite, is likely be a challenge very different from targeting an lncRNA that functions by sequestering proteins through protein-RNA interactions. In fact it is probable that like proteins, different RNAs will be of variable difficulty as targets for small-molecule ligands, regardless of their biological or pharmacological significance. While early results suggest that it is possible to design or identify small molecules that modulate the function of at least some RNAs, many challenges remain, and the question of which RNAs are druggable is far from settled. Here the application of fragment-based approaches may be useful, as the hit rates of fragment-based screens have been used to assess the druggability of protein targets (Zhou and Huang, 2015). Another challenge will be overcoming the problem of specificity and selectivity, which remains a major barrier for RNA-binding molecules. For example, there is currently no general method to probe transcriptome-wide binding of a small molecule, and such an advance would be transformative. However, the implementation of structure-based and high-throughput screening methods described here have proved lucrative at identifying new biologically active small-molecule scaffolds that bind RNA with both high affinity and some specificity. Many of these recently developed small molecules are structurally distinct from historic RNA-binding scaffolds, display good physicochemical properties, and have proved active in cell-based and animal models of disease, supporting the role of RNAs as a therapeutic target. The continued success of these technologies will provide new openings and opportunities for the development of innovative therapeutics that target RNA in the upcoming years.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, the Center for Cancer Research, and the National Cancer Institute, NIH (1 ZIA BC011585 02).
References
- Arenz S., Wilson D.N. Blast from the past: reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol. Cell. 2016;61:3–14. doi: 10.1016/j.molcel.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Barros S.A., Chenoweth D.M. Recognition of nucleic acid junctions using triptycene-based molecules. Angew. Chem. Int. Ed. Engl. 2014;53:13746–13750. doi: 10.1002/anie.201407061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros S.A., Chenoweth D.M. Triptycene-based small molecules modulate (CAG). (CTG) repeat junctions. Chem. Sci. 2015;6:4752–4755. doi: 10.1039/c5sc01595b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros S.A., Yoon I., Chenoweth D.M. Modulation of the E. coli rpoH temperature sensor with triptycene-based small molecules. Angew. Chem. Int. Ed. Engl. 2016;55:8258–8261. doi: 10.1002/anie.201601626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros S.A., Yoon I., Suh S.E., Chenoweth D.M. Bridgehead-substituted triptycenes for discovery of nucleic acid junction binders. Org. Lett. 2016;18:2423–2426. doi: 10.1021/acs.orglett.6b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount K.F., Breaker R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Blount K., Puskarz I., Penchovsky R., Breaker R. Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 2006;3:77–81. doi: 10.4161/rna.3.2.3102. [DOI] [PubMed] [Google Scholar]
- Blount K.F., Wang J.X., Lim J., Sudarsan N., Breaker R.R. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- Blount K.F., Megyola C., Plummer M., Osterman D., O'Connell T., Aristoff P., Quinn C., Chrusciel R.A., Poel T.J., Schostarez H.J. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob. Agents Chemother. 2015;59:5736–5746. doi: 10.1128/AAC.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D., Thermann R., Ostareck-Lederer A., Lewis J.D., Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Bottini A., De S.K., Wu B., Tang C., Varani G., Pellecchia M. Targeting influenza A virus RNA promoter. Chem. Biol. Drug Des. 2015;86:663–673. doi: 10.1111/cbdd.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner J.E., McPherson O.M., Koehler A.N. A method for the covalent capture and screening of diverse small molecules in a microarray format. Nat. Protoc. 2006;1:2344–2352. doi: 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- Bradner J.E., McPherson O.M., Mazitschek R., Barnes-Seeman D., Shen J.P., Dhaliwal J., Stevenson K.E., Duffner J.L., Park S.B., Neuberg D.S. A robust small-molecule microarray platform for screening cell lysates. Chem. Biol. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Breaker R.R., Joyce G.F. The expanding view of RNA and DNA function. Chem. Biol. 2014;21:1059–1065. doi: 10.1016/j.chembiol.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan M.C., Wong C.H. Aminoglycoside array for the high-throughput analysis of small molecule-RNA interactions. Tetrahedron Lett. 2004;45:3639–3642. [Google Scholar]
- Carnevali M., Parsons J., Wyles D.L., Hermann T. A modular approach to synthetic RNA binders of the hepatitis C virus internal ribosome entry site. Chembiochem. 2010;11:1364–1367. doi: 10.1002/cbic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S.W., Gruhl F., Mattick J.S., Dinger M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Z., Sobczak K., Hoskins J., Southall N., Marugan J.J., Zheng W., Thornton C.A., Austin C.P. Two high-throughput screening assays for aberrant RNA-protein interactions in myotonic dystrophy type 1. Anal. Bioanal. Chem. 2012;402:1889–1898. doi: 10.1007/s00216-011-5604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Zhao B.S., He C. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney J.L., Hoskins J., Rzuczek S.G., Thornton C.A., Disney M.D. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney J.L., Stepniak-Konieczna E., Tran T., Yildirim I., Park H., Chen C.Z., Hoskins J., Southall N., Marugan J.J., Patnaik S. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat. Commun. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T.A., Wan L.L., Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressina E., Chen L.H., Abell C., Leeper F.J., Smith A.G. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem. Sci. 2011;2:157–165. [Google Scholar]
- Davidson A., Begley D.W., Lau C., Varani G. A small-molecule probe induces a conformation in HIV TAR RNA capable of binding drug-like fragments. J. Mol. Biol. 2011;410:984–996. doi: 10.1016/j.jmb.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.R., Seth P.P. Therapeutic targeting of HCV internal ribosomal entry site RNA. Antivir. Chem. Chemother. 2011;21:117–128. doi: 10.3851/IMP1693. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov S.M., Johnston-Cox H., Weng Y.H., Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew. Chem. Int. Ed. Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- Dibrov S.M., Ding K., Brunn N.D., Parker M.A., Bergdahl B.M., Wyles D.L., Hermann T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. USA. 2012;109:5223–5228. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding K., Wang A., Boerneke M.A., Dibrov S.M., Hermann T. Aryl-substituted aminobenzimidazoles targeting the hepatitis C virus internal ribosome entry site. Bioorg. Med. Chem. Lett. 2014;24:3113–3117. doi: 10.1016/j.bmcl.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney M.D., Magnet S., Blanchard J.S., Seeberger P.H. Aminoglycoside microarrays to study antibiotic resistance. Angew. Chem. Int. Ed. Engl. 2004;43:1591–1594. doi: 10.1002/anie.200353236. [DOI] [PubMed] [Google Scholar]
- Disney M.D., Labuda L.P., Paul D.J., Poplawski S.G., Pushechnikov A., Tran T., Velagapudi S.P., Wu M., Childs-Disney J.L. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J. Am. Chem. Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- Disney M.D., Yildirim I., Childs-Disney J.L. Methods to enable the design of bioactive small molecules targeting RNA. Org. Biomol. Chem. 2014;12:1029–1039. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Fang X., Stagno J.R., Bhandari Y.R., Zuo X., Wang Y.X. Small-angle X-ray scattering: a bridge between RNA secondary structures and three-dimensional topological structures. Curr. Opin. Struct. Biol. 2015;30:147–160. doi: 10.1016/j.sbi.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K.M., Saunders L.B., Simmons J.K., Leon E., Calabrese D.R., Zhang S., Michalowski A., Gareiss P., Mock B.A., Schneekloth J.S., Jr. Small molecule microarrays enable the identification of a selective, quadruplex-binding inhibitor of MYC expression. ACS Chem. Biol. 2015;11:139–148. doi: 10.1021/acschembio.5b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Gu H., Sudarsan N., Hayakawa Y., Hyodo M., Breaker R.R. Identification of ligand analogues that control c-di-GMP riboswitches. ACS Chem. Biol. 2012;7:1436–1443. doi: 10.1021/cb300138n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka-Marciniak P., Urbanek M.O., Krzyzosiak W.J. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol. Chem. 2012;393:1299–1315. doi: 10.1515/hsz-2012-0218. [DOI] [PubMed] [Google Scholar]
- Garavis M., Lopez-Mendez B., Somoza A., Oyarzabal J., Dalvit C., Villasante A., Campos-Olivas R., Gonzalez C. Discovery of selective ligands for telomeric RNA G-quadruplexes (TERRA) through 19F-NMR based fragment screening. ACS Chem. Biol. 2014;9:1559–1566. doi: 10.1021/cb500100z. [DOI] [PubMed] [Google Scholar]
- Gareiss P.C., Sobczak K., McNaughton B.R., Palde P.B., Thornton C.A., Miller B.L. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J. Am. Chem. Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmann R.F., Gopal A., Athavale S.S., Knobler C.M., Gelbart W.M., Harvey S.C. Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA. 2015;21:877–886. doi: 10.1261/rna.047506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.D., Reyes F.E., Edwards A.L., Batey R.T. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L.R., Disney M.D. Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. Small-molecule inhibitors of microRNA miR-21 function. Angew. Chem. Int. Ed. Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga C.L., Velagapudi S.P., Strivelli J.R., Yang W.Y., Disney M.D., Phinney D.G. Small molecule inhibition of miR-544 biogenesis disrupts adaptive responses to hypoxia by modulating ATM-mTOR signaling. ACS Chem. Biol. 2015;10:2267–2276. doi: 10.1021/acschembio.5b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer M.J., Vaughn I.W., McManus M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Alfonzo J.D. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem. Biol. 2014;21:174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergenrother P.J., Depew K.M., Schreiber S.L. Small-molecule microarrays: covalent attachment and screening of alcohol-containing small molecules on glass slides. J. Am. Chem. Soc. 2000;122:7849–7850. [Google Scholar]
- Hermann T. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Hilimire T.A., Bennett R.P., Stewart R.A., Garcia-Miranda P., Blume A., Becker J., Sherer N., Helms E.D., Butcher S.E., Smith H.C. N-methylation as a strategy for enhancing the affinity and selectivity of RNA-binding peptides: application to the HIV-1 frameshift-stimulating RNA. ACS Chem. Biol. 2015;11:88–94. doi: 10.1021/acschembio.5b00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.A., Neel D.V., Wassaf D., Caballero F., Koehler A.N. Recent discoveries and applications involving small-molecule microarrays. Curr. Opin. Chem. Biol. 2014;18:21–28. doi: 10.1016/j.cbpa.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J.A., Wang H., Fischmann T.O., Balibar C.J., Xiao L., Galgoci A.M., Malinverni J.C., Mayhood T., Villafania A., Nahvi A. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- Kawasumi M., Bradner J., Kim Y., Koehler A., Mazitschek R., Schreiber S., Nghiem P. Small molecule microarrays to discover compounds that modulate cell cycle checkpoint function. J. Invest. Dermatol. 2005;124:A39. [Google Scholar]
- Kim J.N., Blount K.F., Puskarz I., Lim J., Link K.H., Breaker R.R. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem. Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee M.K., Ko J., Park C.J., Kim M., Jeong Y., Hong S., Varani G., Choi B.S. Aminoglycoside antibiotics bind to the influenza A virus RNA promoter. Mol. Biosyst. 2012;8:2857–2859. doi: 10.1039/c2mb25333j. [DOI] [PubMed] [Google Scholar]
- Koehler A.N., Shamji A.F., Schreiber S.L. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J. Am. Chem. Soc. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- Le Grice S.F. Targeting the HIV RNA genome: high-hanging fruit only needs a longer ladder. Curr. Top Microbiol. Immunol. 2015;389:147–169. doi: 10.1007/82_2015_434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.R., Blount K.F., Breaker R.R. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Bottini A., Kim M., Bardaro M.F., Jr., Zhang Z., Pellecchia M., Choi B.S., Varani G. A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem. Commun. (Camb) 2014;50:368–370. doi: 10.1039/c3cc46973e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mason C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lunse C.E., Schmidt M.S., Wittmann V., Mayer G. Carba-sugars activate the glmS-riboswitch of Staphylococcus aureus. ACS Chem. Biol. 2011;6:675–678. doi: 10.1021/cb200016d. [DOI] [PubMed] [Google Scholar]
- Luo Y., Disney M.D. Bottom-up design of small molecules that stimulate exon 10 skipping in mutant MAPT pre-mRNA. Chembiochem. 2014;15:2041–2044. doi: 10.1002/cbic.201402069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansjo M., Johansson J. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 2011;8:674–680. doi: 10.4161/rna.8.4.15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClorey G., Wood M.J. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr. Opin. Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- McNaughton B.R., Gareiss P.C., Miller B.L. Identification of a selective small-molecule ligand for HIV-1 frameshift-inducing stem-loop RNA from an 11,325 member resin bound dynamic combinatorial library. J. Am. Chem. Soc. 2007;129:11306–11307. doi: 10.1021/ja072114h. [DOI] [PubMed] [Google Scholar]
- Naro Y., Thomas M., Stephens M.D., Connelly C.M., Deiters A. Aryl amide small-molecule inhibitors of microRNA miR-21 function. Bioorg. Med. Chem. Lett. 2015;25:4793–4796. doi: 10.1016/j.bmcl.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Naryshkin N.A., Weetall M., Dakka A., Narasimhan J., Zhao X., Feng Z., Ling K.K., Karp G.M., Qi H., Woll M.G. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- Nguyen L., Luu L.M., Peng S., Serrano J.F., Chan H.Y., Zimmerman S.C. Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J. Am. Chem. Soc. 2015;137:14180–14189. doi: 10.1021/jacs.5b09266. [DOI] [PubMed] [Google Scholar]
- Ofori L.O., Hoskins J., Nakamori M., Thornton C.A., Miller B.L. From dynamic combinatorial 'hit' to lead: in vitro and in vivo activity of compounds targeting the pathogenic RNAs that cause myotonic dystrophy. Nucleic Acids Res. 2012;40:6380–6390. doi: 10.1093/nar/gks298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofori L.O., Hilimire T.A., Bennett R.P., Brown N.W., Jr., Smith H.C., Miller B.L. High-affinity recognition of HIV-1 frameshift-stimulating RNA alters frameshifting in vitro and interferes with HIV-1 infectivity. J. Med. Chem. 2014;57:723–732. doi: 10.1021/jm401438g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht S.A., Neidle S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014;24:2602–2612. doi: 10.1016/j.bmcl.2014.04.029. [DOI] [PubMed] [Google Scholar]
- Ott E., Stolz J., Lehmann M., Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- Palacino J., Swalley S.E., Song C., Cheung A.K., Shu L., Zhang X., Van Hoosear M., Shin Y., Chin D.N., Keller C.G. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- Palde P.B., Ofori L.O., Gareiss P.C., Lerea J., Miller B.L. Strategies for recognition of stem-loop RNA structures by synthetic ligands: application to the HIV-1 frameshift stimulatory sequence. J. Med. Chem. 2010;53:6018–6027. doi: 10.1021/jm100231t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim Y.G., Park H.J. Identification of RNA pseudoknot-binding ligand that inhibits the -1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- Parsons J., Castaldi M.P., Dutta S., Dibrov S.M., Wyles D.L., Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 2009;5:823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J., Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- Pushechnikov A., Lee M.M., Childs-Disney J.L., Sobczak K., French J.M., Thornton C.A., Disney M.D. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade N., Boehringer D., Leibundgut M., van den Heuvel J., Ban N. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat. Commun. 2015;6:7646. doi: 10.1038/ncomms8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Luke B. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015;22:853–858. doi: 10.1038/nsmb.3078. [DOI] [PubMed] [Google Scholar]
- Ritchie D.B., Soong J., Sikkema W.K., Woodside M.T. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J. Am. Chem. Soc. 2014;136:2196–2199. doi: 10.1021/ja410344b. [DOI] [PubMed] [Google Scholar]
- Rynearson K.D., Charrette B., Gabriel C., Moreno J., Boerneke M.A., Dibrov S.M., Hermann T. 2-Aminobenzoxazole ligands of the hepatitis C virus internal ribosome entry site. Bioorg. Med. Chem. Lett. 2014;24:3521–3525. doi: 10.1016/j.bmcl.2014.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzuczek S.G., Southern M.R., Disney M.D. Studying a drug-like, RNA-Focused small molecule library identifies compounds that inhibit RNA toxicity in myotonic dystrophy. ACS Chem. Biol. 2015;10:2706–2715. doi: 10.1021/acschembio.5b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon L., Yang S., Al-Hashimi H.M. Advances in the determination of nucleic acid conformational ensembles. Annu. Rev. Phys. Chem. 2014;65:293–316. doi: 10.1146/annurev-physchem-040412-110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2015;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P.P., Miyaji A., Jefferson E.A., Sannes-Lowery K.A., Osgood S.A., Propp S.S., Ranken R., Massire C., Sampath R., Ecker D.J. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J. Med. Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- Shortridge M.D., Varani G. Structure based approaches for targeting non-coding RNAs with small molecules. Curr. Opin. Struct. Biol. 2015;30:79–88. doi: 10.1016/j.sbi.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K., Doudna J.A., Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer A.C., Frank A.T., Kratz J.D., Swanson M.D., Gonzalez-Hernandez M.J., Lee J., Andricioaei I., Markovitz D.M., Al-Hashimi H.M. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztuba-Solinska J., Shenoy S.R., Gareiss P., Krumpe L.R., Le Grice S.F., O'Keefe B.R., Schneekloth J.S., Jr. Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J. Am. Chem. Soc. 2014;136:8402–8410. doi: 10.1021/ja502754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.R., Hergenrother P.J. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Velagapudi S.P., Seedhouse S.J., Disney M.D. Structure-Activity Relationships through Sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew. Chem. Int. Ed. Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S.P., Gallo S.M., Disney M.D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner K.D., Homan P., Weeks K.M., Smith A.G., Abell C., Ferre-D'Amare A.R. Validating fragment-based drug discovery for biological RNAs: lead fragments bind and remodel the TPP riboswitch specifically. Chem. Biol. 2014;21:591–595. doi: 10.1016/j.chembiol.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W., Jr., Swanstrom R., Burch C.L., Weeks K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Wong C.H., Nguyen L., Peh J., Luu L.M., Sanchez J.S., Richardson S.L., Tuccinardi T., Tsoi H., Chan W.Y., Chan H.Y. Targeting toxic RNAs that cause myotonic dystrophy type 1 (DM1) with a bisamidinium inhibitor. J. Am. Chem. Soc. 2014;136:6355–6361. doi: 10.1021/ja5012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Collier M., Loerke J., Ismer J., Schmidt A., Hilal T., Sprink T., Yamamoto K., Mielke T., Burger J. Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J. 2015;34:3042–3058. doi: 10.15252/embj.201592469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon I., Suh S.E., Barros S.A., Chenoweth D.M. Synthesis of 9-substituted triptycene building blocks for solid-phase diversification and nucleic acid junction targeting. Org. Lett. 2016;18:1096–1099. doi: 10.1021/acs.orglett.6b00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.D., Connelly C.M., Grohmann C., Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- Zeiger M., Stark S., Kalden E., Ackermann B., Ferner J., Scheffer U., Shoja-Bazargani F., Erdel V., Schwalbe H., Gobel M.W. Fragment based search for small molecule inhibitors of HIV-1 Tat-TAR. Bioorg. Med. Chem. Lett. 2014;24:5576–5580. doi: 10.1016/j.bmcl.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Huang N. Binding site druggability assessment in fragment-based drug design. Methods Mol. Biol. 2015;1289:13–21. doi: 10.1007/978-1-4939-2486-8_2. [DOI] [PubMed] [Google Scholar]
- Zhou S., Rynearson K.D., Ding K., Brunn N.D., Hermann T. Screening for inhibitors of the hepatitis C virus internal ribosome entry site RNA. Bioorg. Med. Chem. 2013;21:6139–6144. doi: 10.1016/j.bmc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]