Abstract
Lipid droplets are organelles found in most mammalian cells, as well as various plant tissues and yeast. They are composed of a core of neutral lipids surrounded by a membrane monolayer of phospholipids and cholesterol into which specific proteins are embedded. This unit provides protocols for isolating lipid droplets from mammalian cells by discontinuous density gradient centrifugation.
Keywords: Cell cultures, mammalian, preparation of lipid droplets from centrifugation, density-gradient centrifugation, isolation of lipid droplets
INTRODUCTION
Lipid droplets are organelles found in most cultured cells, particularly when the cells have been cultured in medium containing serum or exogenous fatty acids (see Fig. 3.15.1). Due to a relatively low protein content and the presence of a neutral lipid core composed of primarily triacylglycerol or cholesterol esters, lipid droplets are more buoyant than other cellular compartments and can be isolated easily by centrifugation. Brief low-speed centrifugation can rapidly float lipid droplets, but other cellular membranes and particularly mitochondria may adhere to the lipid droplets. This unit describes a method to isolate a highly purified lipid droplet fraction with minimal contaminating membranes (see Basic Protocol). The unit also describes an alternate method to disrupt cells while maintaining the integrity of lipid droplets (see Alternate Protocol), a method for loading cells with lipid (see Support Protocol 1), and two methods for solubilizing lipid droplet membrane proteins (see Support Protocols 2 and 3).
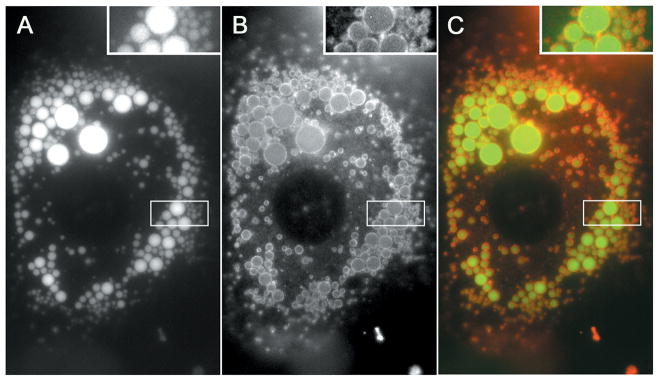
Figure 3.15.1.
Cultured 3T3-L1 adipocyte labeled with fluorescent fatty acids (A and C: green) and stained for perilipin (B and C: red). Cultured 3T3-L1 adipocytes were differentiated for 6 days with the addition of a BODIPY-labeled 12-carbon fatty acid (Molecular Probes®, ThermoFisher Scientific, D-3822, BODIPY® FL C12) for the final 18 hr. Cells were fixed with 2% paraformaldehyde in PBS prior to staining with guinea pig polyclonal antibody to perilipin (Fitzgerald Industries International, 20R-PP004 or Progen Biotechnik, GP29) followed by AlexaFluor 594 goat anti-guinea pig IgG (ThermoFisher Scientific, A-11076). Images were captured with a Zeiss Axioplan-2 microscope equipped with a Hamamatsu Orca CCD camera. For the color version of this figure go to http://www.currentprotocols.com.
BASIC PROTOCOL: ISOLATION OF LIPID DROPLETS FROM CULTURED CELLS BY DENSITY GRADIENT CENTRIFUGATION
Nuclei are removed by low-speed centrifugation and the density of the post-nuclear supernatant is adjusted with sucrose prior to flotation of the lipid droplets through a single discontinuous sucrose gradient. The lipid droplet fraction is characterized by immunoblotting of component proteins.
NOTE: Protease inhibitors (UNIT 3.4) may be included in any or all of the media at the discretion of the investigator.
NOTE: All solutions and glassware should be prechilled to 4°C before the procedure, and kept on ice throughout. Centrifuges, centrifuge rotors, and buckets should be precooled to the same temperature.
Materials
4 to 10 100-mm dishes containing confluent monolayer cells (1–2.5 × 107 cells)
Phosphate-buffered saline (PBS; APPENDIX 2A), ice cold
Hypotonic lysis medium (HLM; see recipe), ice cold
HLM containing 60% and 5% (w/w) sucrose (see recipes), ice cold
Rubber policeman or cell-scraper
15-ml plastic tubes with caps
Potter-Elvehjem tissue homogenizer with loose-fitting Teflon pestle (Wheaton), 4-ml capacity, 0.01 to 0.02 cm clearance
Low-speed refrigerated centrifuge with swinging-bucket rotor and appropriate centrifuge tubes
Beckman or Sorvall ultracentrifuge with SW41Ti or Th-641 swinging-bucket rotor
13.2-ml thin-walled polyallomer or polycarbonate ultracentrifuge tubes
Beckman tube slicer with two metal shim rings and two rubber rings to fit ultracentrifuge tubes
Additional reagents and equipment for SDS-PAGE (UNIT 6.1) and immunoblotting (UNIT 6.2)
Prepare cells
-
1
Wash cells two times with 10 ml ice-cold PBS per dish.
-
2
Using a rubber policeman or cell scraper, scrape cells from all dishes into 10 to 15 ml ice-cold PBS in a single 15-ml plastic tube with cap and pellet cells by centrifuging 10 min at 1000 × g, 4°C.
-
3
Remove the supernatant by aspiration with a pipet (to discard) and gently and thoroughly resuspend the cells in ice-cold HLM by pipetting the cells up-and-down using a pipet tip with a wide opening. Use a volume of about five times the cell pellet volume.
Total volume will be 1.5 to 2.5 ml. -
4
Incubate the suspended cells on ice for 10 min.
Homogenize cells
-
5
Transfer the resuspended cells to a Potter-Elvehjem tissue homogenizer on ice. Keeping the homogenizer on ice, insert the Teflon pestle and slowly homogenize the cells by six to eight gentle strokes with the hand-driven pestle. Transfer the homogenate to a 15-ml tube.
-
6
Centrifuge the cell lysate 10 min at 1000 × g, 4°C.
-
7
Collect the supernatant and the floating fat layer into a separate 15-ml tube.
-
8
Add 1/3 volume of ice-cold HLM containing 60% sucrose (final 20% sucrose), and mix by gentle pipetting using a pipet tip with a wide opening until aggregates of lipid droplets and membranes are finely and thoroughly dispersed.
Isolate lipid droplets
-
9
Layer density adjusted cell lysate into the bottom of a 13.2-ml ultracentrifuge tube for an SW41Ti rotor, or equivalent.
-
10
Gently layer 5 ml ice-cold HLM containing 5% sucrose over sample.
-
11
Gently add 5 to 6.5 ml ice-cold HLM over the sucrose layers to fill the tube.
-
12
Centrifuge tube 30 min at 28,000 × g, 4°C. Allow rotor to coast to a stop.
A balance tube containing equivalent volumes of the solutions and equal mass will be required if a single sample is prepared.
Collect lipid droplets
-
13
Remove ultracentrifuge tubes from ultracentrifuge rotor and place into Beckman tube slicer with blade placement 5 to 8 mm below the floating opaque lipid droplet layer. Slice tube firmly and steadily.
-
14
Collect the lipid droplet fraction from the top chamber of the tube slicer using ice-cold HLM to rinse residual lipid droplets from the sides of the tube and surface of the cutting blade.
Lipid droplet purity may be improved by additional wash steps; transfer the lipid droplet fraction to a microcentrifuge tube, centrifuge 10 minutes at maximum speed at 4°C and remove the infranatant from beneath the floating lipid droplet layer using a gel-loading pipette tip or wide gauge needle attached to a syringe. Resuspend lipid droplets in ice-cold HLM and repeat centrifugation step, as needed.
Characterize lipid droplets
-
15
Check recovery and purity of an aliquot of the lipid droplet fraction by SDS-PAGE (UNIT 6.1) of solubilized proteins (see Support Protocol 2) and immunoblotting (UNIT 6.2), assaying for immunoreactivity against lipid droplet–associated proteins including perilipin 2 (~47 kDa for mouse; previously called adipophilin or ADRP) for all types of cells except differentiated adipocytes, and perilipin 1A (~57 kDa for mouse) for differentiated adipocytes.
The sizes of the two proteins perilipin 2 and perilipin 1A provided here, ~47 kDa and ~57 kDa, respectively, pertain to mouse proteins. These protein sizes vary between species. -
16
Check for contamination of lipid droplet fractions with other membranes by immunoreactivity with antibodies specific for various intracellular membrane marker proteins.
ALTERNATE PROTOCOL: ISOLATION OF LIPID DROPLETS WITH LYSIS OF CELLS USING A CELL DISRUPTION BOMB
Nitrogen cavitation is particularly effective in lysing cells while keeping lipid droplets intact.
Additional Materials (also see Basic Protocol)
Suspended cells in HLM (see Basic Protocol, step 4)
45-ml cell disruption bomb (Parr)
15- and 50-ml plastic tubes
Transfer suspended cells in HLM into the chamber of a cell disruption bomb. Pressurize the chamber with nitrogen at 450 psi for 15 min.
Release the sample dropwise and collect in a 50-ml plastic tube.
Centrifuge tube 5 min at 100 × g, 4°C, to collect the sample at the bottom of the tube; transfer to a 15-ml plastic tube with cap.
Continue with Basic Protocol, step 6.
SUPPORT PROTOCOL 1: LIPID LOADING OF CULTURED CELLS
Many cultured cells store very low mass of neutral lipids in lipid droplets. To promote the synthesis of triacylglycerols and the formation of lipid droplets, the culture medium can be supplemented with 100 μM to 1 mM fatty acids complexed to albumin. Oleic acid is commonly used for lipid loading of cells because it is a good substrate for triacylglycerol biosynthesis (Coleman and Lee, 2004). The most critical step of the procedure is to assure complete complexation of fatty acids to the albumin to prevent detergent effects of unbound fatty acids. One mole of albumin may bind up to 7 mol of fatty acids, depending upon the acyl chain length of the fatty acid (Petitpas et al., 2001); the accompanying procedure guides complex formation of 6 mol of fatty acids to 1 mol of bovine serum albumin. The starting albumin must be fatty acid–free to avoid the addition of an excess of fatty acids. The formation of complexes is accompanied by a visible clearing of the solution; cloudy solutions contain aggregates of uncomplexed fatty acids.
Materials
Fatty acid–free bovine serum albumin
0.1 M Tris·Cl, pH 8.0
Oleic acid
50-ml screw-capped polypropylene tubes
Rotisserie shaker
0.2- or 0.45-μm filter unit
Dissolve 3.36 g fatty acid–free bovine serum albumin in 24 ml of 0.1 M Tris·Cl, pH 8.0.
-
Transfer 84 mg oleic acid into a clean 50-ml screw-capped tube using a pipet tip with a wide opening.
A wide-opening tip should be used because the oleic acid is viscous and difficult to pipet. -
Add 24 ml of albumin solution to the oleic acid, and mix using a rotisserie shaker, or by gentle inversion and swirling to avoid excessive foaming.
Complex formation is complete when cloudiness in the solution disappears and there is no visible uncomplexed oleic acid on the side or bottom of the tube. Sterile filter the solution through a 0.2- or 0.45-μm filter unit.
Add solution to culture medium to a final concentration of 100 μM to 1 mM oleic acid.
SUPPORT PROTOCOL 2: SOLUBILIZATION OF LIPID DROPLET–ASSOCIATED PROTEINS FOR IMMUNOBLOTTING
Lipid droplet fractions contain a low mass of protein, but high relative mass of neutral lipids, including either triacylglycerols or cholesterol esters, in addition to phospholipids. The high lipid content of the samples interferes with the resolution of proteins by SDS-PAGE. When the lipid droplet fractions are fresh and have not been frozen, the component proteins can be solubilized using detergent solutions with warming and sonication of the sample.
Materials
10% (w/v) sodium dodecyl sulfate (SDS; see recipe)
Fresh lipid droplet fraction (see Basic Protocol)
2× SDS sample buffer (for discontinuous systems; see UNIT 6.1)
Sonicating water bath with adjustable temperature
Vortex mixer
1.5-ml microcentrifuge tubes
Gel-loading pipet tip or wide-gauge needle attached to a small syringe
Additional reagents and equipment for a discontinuous SDS-PAGE gel (see UNIT 6.1)
-
Add 1 vol of 10% SDS to the fresh (not frozen) lipid droplet fraction in HLM. Incubate for at least 1 hr at 37°C in a sonicating water bath. Remove the sample every 5 to 10 min and agitate on a vortex mixer before returning to the bath.
Raising the temperature of the water bath to 60°C or extending the length of the incubation period may help to solubilize lipid droplet-associated proteins. Transfer the sample to a 1.5-ml microcentrifuge tube and microcentrifuge 10 min at maximum speed, room temperature.
-
Use a gel-loading pipet tip or a wide-gauge needle attached to a small syringe to collect the infranatant containing the solubilized proteins from beneath the floating lipid layer without removing any of the opaque floating lipid or any precipitated insoluble material from a visible pellet. Transfer to a new 1.5-ml microcentrifuge.
If the floating lipid layer becomes disrupted, repeat microcentrifugation to consolidate the lipid layer. Add an equivalent volume of 2× SDS sample buffer, and boil the sample for 10 min prior to loading onto a discontinuous SDS-PAGE gel (see UNIT 6.1).
SUPPORT PROTOCOL 3: DELIPIDATION AND SOLUBILIZATION OF LIPID DROPLET–ASSOCIATED PROTEINS
Alternatively, frozen lipid droplet fractions, or fractions containing excessive lipid can be delipidated using solvents, and the precipitated proteins solubilized in concentrated detergent solutions with warming and sonication. The extent of solvent delipidation required is dependent upon the lipid content of the sample; whereas most samples require only a single delipidation step, lipid droplets isolated from adipocytes and highly lipid-enriched cells require multiple solvent delipidation steps.
Materials
Acetone, −80°C and room temperature
Frozen lipid droplet fraction, thawed
1:1 (v/v) acetone/ether
Ether
2× SDS sample buffer (for discontinuous systems; see UNIT 6.1)
Extra reducing reagent (e.g., β-mercaptoethanol or dithiothreitol)
Polypropylene screw-capped centrifuge tubes (Sarstedt)
High-speed refrigerated centrifuge with Sorvall SS34 rotor and tube adapter sleeves, or equivalent
Sonicating water bath with adjustable temperature
1.5-ml microcentrifuge tubes
NOTE: All procedures using organic solvents should be carried out in a fume hood. Tubes should be tightly capped before removing samples from the fume hood for incubation or centrifugation steps. Glass pipets and storage containers should be used to transfer solvents, because disposable polystyrene laboratory pipets will dissolve in many organic solvents. Solvent waste should be disposed of in accordance with institutional policy.
Delipidate sample with acetone
-
1
Add at least 10 vol of cold acetone that has been stored at −80°C to the thawed or fresh lipid droplet fraction in a screw-capped polypropylene tube. Cap the tube and mix the sample thoroughly by inversion.
-
2
Incubate the sample overnight at −20°C, or for 4 hr on dry ice.
-
3
Mix the sample by inversion and centrifuge 1 hr at 4300 × g, 4°C.
-
4
Carefully remove acetone from the loose pellet by pouring off the solvent, or removing with a glass pipet. Discard the acetone in a suitable container.
If samples do not require additional delipidation
-
5a
Remove residual solvent residue from the pellet using a gentle stream of nitrogen in a fume hood. Continue to step 12.
If additional delipidation is required (adipocyte lipid droplet fractions and samples yielding large fluffy appearing pellets)
-
5b
Check the tube carefully for stress fractures. Add 10 vol of room temperature acetone to the protein pellet and vortex to mix. Centrifuge 30 min at 4300 × g, 4°C.
-
6b
Carefully remove acetone from the loose pellet by pouring off the solvent, or removing with a glass pipet. Discard the solvent in an appropriate container.
-
7b
Add 10 vol of 1:1 acetone/ether to the protein pellet at room temperature and vortex to mix. Centrifuge 30 min at 4300 × g, 4°C.
-
8b
Carefully remove the solvent from the loose pellet. Discard the solvent in an appropriate container.
-
9b
Add 10 vol ether to the pellet at room temperature and vortex to mix. Centrifuge 30 min at 4300 × g, 4°C.
-
10b
Carefully remove the solvent from the loose pellet. Discard the solvent in an appropriate container.
-
11b
Remove residual solvent residue using a gentle stream of nitrogen in a fume hood.
Prepare samples for SDS-PAGE gel
-
12
Add 2× SDS sample buffer to the dry pellet using the largest volume that will fit into sample wells of gel; do not further dilute 2× SDS sample buffer to a 1× concentration.
-
13
Incubate sample 4 to 6 hr at 60°C in a sonicating water bath. Remove the sample every 5 to 15 min and agitate vigorously using a vortex mixer before returning to the water bath.
-
14
Transfer the sample to a 1.5-ml microcentrifuge tube and microcentrifuge 10 min at 14,000 × g, room temperature.
The presence of a pellet indicates incomplete solubilization of proteins. Repeat sonication at 60°C, if necessary. -
15
Add sufficient additional reducing reagent (e.g., β-mercaptoethanol or dithiothreitol) to replace the majority of reducing reagent in the initial volume of 2× SDS sample buffer (much of the reagent will have been lost during solubilization of the proteins). Load samples onto an SDS-PAGE gel in 2× SDS sample buffer without dilution. Run the gel.
All samples and molecular weight standards must contain the same volume and 2× concentration of SDS sample buffer.Samples may be boiled prior to loading, or loaded onto gels without further processing.A full-size SDS-PAGE gel is recommended rather than a mini-gel due to the larger size of the sample wells; it is advantageous to use as large a volume of 2× SDS sample buffer as possible to fully solubilize delipidated lipid droplet proteins. Failure to adequately delipidate the samples will result in one of two possible outcomes: (1) proteins within the samples will be retarded in the wells or stacking gel, and will not migrate sufficiently into the gel, or (2) vertical streaking or smearing of the samples will occur, yielding poor resolution of protein bands. Insufficient solubilization of the samples will yield a low signal of lipid droplet–associated proteins.See Figure 3.15.2 for a gel showing proteins associated with lipid droplets.
Figure 3.15.2.
Coomassie Blue-stained proteins of lipid droplets isolated from 3T3-L1 adipocytes incubated under basal, lipid-storing conditions (A) and lipolytically stimulated conditions (B). Positions of molecular mass markers and stained bands are depicted on the left and right sides of the panels, respectively. Figure reprinted with permission of The Journal of Biological Chemistry, from “Proteomic Analysis of Proteins Associated with Lipid Droplets of Basal and Lipolytically Stimulated 3T3-L1 Adipocytes,” Dawn L. Brasaemle, Georgia Dolios, Lawrence Shapiro, and Rong Wang, Vol. 279 (2004) 46835–46842 (original manuscript); Correction Vol. 280 (2005) 4004; permission conveyed through Copyright Clearance Center, Inc.
REAGENTS AND SOLUTIONS
Use deionized or distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see SUPPLIERS APPENDIX.
HLM containing 5% (w/w) sucrose
2.54 g sucrose
48.34 g HLM (see recipe)
Store 1 to 2 days at 4°C
HLM containing 60% (w/w) sucrose
38.6 g sucrose
25.73 g HLM (see recipe)
Store 1 to 2 days at 4°C
Hypotonic lysis medium (HLM)
1 ml 1 M Tris·Cl, pH 7.4 (20 mM final)
100 μl 0.5 M EDTA (1 mM final)
1 ml 0.5 M sodium fluoride (10 mM final; optional)
Protease inhibitors (see UNIT 3.4)
H2O to 50 ml
-
Prepare fresh and keep on ice
Sodium fluoride is a general phosphatase inhibitor.
Sodium dodecyl sulfate (SDS) 10% (w/v)
10 g SDS
H2O to 100 ml
Store 1 month at room temperature
COMMENTARY
Background Information
Most cultured cells and many types of cells within tissues have lipid droplets composed of a core of neutral lipids surrounded by a membrane monolayer of phospholipids and cholesterol, into which proteins are embedded. The majority of cells have a few tiny lipid droplets of 0.5 to 5 μm in diameter and containing cores of primarily cholesterol esters, whereas adipocytes store triacylglycerol in large lipid droplets that can reach diameters of 100 μm. In adipocytes and muscle cells, triacylglycerol stored in lipid droplets serves as an important source of energy, whereas in the majority of cells, stored cholesterol esters serve a role in the maintenance of cellular cholesterol homeostasis and are used for membrane synthesis and repair. In steroidogenic cells found in the adrenal cortex, testes, and ovaries, stored cholesterol esters are a source of substrate for steroid hormone synthesis. In stellate cells of the liver and in the retina of the eye, lipid droplets are enriched with retinyl esters.
Two or more members of the perilipin family of lipid droplet–associated proteins (translated from genes for perilipins 1 through 5) are associated with lipid droplets in most cells of vertebrates. For most types of cells, perilipin 2 is a reliable marker for the recovery of lipid droplets; however, perilipin 1 is a better marker for lipid droplets from adipocytes of white or brown adipose tissue or cultured adipocytes.
Additionally, over the past sixteen years, research efforts have established the presence of multiple enzymes required for the biosynthesis of sterols, neutral lipids, and phospholipids on lipid droplets (reviewed in Goodman, 2009; Hodges and Wu, 2010; Schittmayer and Birner-Gruenberger, 2009; Wifling et al., 2014; Yang et al., 2012).; Studies in cells and animals have demonstrated that several lipid droplet-associated proteins are key regulators of energy metabolism.
In general, the culture medium used to promote the growth of most cell lines contains a relatively low concentration of fatty acid and cholesterol substrates for neutral lipid synthesis. Under these conditions, most cell lines display few or no lipid droplets per cell, when visualized by staining of cells with the lipophilic dyes Nile Red (Greenspan et al., 1985), or Bodipy 493/503 (Gocze and Freeman, 1994). The addition of either decimillimolar concentrations of fatty acids complexed to albumin, or a combination of fatty acids bound to albumin and cholesterol delivered via liposomes or cyclodextrins promotes increased synthesis of neutral lipids and subsequent storage of these lipids in lipid droplets.
The isolation of lipid droplets relies upon the buoyant density of lipid droplets, which is <1 g/cm3. Two low-speed centrifugation steps and a single ultracentrifugation step using a discontinuous density gradient will collect >95% of the lipid droplets from a cell lysate. The isolated lipid droplets may be used as a source of lipid substrates for studying the activities of lipid metabolic enzymes. To date, no specific enzyme activities have been localized exclusively to lipid droplets, although isolated lipid droplets can provide a source of lipid metabolic enzymes. Multiple studies have shown that lipid droplets have transient contact with the endoplasmic reticulum, and recent studies have demonstrated the movement of membrane-associated proteins between these compartments (Wifling et al., 2013; Poppelreuther et al., 2012, Brasaemle and Wolins, 2012). Moreover, it is likely that lipid droplets transiently contact the mitochondria to exchange fatty acids (Rambold et al., 2015). The transient contact of lipid droplets with other subcellular compartments complicates the definition of the lipid droplet compartment, since many lipid droplet-associated proteins are also found in the endoplasmic reticulum. Similarly, isolating lipid droplets completely free of mitochondrial proteins may not be possible, if the membranes are sometimes continuous. Nonetheless, the perilipin family proteins, perilipins 1 and 2, are the most lipid droplet-specific proteins identified to date and are thususeful markers of the lipid droplet compartment in immunoblotting and indirect immunofluorescence applications. However, the appearance of small quantities of these perilipins in other membrane compartments during subcellular fractionation procedures may not always be indicative of a failure to consolidate the lipid droplet compartment. The use of isolated lipid droplet fractions for proteomics studies necessitates the washing of the initial lipid droplet fraction through several additional centrifugation steps to remove traces of contaminating membranes. The described methods may also be scaled up and used to isolate lipid droplets from tissue samples, following the digestion of the tissue to yield a uniform suspension of intact, disaggregated cells.
Critical Parameters and Troubleshooting
The isolation of lipid droplets by centrifugation is a relatively simple procedure. A very low protein-to-lipid ratio renders lipid droplets more buoyant than all other subcellular structures; lipid droplets can be separated from more dense subcellular compartments using discontinuous density gradients. The most critical step in the isolation of lipid droplets is making an appropriate choice for the method of cell disruption to keep the droplets intact, particularly when working with cells containing very large lipid droplets. Gentle disruption of cells is required to preserve lipid droplet structure. The use of rotor-stator homogenizers or sonication to lyse cells will disrupt and emulsify lipid droplets. Likewise, the extrusion of cells or isolated lipid droplets through small openings exerts sufficient shear force to disrupt lipid droplets. Gentle homogenization using a hand-operated homogenizer with a loose-fitting Teflon pestle, or the use of a nitrogen cavitation bomb will yield better results.
The buoyant density of lipid droplets facilitates flotation during centrifugation of any aqueous medium. The use of solutions lacking electrolytes may reduce the aggregation of lipid droplets with other subcellular organelles; the use of hypotonic solutions does not compromise the integrity of lipid droplets because they have no aqueous compartment. Since segments of endoplasmic reticulum and numerous mitochondria are often closely apposed to the droplets in intact cells, isolated lipid droplet fractions contain low levels of these membranes. Contamination of lipid droplets with these membranes can be minimized by layering density-adjusted cell lysates beneath one or two layers of decreasing density in a discontinuous gradient prior to centrifugation. The movement of the lipid droplets upwards through the layers of the gradient reduces the adherence of contaminant membranes to the droplets and resolves the droplets from soluble proteins. Further removal of contaminating membranes may be enhanced by flotation of the isolated lipid droplets through additional discontinuous density gradients, which may include solutions containing a low concentration of glycerol, 100 mM to 1 M sodium chloride, or 100 mM sodium carbonate, pH 11.5 (Fujiki et al., 1982). The use of sodium carbonate wash solutions, however, may remove loosely adherent proteins that normally localize to the outer membrane monolayer of lipid droplets, and may denature the proteins. While lipid droplets can be isolated using tubes with volumes smaller than those described, and with correspondingly reduced volumes of the gradient solutions, higher levels of contaminating membranes may be floated with the lipid droplet fraction. Wide-diameter tubes are not recommended for the isolation of lipid droplets from relatively small numbers of cells, because the lipid droplet layer will be diffuse and more easily disrupted during sample collection. Additionally, swinging-bucket rotors are recommended to band the floating lipid droplets compactly at the top of the tube; rotors should be allowed to coast to a stop through the final deceleration to minimize disruption of the lipid droplet layer.
Following centrifugation, lipid droplets will appear as a milky layer at the top of the tube. If there is a clear oily layer at the top of the tube, the lipid droplets have broken during sample preparation; this is indicative of a poor isolation. Gentle homogenization with fewer strokes and slower insertion of the pestle will reduce disruption of the lipid droplets. There will be a pellet containing dense membranes at the bottom of the tube and one or two translucent bands of membranes at the interfaces of the density phases. If these bands appear very cloudy and white, this indicates breakage and emulsification of the lipid droplets, and a poor preparation of lipid droplets. A portion of the floating droplet fraction will adhere to the sides of the tube, and to the surfaces of any pipetting device used to collect the fraction. Additionally, if the layer is disrupted during handling of the tube or collection of the samples, some of the droplets may swirl down into the top layer of the gradient, making sample collection more difficult.
The best way to collect the majority of the lipid droplet fraction in a small volume is to use a Beckman tube slicer in combination with thin-walled polyallomer or polycarbonate tubes. The tube is inserted into the tube slicer to position the cutting blade several millimeters below the lipid droplet layer. The steady and firm insertion of the blade through the tube isolates the lipid droplet layer in a small volume of the top gradient solution in the sliced top portion of the tube. Rubber rings within the tube slicer form a seal with the blade, keeping the top solution from leaking out. Use of a tube slicer facilitates the most efficient collection of the lipid droplets in a minimal volume, and it permits rinsing of the surfaces of the blade and tube with additional solution to more completely collect the fraction. When a tube slicer is unavailable, the lipid droplet fraction can be collected using either a pipetting device or a syringe with a wide-bore needle; however, it is more difficult to collect the entire lipid droplet layer in a minimal volume. Additionally, there will be significant loss of lipid droplets that are adherent to the sides of the tube. Gradient collectors that puncture the bottoms of tubes and collect the densest fractions first are unsuitable for the collection of lipid droplets, because the majority of the lipid droplets will adhere to the sides of the tube as the volume of liquid drops.
Lipid droplets from chordates are characterized by their content of neutral lipids and by the presence of one or more members of the perilipin family of proteins. The neutral lipid content may be assessed by quantitative thin layer chromatography (Brasaemle et al., 1997a) of solvent extracts of lipids (Bligh and Dyer, 1959), or by the adaptation of commercially available clinical assays for the quantitation of triacylglycerol (Schwartz and Wolins, 2007) or cholesterol for use with extracted lipids. The content of perilipin family proteins is best assessed by immunoblotting of proteins extracted from lipid droplet fractions. Since lipid droplets have an extremely low content of protein relative to lipid, it is imperative to remove the excess lipid before applying samples to SDS-PAGE gels (see Support Protocols 2 and 3). Detergent solubilization of proteins in preparation for SDS-PAGE yields the best recovery when using freshly isolated lipid droplet fractions; the recovery of perilipin family proteins from frozen and thawed samples of lipid droplets is significantly reduced. A buffered solution containing 1% Triton X-100 and 0.5% sodium deoxycholate can substituted for the SDS-containing sample buffer, but the yield of lipid droplet-associated proteins may be reduced. Lipid droplet fractions that have been frozen and thawed can be delipidated with solvents and the proteins solubilized in 2× concentrated sample buffer; the higher content of SDS and reducing reagent in undiluted 2× sample buffer helps to solubilize the relatively insoluble component proteins. Effective solubilization of precipitated proteins requires extensive warming and sonication of the samples; however, the use of a probe sonicator will cause foaming of the sample and should be avoided. Solubilization of samples for several hours at 60°C with sonication in a bath sonicator is more effective than brief boiling of the samples. Samples should be loaded on gels immediately after the solubilization steps to avoid precipitation of the component proteins. For most types of cells, immunoblotting for perilipin 2 will identify lipid droplets; however, immunoblotting for perilipin 1 is required for lipid droplets isolated from mature adipocytes, since perilipin 2 is excluded from the droplets when the expression of perilipin 1 is induced during adipose differentiation (Brasaemle et al., 1997b).
Anticipated Results
This procedure will yield a lipid droplet fraction that contains >95% of the total neutral lipid content of the cells, and >95% of perilipins 1 or 2. The lipid droplet fraction will contain an extremely low protein mass relative to that of lipid; the relative protein mass will vary with the size of the lipid droplet and type of cell used. The lipid droplet fraction will also contain low levels of contamination with mitochondria and membrane fragments from endoplasmic reticulum and plasma membrane. Typical preparations of adipocyte lipid droplets using this procedure contain ≤5% of the total content of calnexin, a marker for endoplasmic reticulum, when assessed by immunoblotting. The relative purity of the lipid droplet fraction will be influenced by several factors including the size of the lipid droplets and the number of lipid droplets in the cell lysate. Lower levels of contaminants may be anticipated when isolating lipid droplets from cells that have very small lipid droplets, and when centrifugation of the cell lysates yields a distinct, but relatively thin lipid layer.
If the isolated lipid droplets are being used to establish the relative content in lipid droplets of a protein that is found in other subcellular compartments, then it is essential to characterize the levels of contaminant membranes using either immunoblotting methods, or marker enzyme assays. The use of mass spectrometry to identify protein components of subcellular fractions is significantly more sensitive than immunoblotting methods and will likely reveal levels of contaminant proteins in lipid droplet fractions that have escaped detection by immunoblotting. When analyzing the distribution of a protein that localizes to both lipid droplets and another subcellular compartment, care must be taken to make appropriate comparisons. Lipid droplets contain an extremely low protein content, whereas other membrane compartments are relatively protein dense. Consequently, proteins in the lipid droplet fraction comprise a very small fraction of the total protein content of the cell. Thus, comparing lipid droplets to other membrane fractions by loading equal total protein content for each of the various compartments onto a protein gel is inappropriate, as it will require the use of lipid droplets from 20- to 100-fold greater cell mass than for the other subcellular compartments. This will enhance the detection of contaminating proteins that actually reside in other compartments within the lipid droplet fraction. It is more appropriate to compare a volume of the lipid droplet fraction (and each of the other subcellular compartments) that represents an equivalent fraction of the whole cell lysate. Thus, contaminating proteins will be minimally detected in the lipid droplet fraction, and bona fide lipid droplet-associated proteins will be detected proportionally to their abundance in the lipid droplet fraction.
Time Considerations
The isolation of intact lipid droplets requires fresh and not frozen cell or tissue samples. It is necessary to proceed from harvest of cells through the collection of lipid droplets without storage of samples at intermediate steps of the procedure. Since the centrifugation steps are relatively brief, all steps from the harvest of cells through the initial collection of the lipid droplet fraction can be completed within 4 hr. Additional washes of the lipid droplet fraction should be done on freshly isolated material, immediately following the initial isolation. The lipid droplet fraction can then be stored for several hours at 4°C, or several weeks at −80°C.
Assessment of lipid droplet components can be performed upon frozen and thawed samples of isolated lipid droplets, although recovery of proteins using the detergent solubilization procedure will be reduced for samples that have been stored at −80°C. Protein precipitation by solvent delipidation, and the subsequent solubilization of the proteins will require at least 8 hr, if a one-step delipidation is completed. SDS-PAGE may be completed in 2 to 4 hr and transfer of proteins to membranes and subsequent immunoblotting takes 10 to 24 hr.
Acknowledgments
D.B. was supported by NIH grant NIH R01 DK054797 and N.W. was supported by NIH grant NIH R01 DK088206.
Footnotes
Contributed by Dawn L. Brasaemle, Rutgers, The State University of New Jersey, New Brunswick, New Jersey
Nathan E. Wolins, Washington University School of Medicine, St. Louis, Missouri
Literature Cited
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Med Sci. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Barber T, Kimmel AR, Londos C. Post-translational regulation of perilipin expression. Stabilization by stored intracellular neutral lipids. J Biol Chem. 1997a;272:9378–9387. doi: 10.1074/jbc.272.14.9378. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is a ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997b;38:2249–2263. [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. Packaging of Fat: An Evolving Model of Lipid Droplet Assembly and Expansion. J Biol Chem. 2012;287:2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocze PM, Freeman DA. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17:151–158. doi: 10.1002/cyto.990170207. [DOI] [PubMed] [Google Scholar]
- Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50:2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: A selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitpas I, Grune T, Bhattacharya AA, Curry S. Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids. J Mol Biol. 2001;314:955–960. doi: 10.1006/jmbi.2000.5208. [DOI] [PubMed] [Google Scholar]
- Poppelreuther M, Rudolph B, Du C, Großmann R, Becker M, Thiele C, Ehehalt R, Füllerkrug J. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res. 2012;53:888–900. doi: 10.1194/jlr.M024562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittmayer M, Birner-Gruenberger R. Functional proteomics in lipid research: lipases, lipid droplets and lipoproteins. J Proteomics. 2009;72:1006–1018. doi: 10.1016/j.jprot.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Schwartz DM, Wolins NE. A simple and rapid method to assay triacylglycerol in cells and tissues. J Lipid Res. 2007;48:2514–20. doi: 10.1194/jlr.D700017-JLR200. [DOI] [PubMed] [Google Scholar]
- Wifling F, Haas JT, Walther TC, Farese RV., Jr Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wifling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Jr, Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]