Summary
Lung squamous cell carcinoma (LSCC) is a devastating malignancy with no effective treatments, due to its complex genomic profile. Therefore, preclinical models mimicking its salient features are urgently needed. Here we describe mouse models bearing various combinations of genetic lesions predominantly found in human LSCC. We show that SOX2 but not FGFR1 overexpression in tracheobronchial basal cells combined with Cdkn2ab and Pten loss results in LSCC closely resembling the human counterpart. Interestingly, Sox2;Pten;Cdkn2ab mice develop LSCC with a more peripheral location when Club or Alveolar type 2 (AT2) cells are targeted. Our model highlights the essential role of SOX2 in commanding the squamous cell fate from different cells of origin and represents an invaluable tool for developing better intervention strategies.
Graphical Abstract

Highlights
-
•
Pten;Cdkn2ab loss causes a diversity of tumors in tracheobronchial basal cells
-
•
SOX2 overexpression is crucial in promoting LSCC upon loss of Pten;Cdkn2ab
-
•
Sox2;Pten;Cdkn2ab mice develop LSCC from Basal, Alveolar type 2, and Club cells
-
•
Mouse LSCC phenotypically and molecularly resembles the human counterpart
Ferone et al. establish mouse models bearing various combinations of genetic alterations commonly found in human lung squamous cell carcinoma and show that SOX2, but not FGFR1, overexpression induces the squamous cell fate from different cells of origin carrying the same combined Cdkn2ab and Pten loss.
Significance
LSCC is a devastating disease for which more effective treatments are urgently needed. Therefore, representative models reproducing its salient features are of pivotal importance. We show that Pten- and Cdkn2ab-deficient tracheobronchial basal cells serve as a highly effective cell of origin of diverse lung tumors. We show that SOX2, but not FGFR1, overexpression drives Pten- and Cdkn2ab-deficient heterogeneous tumors into LSCC, in line with the frequent overexpression of SOX2 in human LSCC. We show that multiple cell types in lung can give rise to LSCC, with different growth patterns in the central and peripheral lung.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with more than one-quarter (27%) of deaths among both men and women (Siegel et al., 2015). Non-small cell lung cancer, which includes adenocarcinoma (ADC), squamous cell carcinoma (SCC), and large cell carcinoma, is the predominant histological type, accounting for more than 80% of cases. More than 50% of cases present a mixed histological phenotype, indicative of the marked heterogeneity in lung cancer (Travis, 2002, Walker, 2008). With more than 400,000 deaths worldwide each year (Cancer Genome Atlas Research Network, 2012, Jemal et al., 2011) SCC accounts for 30% of all lung cancer cases, ranking second only to ADC. These two major subtypes are associated with distinct molecular abnormalities (Cancer Genome Atlas Research Network, 2012, Cancer Genome Atlas Research Network, 2014) and are thought to have distinct cells of origin (Giangreco et al., 2007, Sutherland and Berns, 2010, Sutherland et al., 2011). The knowledge of the molecular mechanisms driving lung ADC (LADC) has resulted in targeted treatments, not yet found for lung SCC (LSCC).
In the comprehensive analysis of human LSCC recently reported by The Cancer Genome Atlas (TCGA) network, LSCC samples displayed a large variety of DNA alterations clustered in four well-defined pathways: squamous differentiation pathway (SOX2, NOTCH), phosphoinositol 3-kinase (PI3K) pathway (FGFR1, PTEN), cell-cycle regulation (CDKN2A, RB1, TP53), and oxidative stress response (CUL3, KEAP1, NFE2L2) (Cancer Genome Atlas Research Network, 2012). The higher molecular complexity and the absence of predominant actionable driver alterations (Bass et al., 2009, Cancer Genome Atlas Research Network, 2012, Weiss et al., 2010) have hampered the development of preclinical models that mimic human LSCC at both genotype and phenotype levels. Single oncogenic alterations introduced in the mouse failed to drive the development of LSCC while mostly promoting LADC (Lu et al., 2010, Malkoski et al., 2014).
The first model of LSCC morphologically resembling the human counterpart is based on the deregulation of IKKα, a protein kinase, whose reduced activity was already associated with SCC development in skin and oral cavity but rarely in lung (Liu et al., 2012). A dominant negative mutant form of IKKα causes SCC in a variety of tissues but at a very low frequency in lung (Xiao et al., 2013). Prolonged survival by restricting the dominant negative effect to the lungs did, however, increase the frequency of LSCC. Two conditional models recently described are based on Lkb1 loss, which is found to be inactivated in approximately 2% of human LSCC (Cancer Genome Atlas Research Network, 2012, Travis, 2002). In one model, which combines Lkb1 and Pten deletion, mice develop LSCC morphologically resembling the human counterpart with a latency of 40–50 weeks (Xu et al., 2014). In the other model, mice harboring a conditional deletion of Lkb1 develop LSCC, and in a few cases LADC, following intranasal infection with a lentivirus carrying SOX2 and PGK-Cre-recombinase; the latency is shorter (6–10 months) due to the concomitant overexpression of SOX2, and the penetrance of tumor formation is 40% (Mukhopadhyay et al., 2014).
Although the combination of genetic alterations is critical for the tumor phenotype, increasing evidence also points to the cell of origin as an important factor in determining tumor characteristics (Sutherland et al., 2011, Sutherland et al., 2014, Visvader, 2011). LSCC was thought to mainly arise in the upper airways, but according to recent reports peripheral LSCC is becoming as frequent as the central type (Funai et al., 2003, Hayashi et al., 2013, Sakurai et al., 2004, Yousem, 2009). The multiple locations may have therapeutic implications if peripheral and central LSCC have a different cells of origin and, therefore, different growth patterns.
Trachea, mainstem bronchi, and the most proximal region of the intralobular airway are lined by a pseudostratified columnar epithelium composed of Basal, Ciliated, Neuroendocrine, and Club secretory cells. Basal cells serve as tissue-specific stem cells for the tracheobronchial compartment, since they can both self-renew and give rise to Club and ciliated epithelial cells (Hong et al., 2004, Rock et al., 2009). They express high levels of the transcription factor p63, which is required for development of the trachea (Daniely et al., 2004), and cytokeratin 5 (K5) and 14 (K14). Their expression profile (p63, K5) and their stem cell properties make them a likely candidate for the cell of origin of LSCC. Club cells are more abundant and line the bronchi and bronchioles. They can both self-renew and generate ciliated cells both under homeostatic conditions and in response to epithelial injury (Rawlins et al., 2009). The most distal region of the lung is organized into a complex system of alveoli, composed of alveolar type 1 (AT1) and 2 (AT2) cells. The latter are considered to be the major stem cells of the alveolar epithelium, based upon their ability to self-renew and give rise to AT1 cells (Adamson and Bowden, 1974, Evans et al., 1975). Club cells and AT2 cells are both indicated as cells of origin of lung LADC (Sutherland et al., 2014). In this study, we define the impact of the cell of origin on LSCC development.
Results
Targeted Introduction of LSCC Recurrent Aberrations by Recombinant Adenoviral Vectors
We have previously described a series of adenoviral vectors that drive Cre-recombinase to Club and AT2 cells in the adult mouse lung and have demonstrated that they are robust tools for the assessment of the cell of origin of lung cancer (Sutherland et al., 2011, Sutherland et al., 2014). We applied this same approach to target basal progenitor cells. We utilized the promoter region of K14 or K5 to direct Cre-recombinase to basal progenitor cells (see Supplemental Experimental Procedures for details).
To assess the specificity and efficiency of Ad5-K14-Cre and Ad5-K5-Cre, we infected primary keratinocytes and mouse embryonic fibroblasts (MEFs) isolated from mT/mG mice, a Cre reporter mouse strain that expresses Tomato (mT) prior to Cre-mediated excision and membrane-targeted GFP (mG) upon excision (Muzumdar et al., 2007) (Figures S1A and S1B). Both Ad5-K14-Cre and Ad5-K5-Cre efficiently delivered and activated Cre-recombinase expression in keratinocytes, as indicated by GFP expression (Figure S1A), but not in MEFs (Figure S1B). We used Ad5-CMV-Cre as positive control of infection and Ad5-SPC-Cre (Sutherland et al., 2011) as negative control, since the promoter of SPC drives Cre expression only in AT2 cells. The result was confirmed by western blot analysis (Figure S1C).
To validate the specificity of adenovirus promoter targeting in vivo, we intratracheally injected mT/mG mice with a high titer of either Ad5-K14-Cre or Ad5-K5-Cre, and performed GFP staining on trachea and lungs collected 3 weeks after infection to identify switched cells. mT/mG mice were pretreated with naphthalene, which depletes Club secretory cells (Hong et al., 2004), facilitating the access to tracheobronchial basal cells. Indeed, GFP staining of tracheas isolated from mT/mG mice treated with corn oil (vehicle control) was negative (Figure 1A), indicating that under steady-state conditions the tracheobronchial epithelium is relatively refractory to adenoviral infection. This is also the case when Cre is under the direction of the cytomegalovirus (CMV) promoter (Figure 1A). In stark contrast, however, Ad5-CMV-Cre, Ad5-K14, and Ad5-K5-Cre showed a high level of infection of basal cells when injected following naphthalene administration (Figure 1A). In the lung, Cre-recombinase-mediated switching occurred in the alveolar compartment of mice intratracheally injected with Ad5-CMV-Cre, regardless whether they were pretreated with naphthalene or corn oil (Figure 1B). Ad5-K5-Cre and Ad5-K14-Cre were not able to drive Cre expression in any of the peripheral lung cell populations when pretreated with corn oil (Figure 1B), while naphthalene treatment gave rise to a readily detectable population of switched cells along the bronchial lumen (Figure 1B).
Figure 1.
In Vivo Characterization of Basal Cell-Specific Adenoviral Vectors Using mT/mG Reporter Mice
GFP IHC staining of longitudinal sections of tracheas (A) and lungs (B) isolated from mT/mG reporter mice 3 weeks after intratracheal injection of Ad5-CMV-Cre, Ad5-K14-Cre, and Ad5-K5-Cre following pretreatment with corn oil or naphthalene as indicated. Scale bars, 100 μm (A) and 200 μm (B). Scale bars in insets, 50 μm (B). See also Figure S1.
To demonstrate that GFP was specifically expressed by basal cells, we performed dual immunofluorescence (IF) staining for GFP and K14, K5, or CC10 on tracheal sections collected at 7 days after intratracheal injection of either Ad5-K14-Cre or Ad5-K5-Cre in mice previously treated with naphthalene (Figure S1D). The co-expression of GFP with both K14 and K5, but not CC10, indicated that Cre is specifically activated in basal cells (Figure S1D). At 21 days the GFP staining was also found in cells overlaying the basal cells, consistent with prior lineage-tracing experiments (Hong et al., 2004, Rock et al., 2009) (Figure S1E). GFP staining on trachea and lungs of mT/mG mice collected 2, 4, 6, 8, and 15 months after virus injection upon naphthalene treatment indicated that the specificity is retained over an extended period of time (Figure S1F). We analyzed 11 mT/mG mice, which did not show any neoplastic lesions, indicating that naphthalene does not induce tumors. Taken together, our data indicate that Ad5-K14-Cre and Ad5-K5-Cre efficiently and specifically target tracheobronchial basal cells following naphthalene treatment.
To recapitulate the genomic complexity of LSCC, we developed a mouse model of LSCC based on the deregulation of multiple pathways involved in LSCC initiation and progression. We generated compound conditional mutant mice for: (1) overexpression of SOX2, a transcription factor involved in squamous differentiation by suppressing the Notch pathway and whose gene is one of the most frequently amplified in SCC; (2) overexpression of FGFR1, a tyrosine kinase receptor overexpressed in 27% of LSCC cases and as such a possible therapeutic target; (3) deletion of PTEN, which encodes a phosphatase that antagonizes the PI3K signaling pathway and found to be altered in 24% of LSCC cases; and (4) deletion of the CDKN2AB locus that encodes for p16INK4A, p15INK4B, and p14ARF, three cell-cycle inhibitors with a well-established tumor-suppressor function and inactivated in 72% of LSCC cases (Cancer Genome Atlas Research Network, 2012). In the absence of p16INK4A, p15INK4B can provide a backup function, thereby mitigating the effects of p16INK4A loss (Krimpenfort et al., 2007). Consequently, loss of both CDKN2A and CDKN2B has more profound effects and is likely the reason why both are often co-deleted in LSCC. These genetic alterations collectively cover a large fraction of the human cases (Figure 2A).
Figure 2.
Generation of Mouse Models that Recapitulate the Genomic Complexity of Human LSCC
(A) Genetic alterations of CDKN2A, CDKN2B, PTEN, FGFR1, and SOX2 in the 178 human LSCC specimens in the TCGA dataset. Exon skipping and DNA methylation for the CDKN2A locus are excluded.
(B) Schematic representation of the experimental design used for all cohorts of mice: PC, Fgfr1PC, and Sox2PC. Mice are treated with naphthalene on day 0 and intratracheally injected with Ad5-K14-Cre or Ad5-K5-Cre on day 3.
(C) Representative H&E staining on various lung lesions of PC mice. Scale bars, 100 μm.
See also Table S1.
Mice carrying the conditional allele of either Pten (Ptenflox/flox) or Cdkn2ab have been previously generated in our laboratory (Krimpenfort et al., 2007, Marino et al., 2002). Therefore, we generated mice carrying the combined conditional deletions of Pten and Cdkn2ab (hereafter PC mice). PC mice were crossed to mT/mG reporter mice to identify cells in which the alleles were actually recombined. To determine whether their inactivation in basal cells is sufficient to induce LSCC, we intratracheally injected 6- to 8-week-old PC mice with either Ad5-K14-Cre or Ad5-K5-Cre following naphthalene treatment (Figure 2B). Mice were euthanized when they started to show signs of disease (e.g., shortness of breath, weight loss), and trachea and lungs were sampled for histopathological analysis. PC mice developed tumors with a latency of 10–15 months and a frequency of 55% (Table S1). The penetrance was calculated by scoring the mice based on GFP staining. Mice negative for GFP staining were not included because this would imply the absence of recombination. At 6 or 9 months the penetrance was very low (1/7 and 1/9, respectively). At 10–15 months tumor penetrance increased (10/19; Table S1) and a wide spectrum of tumor lesions was found (Figure 2C and Table S1). No obvious differences between Ad5-K14-Cre- or Ad5-K5-Cre-injected PC mice were found. Taken together, these data demonstrated that tracheobronchial basal cells can be efficiently transformed by loss of Pten and Cdkn2ab, giving rise to a variety of tumors.
FGFR1 Overexpression Causes Neoplasia with Sporadic Squamous Differentiation
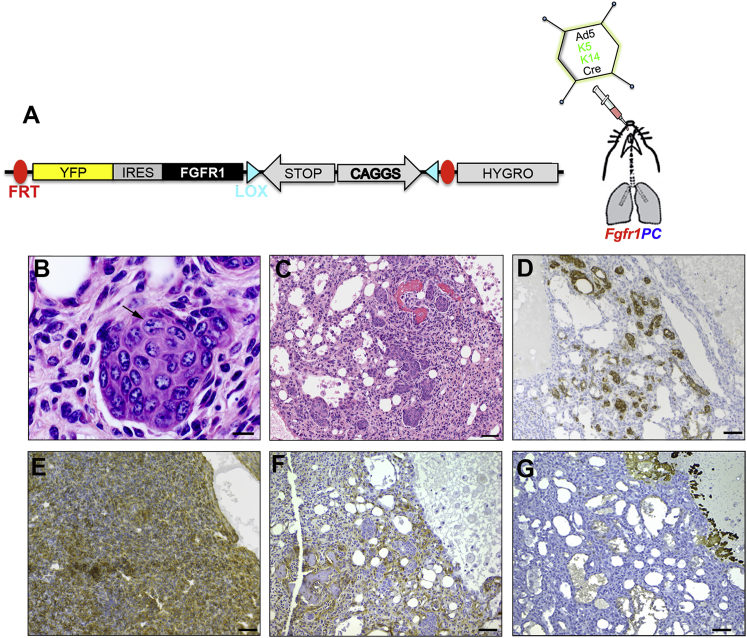
To directly test the consequences of overexpressing fibroblast growth factor receptor 1 (FGFR1) in vivo, we targeted a bicistronic cDNA cassette containing a mutant active form of FGFR1 (K656E) and the YFP to the Col1a1 locus (Figure S2A). Validation of the recombinant event in embryonic stem cells (ESCs) is described in Supplemental Experimental Procedures and in Figures S2B and S2C. We crossed LSL-Fgfr1K656E mice with PC mice and with mT/mG reporter mice (hereafter Fgfr1PC; Figure 3A). Fgfr1PC mice pretreated with naphthalene and intratracheally injected with either Ad5-K14-Cre or Ad5-K5-Cre developed tumors with a latency of 1.5–6.5 months and a frequency of 76% (16/21; Table S1).
Figure 3.
Combined Overexpression of FGFR1 Partially Drives Squamous Differentiation within Heterogeneous Neoplastic Lesions
(A) Schematic representation of the Col1a1 locus targeted with LSL-Fgfr1K656E transgene before Cre infection. Fgfr1PC mice were infected with basal specific adenoviruses.
(B and C) H&E staining of lung sections showing squamous differentiation with nest of cells with evident intercellular bridges (arrow in B) in circular arrangement (C).
(D–G) IHC analysis of lung sections possessing tumors with squamous differentiation, for K5 (D), GFP (E), FGFR1 (F), and p63 (G).
Scale bars, 10 μm (B) and 50 μm (C–G). See also Figure S2 and Table S1.
We observed bronchoalveolar adenoma, polyp-like spindle cell neoplasia, ADC, chondroid sarcoma, adenofibroma, and osteosarcoma (Figure S2D and Table S1). In a few cases sporadic squamous cell differentiation was observed within the heterogeneous lesion (19%; Figure 3 and Table S1). In general, tumors were invasive and did clearly arise from the bronchial lining (Figure S2D). Lesions containing squamous differentiation were arranged as nests of cells in circular structures (Figures 3B and 3C) strongly positive for K5 (Figure 3D). Immunohistochemistry (IHC) analysis showed that the lesions were positive for GFP and FGFR1, as expected (Figures 3E and 3F); however, they were negative for p63 (Figure 3G). Taken together, our results indicate that FGFR1 overexpression transforms Pten- and Cdkn2ab-deficient tracheobronchial basal cells at a higher frequency and at a faster rate. Tumors are heterogeneous with only sporadic squamous differentiation.
SOX2 Serves as a Critical Switch to Drive Heterogeneous Lesions into LSCC
To overexpress SOX2 in vivo, we used an approach similar to that described above for FGFR1 using cDNA for mouse SOX2 and CFP (Figure S2A). Successful targeting of ESCs and expression of LSL-Sox2 allele in vivo are described in Supplemental Experimental Procedures and Figures S2B, S2C, and S2E. We crossed LSL-Sox2 mice with PC mice, and with mT/mG reporter mice (hereafter Sox2PC; Figure 4A). Seven to nine months after basal cell virus inoculation, 73% of Sox2PC mice developed multiple LSCC that ranged from moderately to well differentiated (Figure 4 and Table S1). The large tumors expanded in the alveolar compartment (Figure 4B). Squamous cell differentiation was evident from the keratin pearls and the prominent cornification in advanced lesions (Figure 4B). The cornification was accompanied by infiltration of inflammatory cells and large areas of necrosis. LSCC was morphologically characterized by irregular nests or large nodules (Figure 4C). In some cases, although the squamous cell differentiation was predominant, small areas of ductal/glandular/mucinous differentiations were observed (data not shown).
Figure 4.
SOX2 Promotes the Switch from Heterogeneous Neoplastic Lesions to Typical LSCC
(A) Schematic representation of the Col1a1 locus targeted with LSL-Sox2 transgene before Cre infection. Sox2PC mice were generated and infected with basal specific adenoviruses.
(B and C) H&E staining on mouse lung sections collected 7–9 months after injection. The tumor showed prominent cornification (arrow in B), keratin pearls, and inflammatory infiltration (arrows in C).
(D) H&E staining of well-differentiated human LSCC.
(E) IHC staining on human and mouse lung sections with the indicated histopathological markers.
(F) IHC staining for GFP, K14, and K5 on mouse lung sections.
(G and H) Real-time RT-PCR of Sox2 endogenous (Sox2 end) and exogenous (SOX2 exo) levels performed on tumor tissues isolated from the indicated mouse tumor samples (G) and MEFs untreated (UNT) or treated with tamoxifen (TAM) (H). Data represent means ± SD. Samples are normalized by using actin RNA level.
Scale bars, 200 μm (B) and 50 μm (C–F). See also Figures S2 and S3; Tables S1 and S2.
Twelve cases of human LSCCs were selected for a comparative study of the Sox2PC mouse model. The selected cases displayed a wide spectrum of differentiation features of LSCCs, from well-differentiated carcinomas showing a pavement-like arrangement of cells with keratinization (Figure 4D) to poorly differentiated carcinomas with comedo-like necrosis. Importantly, 11 of the 12 cases of human tumors expressed SOX2 (Figure 4E and Table S2). All human cases showed positive IHC for p63 but were negative for TTF-1, a marker used to distinguish LSCC from LADC (Figure 4E and Table S2). In line with this, Sox2PC mouse lesions showed positive staining for SOX2 and p63 and were negative for TTF-1 (Figure 4E and Table S2). We performed GFP staining to confirm that LSCC arose from cells that had switched the conditional alleles (Figure 4F). GFP staining strongly overlapped with K14 and K5 staining (Figure 4F).
In the remaining 27% of cases, Sox2PC mice showed lesions of atypical hyperplasia commonly in bronchi (Figure S3A), but also in proximal and distal bronchioles, as indicated by GFP staining (Figures S3A and S3B; Table S1). They were positive for GFP, SOX2, K5, and to a lesser extent p63, indicative of the early commitment toward squamous differentiation (Figure S3B). We performed histological examination of all organs and tissues for six Sox2PC mice to identify putative metastasis. We observed SCC in the left atrium of the heart only in one case (Figure S3C). In the same mouse we found large lesions of SCC in the anterior part of the nasal cavity (Figure S3D).
We also measured the RNA expression level of the endogenous and exogenous SOX2 in the tumor area by performing real-time RT-PCR analysis using oligonucleotide primers annealed with the 3′ UTR (Sox2 end) and with the IRES (SOX2 exo). LSCC isolated from Sox2PC mice showed high expression of the exogenous SOX2, whereas the endogenous form was undetectable (Figure 4G). We used as control LADC samples isolated from Kras;p53 mice and PC mice, where we could only detect a low level of endogenous Sox2 expression (Figure 4G). The levels of SOX2 expression were an order of magnitude higher than what we could observe in LSL-Sox2;CreERT2 MEFs upon tamoxifen treatment (Figure 4H). As described above, combined Pten and Cdkn2ab loss gives rise to heterogeneous neoplastic lesions; however, in Sox2PC mice none of these heterogeneous lesions were found. These data indicate that SOX2 overexpression drives a homogeneous LSCC phenotype resembling the histopathological marker profile and the morphology of the human counterpart.
An Epidermal Transcription Program Differentiates Human and Mouse LSCC from LADC
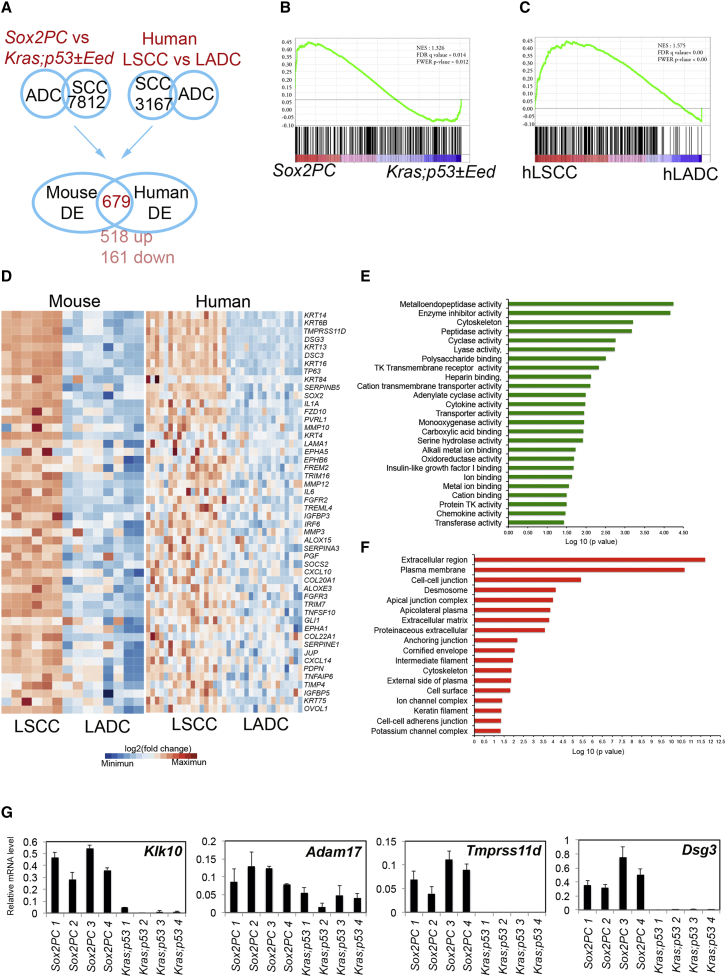
To molecularly characterize our mouse model in an unbiased manner, we performed genome-wide transcriptional analysis of LSCC of Sox2PC (LSCC) mice and LADC of Kras;p53 mice with and without Eed (Kras;p53±Eed). Initially, we compared the mouse transcriptional landscapes of six Sox2PC LSCC with those of eight Kras;p53±Eed LADC. In parallel, the gene expression profiles of 18 human LSCC with SOX2 amplification and CDKN2AB deletion, with or without PTEN deletion from the TCGA dataset (Cancer Genome Atlas Research Network, 2012), were compared with 17 human primary LADC (Cancer Genome Atlas Research Network, 2014) (Table S3) to generate a list of genotype-specific LSCC genes. We identified 3,167 differentially expressed (DE) genes between human LSCC and LADC by applying a log2 (fold change) of ≥ ±1 and a false discovery rate of ≤0.05. Of these, 1,599 genes were upregulated in LSCC tumors and 1,568 were downregulated. A comparison between Sox2PC and Kras;p53±Eed tumor cells yielded 7,812 DE genes, of which 5,208 were upregulated and 2,604 downregulated. Importantly, 679 genes were DE genes in both species (518 upregulated and 161 downregulated; Figures 5A and S4A). Gene set enrichment analysis (GSEA) showed a correlation between genes that typify LSCC in mouse and human over LADC (Figures 5B and 5C).
Figure 5.
Gene Expression Profile Analysis
(A) Venn diagram showing the commonly DE genes between mouse and human LSCC found by comparing DE genes between Sox2PC versus Kras;p53±Eed and human LSCC versus LADC.
(B and C) GSEA for genes upregulated in Sox2PC (B) and hLSCC (C) compared with Kras;p53±Eed and hLADC, respectively.
(D) Hierarchical clustering of upregulated genes in both mouse and human LSCC, which are enriched for a squamous differentiation signature.
(E and F) Significant GO categories for molecular functions (E) and cellular components (F).
(G) Real-time RT-PCR of the indicated genes performed on tumor tissues isolated from the indicated mouse tumor samples. Data represent means ± SD. Samples are normalized by using actin RNA level.
Among the shared upregulated genes, gene ontology (GO) analysis revealed enrichment for 140 genes implicated in squamous differentiation (p = 1.66 × 10−6), including SOX2, TP63, NOTCH3, and several keratins involved in squamous carcinoma (KRT14, KRT6B, KRT16, KRT84, and KRT4) (Figure 5D). The most enriched molecular functions were linked to metalloendopeptidases and proteases, a hallmark of epidermal cells (Figure 5E). The most enriched GO categories were associated with cellular components transcriptionally regulated by p63, such as cell adhesion and, in particular, desmosomes (Figure 5F). Metalloendopeptidase genes included Klk10 and Adam17, and protease genes included Tmprss11d (Figure 5G), whose RNA levels were confirmed by real-time RT-PCR to discriminate mouse LSCC (Sox2PC) and LADC (Kras;p53). We also confirmed the differential expression of Dsg3 that encodes an important component of desmosomes (Figure 5G). Desmosomal components found to be upregulated in Sox2PC tumors included direct p63 target genes such as Dsg1 and Dsc3 (Ferone et al., 2013). Other p63 direct targets such as Fgfr2 and Irf6, with an important role in the homeostasis of squamous epithelia but deregulated in the presence of p63 mutations (Ferone et al., 2012, Thomason et al., 2010), were also upregulated (Figure 5D).
As a complementary subtype assignment, we compared our mouse LSCC gene expression data with the four previously identified LSCC subtypes (Wilkerson et al., 2010). Gene sets specific for each subtype including basal, primitive, classical, and secretory were obtained from Wu et al. (2013). LSCC from Sox2PC mice was enriched for all four subtypes (Figure S4B). Taken together, our results indicate that a p63-mediated epidermal transcriptional program distinguishes human and mouse LSCC from their species-matched LADC counterpart.
Tumor Microenvironment in the Development of LSCC
Inflammatory infiltrates were marginal in PC and Fgfr1PC mice compared with Sox2PC mice, where large areas of neutrophil infiltrations were clearly evident at histology (Figure 4B). Interestingly, we found a sharp distinction between Sox2PC and PC or Fgfr1PC mice with respect to the presence of tumor-associated macrophages (TAMs) and tumor-associated neutrophils (TANs), the two major components of tumor inflammatory infiltrates (Murdoch et al., 2008). IHC analysis showed that TANs were abundantly present in LSCC of Sox2PC mice, as indicated by the expression of LY6G (Figure 6A and Table S4) (Xu et al., 2014). To confirm this result, we performed IHC analysis with antibodies against myeloperoxidase (MPO). MPO is an antimicrobial enzyme most abundantly expressed by TANs compared with naive neutrophils (Youn et al., 2012) and, as expected, was overly present in the tumor lesions of Sox2PC mice (Figure 6A). TAMs, identified by F4/80, were poorly represented in LSCC lesions of Sox2PC mice as compared with TANs (Figure 6A). Conversely, in the heterogeneous lesions of PC mice TANs were completely absent as indicated by the negative staining for LY6G and MPO (Figure 6A), whereas TAMs were the only inflammatory infiltrate found (Figure 6A). Surprisingly the squamous lesions of Fgfr1PC mice were also negative for LY6G and MPO (Figure 6A), in favor of a role for SOX2 in the activation of TANs. However, they showed low-level expression of F4/80 (Figure 6A), indicative of TAMs.
Figure 6.
Tumor Microenvironment in the Three Cohorts of Mice and Comparative Analysis of Human and Mouse LSCC
(A) IHC analysis performed on adult lung tissues of Sox2PC, PC, and Fgfr1PC mice, using the indicated antibodies against markers of inflammatory cells.
(B) IHC analysis performed on human and mouse LSCC for the indicated markers of immune cells.
Scale bars, 200 μm (A) and 50 μm (B). Scale bars in insets, 50 μm (A). See also Table S4.
We then focused on the analysis of immune populations present in LSCC of Sox2PC mice compared with human LSCC. The neutrophil infiltrations were present in all cases of human LSCC analyzed (Table S4). Moreover, we found a striking similarity in the highly immunogenic environment present in both species, as indicated by CD4- and CD8-positive infiltrating cells, which were readily observed in tumor stroma and tumor nests of both species (Figure 6B and Table S4). HIF1-α, a transcriptional regulator of cellular response to hypoxia, was detected to various extents in the tumor cells of both human and mouse LSCC (Figure 6B and Table S4). Interestingly we also found a high level of PD-L1 expression, which showed membranous staining in tumor cells of both human and mouse LSCC (Figure 6B and Table S4), although the percentage of positive cells in human varied from 0.5% to 80% (Table S4). PD-1, the receptor of PD-L1, was mainly observed in clusters of immune cells in tumor stroma (Figure 6B and Table S4). No differences were detected between peripheral or central LSCC (Table S4).
Sox2PC Mice Develop Peripheral LSCC when AT2 or Club Cells Are Targeted
To investigate whether other cell types can serve as the cell of origin of LSCC, we targeted Club and AT2 cells of Sox2PC mice. We employed the promoter region of the mouse Cc10 that is expressed by tracheal and bronchiolar Club cells, including bronchoalveolar stem cells (BASCs) (Kim et al., 2005) and the promoter region of the mouse Surfactant Protein C gene (Spc) that is expressed by AT2 cells present in the alveoli and by BASCs. We injected 6- to 8-week-old Sox2PC mice intratracheally with either Ad5-CC10-Cre or Ad5-SPC-Cre (Sutherland et al., 2011). Strikingly, all mice developed tumors around 7–8 months after Cre activation, which were well to moderately differentiated LSCC in more than 90% of cases (Figure S5A and Table S5). Histologically, these tumors showed the hallmarks of LSCC differentiation such as keratin pearls, cornification, necrosis, and inflammation (Figure S5A) and were positive for SOX2, p63, and K5 and negative for TTF-1 (Figure S5B and Table S2). The early lesions were detected by GFP staining, which overlapped with SOX2 and to a lesser extent with K5 and p63 (Figure S5C).
Although the histopathological profiles were perfectly overlapping, the size, number, and location of the lesions differed between the two groups. The number of carcinomas versus early lesions was substantially higher in Ad5-CC10-Cre-injected mice compared with Ad5-SPC-Cre (Figure 7A). The difference was found to be significant by a two-way ANOVA test (Figure 7B). This finding indicates that only a subset of early lesions originating from AT2 cells progresses to LSCC, whereas this process is more efficient when Club cells are targeted. Single cells or small clusters of switched cells in the alveolar compartment of Ad5-SPC-Cre-injected mice were also found to be positive for CC10 as indicated by the dual GFP-CC10 IF (Figure 7C and Table S5), and might represent the previously described subpopulation of AT2 cells expressing a low level of CC10 (Rawlins et al., 2009). When mice were injected with Ad5-CC10-Cre, switched single cells or small clusters were often observed in bronchioles (Figure 7C), at the bronchoalveolar duct junctions (BADJ) and in few cases also in the alveolar compartment (data not shown), where a subpopulation of AT2 cells expressing a low level of CC10 was noted (Rawlins et al., 2009). Early lesions of either injection in mice strongly retained CC10 expression, as showed by GFP-CC10 dual IF (Figure 7D) and, in the case of Ad5-SPC-Cre-injected mice, lost the expression of SPC while progressing, as indicated by SPC-CC10 dual IF (Figure 7D and Table S5). During progression of early lesions to carcinoma, the expression of CC10 is replaced by K5, the two cell markers being mutually exclusively expressed in the lesions of both Ad5-SPC-Cre- and Ad5-CC10-Cre-injected mice (Figure 7E and Table S5). The transition of CC10+ to K5+ cells is complete in advanced LSCC of both Ad5-SPC-Cre- and Ad5-CC10-Cre-injected mice (Figure 7F). Taken together, our data indicate that SOX2 overexpression in combination with Pten and Cdkn2ab deletion is able to drive AT2 and Club cells toward LSCC. Tumor progression is clearly more efficient when Club cells are targeted. In both situations the location of the lesions is peripheral, resembling the human peripheral subtype of LSCC.
Figure 7.
Club and AT2 Cells Are Transformed to Give Rise to LSCC in Sox2PC Mice
(A) Scan images of GFP staining performed on lungs isolated from Sox2PC mice injected with the indicated adenoviruses.
(B) Quantification of the comparison of early lesions versus carcinoma in Ad5-SPC-Cre- and Ad5-CC10-Cre-injected mice (Ad-SPC-Cre, p < 0.0001; Ad5-CC10-Cre, p < 0.05). Data represent means ± SD.
(C–F) Dual IF staining with the indicated markers, performed on lung tissues isolated from Sox2PC mice injected with the indicated adenoviruses, representative of single/small clusters of targeted cells (C), early lesions (D), intermediate lesions (E), and advanced LSCC (F).
Scale bars, 20 μm (C, D) and 50 μm (E, F).
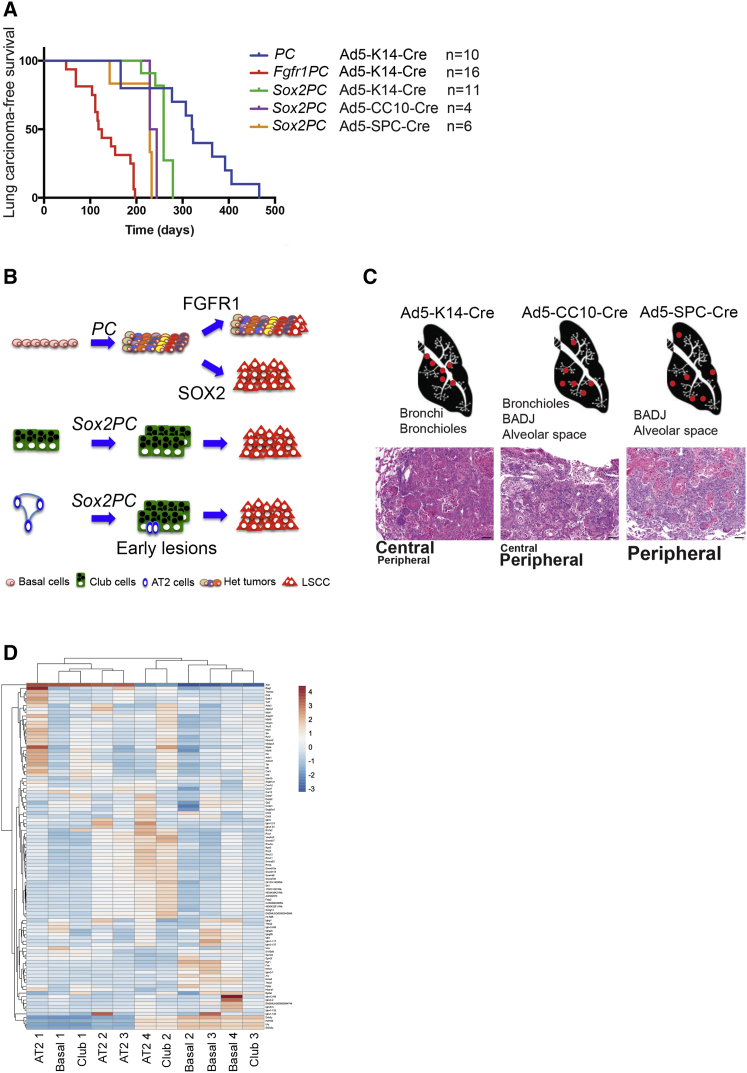
By comparing the tumor-free survival of PC mice with Fgfr1PC and Sox2PC mice, injected with Ad5-K14-Cre, we found that both FGFR1 and SOX2 are able to accelerate tumor onset (Figure 8A and Table S1). The tumor-free survivals of Sox2PC mice injected with Ad5-K14-Cre, Ad5-SPC-Cre, or Ad5-CC10-Cre are comparable, ranging between 7 and 9 months. In our model, Pten and Cdkn2ab deletion provides basal cells with the capability to transform in multiple heterogeneous lesions. FGFR1 overexpression accelerates tumorigenesis without imposing a particular differentiation pattern, resulting only in occasional squamous differentiation. In contrast, SOX2 overexpression remarkably imposes a squamous phenotype to Basal, Club, and AT2 cells (Figure 8B).
Figure 8.
FFGR1 and SOX2 Are Both Tumor Drivers but Only SOX2 Imposes an LSCC Phenotype
(A) Lung carcinoma-free survival curve of mice with the indicated genotypes.
(B) Schematic representation of tumors arising in mice with different genetic lesions, activated in distinct lung cell types: LSCC arises from PC mice following overexpression of SOX2 but not FGFR1. Sox2PC mice develop LSCC also from AT2 and Club cells.
(C) LSCC arises in different locations according to the targeted cell population. Scale bars, 50 μm.
(D) Hierarchical clustering of transcriptional profiling of the indicated Basal-, AT2-, and Club-derived LSCC samples. One hundred genes with the largest variability across samples were used. Columns are for samples and rows are for genes.
Early lesions arise in bronchi and bronchioles when Sox2PC mice are injected with Ad5-K14-Cre; bronchioles, BADJ, and, to a lesser extent, alveolar space when they are injected with Ad5-CC10-Cre, and BADJ and alveolar space when injected with Ad-SPC-Cre (upper panel, Figure 8C). The morphology of LSCC originating from different cells is identical, but the location is in line with the different cell of origin (lower panel, Figure 8C). We performed RNA-sequencing (RNA-seq) analysis and compared the gene expression profiles of Basal-, AT2-, and Club-derived LSCC. Our analysis revealed a strong correlation between their expression profiles, with only 60 genes found to be DE in AT2-derived LSCC versus Basal-derived LSCC, and as little as 28 differentially expressed genes in Club-derived LSCC versus Basal-derived LSCC (Figure 8D and data not shown). These results suggest that enforced expression of SOX2, rather than the cell of origin, dictates the resulting tumor phenotype. We suggest that human peripheral LSCC might also arise from peripheral SPC and CC10 expressing cells.
Discussion
In this study we demonstrate that Pten- and Cdkn2ab-deficient tracheobronchial basal cells can be efficiently transformed into a variety of lung tumors. The additional overexpression of SOX2, but not FGFR1, results in a homogeneous LSCC phenotype closely resembling the human counterpart. Genome-wide transcriptional profiles of tumors from Sox2PC and Kras;p53±Eed mice revealed the differential regulation of gene categories that typify human LSCC over LADC, indicating that SOX2 imposes squamous differentiation most likely by activating a p63-mediated transcriptional epithelial program. Critically, SOX2 overexpression is also able to impose an LSCC phenotype to Club and AT2 cells, indicating the potency of SOX2 in directing transdifferentiation.
The gene promoters we used to target basal cells are already well described as recapitulating the gene expression of K5 and K14 (Jonkers et al., 2001, Ramirez et al., 2004), which are established markers of basal cells of stratified (skin) and pseudostratified epithelia (trachea). Nevertheless, loss of Pten and Cdkn2ab gave rise to a variety of tumors including sarcomas, osteomas, and histiocytic lesions. IHC analysis for a panel of EMT markers including E-cadherin (ECAD), keratin 8 (K8), and vimentin (VIM) showed residual expression of ECAD and K8 in some mesenchymal lesions strongly expressing VIM. In other cases the epithelial compartment co-existed with a distinct mesenchymal compartment, pointing to the presence of multiple populations of basal cells with different lineage commitments. The genotype including Cdkn2ab deletion might play an important role in this high plasticity, which becomes restricted to SCC only by forced SOX2 expression.
Both SOX2 and FGFR1 are frequently amplified in LSCC, in some cases in the absence of mutations in the CDKN2AB and PTEN (Cancer Genome Atlas Research Network, 2012). This led to the hypothesis that they are critical driver oncogenes in this malignancy. Our data support a role for FGFR1 in accelerating tumor development without forcing the cells toward a particular tumor subtype. In contrast, SOX2 appears critical in driving cells toward an aggressive and penetrant LSCC phenotype. It will be interesting to assess in future experiments whether the combination of FGFR1 and SOX2 results in exclusive LSCC with a much shorter latency period than observed with SOX2. It will also be important to determine how SOX2 amplification synergizes with aberrations in TP53 (cell cycle) and NFE2L2 (oxidative stress response) that are also frequently observed in LSCC (Cancer Genome Atlas Research Network, 2012).
The most remarkable feature of LSCC is the amplification on chromosome 3q26 (Bass et al., 2009, Hussenet et al., 2010, Tonon et al., 2005) where both SOX2 and TP63 lie. More refined genomic analyses have shown that SOX2 is the focus of high-level amplifications in lung and esophageal SCC (Bass et al., 2009, Liu et al., 2013). SOX2 is actually amplified and/or overexpressed in 63% of human LSCC (Cancer Genome Atlas Research Network, 2012) but 80%–90% of LSCC expresses SOX2 at the protein level (Brcic et al., 2012, Sholl et al., 2010, Tsuta et al., 2011), suggesting that multiple biological mechanisms can ensure its activation. The homogeneous LSCC phenotype of Sox2PC mice is in line with this and suggests that human LSCC in which SOX2 is not amplified might carry other driver mutations with a similar effect. The expression patterns of SOX2 and p63 clearly overlap; both proteins showed heterogeneous staining. We showed that this heterogeneity is maintained in both human and mouse LSCC regardless of whether the location of the tumor is peripheral or central. Importantly, the co-expression with p63, a known stem cell marker of stratified epithelia (Senoo et al., 2007), suggests a critical role for SOX2-expressing cells in tumor maintenance. This observation is in line with previous studies performed on human skin SCC in which SOX2-expressing cells marked cancer stem cells able to initiate and propagate the tumor (Boumahdi et al., 2014). The transcriptional analysis of LSCC from Sox2PC mice confirmed the strong interplay between p63 and SOX2. Further analysis, for example chromatin immunoprecipitation sequencing using antibodies against either p63 or SOX2 performed on early and late LSCC lesions from Sox2PC mice, could reveal the sequence of events driving the development of LSCC.
We observed a highly immunogenic environment in both human and mouse LSCC. Tumor development is often associated with the local accumulation of myeloid-derived suppressor cells (MDSC), which represent a heterogeneous population of cells including polymorphonuclear neutrophils, monocytes, macrophages, myeloid precursor cells, and dendritic cells, that support tumor growth by suppressing anti-tumor T cell responses. Tumor regression might be achieved by targeting MDSC in LSCC (Chioda et al., 2011, Condamine and Gabrilovich, 2011, Filipazzi et al., 2012, Youn and Gabrilovich, 2010, Youn et al., 2008). Moreover, we found consistently high levels of PD-L1 in LSCC of Sox2PC mice whereas in human LSCC its expression was rather variable. This is not surprising, given the high complexity of human LSCC genome compared with the Sox2PC mouse model in which mice carry the same combination of genetic lesions. However, both human and Sox2PC mouse LSCC consistently express PD-1. Since PD-L1 and PD-1 blockage is an appealing cancer therapy (Zou et al., 2016), the Sox2PC mouse is an appealing model for the assessment of immunotherapeutic parameters.
In the LSCC model in which SOX2 was delivered by intranasal lentivirus administration on an Lkb1 null background, mice developed tumors in 40% of cases and this percentage included a few LADC (Mukhopadhyay et al., 2014). In another model in which SOX2 was overexpressed in either AT2 or Club secretory cells, mice developed well-differentiated LADC but not LSCC (Lu et al., 2010). When targeting AT2 and Club cells, Sox2PC mice exclusively developed LSCC. These results indicate that the context of SOX2 overexpression with other cooperating mutations is decisive in driving these different cell types toward LSCC transformation. Despite the different location RNA-seq data showed strong similarities, with only a small number of genes being DE in LSCC arising from Club and AT2 cells versus basal cells. In future experiments we will address whether any of these can serve as a marker of the cell of origin. Numerous early lesions do not progress to LSCC when AT2 cells are targeted. Although the targeted cells were primarily SPC-expressing cells, some of them also showed CC10 expression. It is possible that the subpopulation of double CC10+SPC+ cells described in the alveolar compartment and at the BADJ (Rawlins et al., 2009) have served as the cell of origin of LSCC when AT2 cells are targeted. The CC10+ lesions then progress further, losing CC10 expression and acquiring squamous markers. In a way we witness here the reverse transdifferentiation as described earlier for conditional KrasG12D mice in which CC10 targeting resulted in the transdifferentiation of SOX2+CC10+ cells toward SPC+ adenocarcinomas (Sutherland et al., 2014). There is an ongoing debate about whether peripheral and central LSCC have different cells of origin, as this might be associated with different responses toward therapy (Funai et al., 2003, Hayashi et al., 2013, Sakurai et al., 2004, Yousem, 2009). Our results point to the likelihood that central and peripheral LSCC have different cells of origin but are otherwise very similar.
In conclusion, we have generated a mouse model that reproduces the salient features of human LSCC, expressing a set of LSCC-specific markers and showing infiltration by distinct subsets of inflammatory cells. Since this mouse model is based on the mutations frequently found in human LSCC, it is likely that identical or, at least, very similar signaling pathways are involved. This makes this model particularly valuable for exploring the effects of additional lesions and as a model to design and assess the value of intervention strategies. Furthermore, the set of mutations we used can impose both peripheral and central LSCC, dependent on the cell types targeted. It will be interesting in future experiments to investigate whether the LSCC retain any molecular markers from their cell of origin and whether this provides new inroads for targeted therapies.
Experimental Procedures
More detailed procedures can be found in Supplemental Experimental Procedures.
Mouse Strains and Human Samples
All experiments involving animals were performed in accordance with Dutch and European regulations on care and protection of laboratory animals and have been approved by the local animal experiment committee at Netherlands Cancer Institute, DEC NKI (OZP ID 12051). Mice were housed under standard condition of feeding, light, and temperature, with free access to food and water.
Ptenflox/flox (Marino et al., 2002), Cdkn2ab flox/flox (Krimpenfort et al., 2007), Kras;p53±Eda (Jackson et al., 2001, Jonkers et al., 2001, Serresi et al., 2016), and mT/mG (Muzumdar et al., 2007) mice have been previously described. LSL-Sox2 and LSL-Fgfr1K656E mice were generated using the Flp-RMCE technology (see Supplemental Experimental Procedures for details). All animals were maintained on an FVB background (backcrossed from strains generated from 129 Ola ESCs).
We collected 12 primary LSCC resections from the archive of the pathology department of the Antoni van Leeuwenhoek hospital. The study CFMPB459 entitled “Genomic and Immunohistochemistry Profiling of Human Squamous Cell Carcinoma” has been reviewed and approved by the NKI Institutional Review Board (IRB). The IRB approval procedure for “secondary use” of human material is according to Dutch legislation (code of conduct). All tumor samples were reviewed by at least two expert pathologists.
Intratracheal Adenovirus Instillation
Mice were treated with cyclosporine A (Novartis) orally in the drinking water, 1 week prior and 2–3 weeks following adenovirus injection. Three days before the infection, mice were intraperitoneally injected with naphthalene (250 mg/kg) or an equal amount of corn oil. On the fourth day they were intratracheally injected with 20 μL of 1 × 1010 pfu/mL purified Ad5-K14-Cre or Ad5-K5-Cre viruses (see Supplemental Experimental Procedures for details). Mice injected with either Ad5-SPC-Cre or Ad5-CC10-Cre were only treated with cyclosporine A 1 week prior to and 2–3 weeks following adenovirus injection.
Histology and Immunohistochemistry
For histological analysis, lungs were inflated and fixed for 24 hr with ethanol/acetic acid/formalin (EAF). Trachea was collected separately and also fixed EAF. Fixed tissues were subsequently dehydrated and embedded in paraffin, and sections of 2–4 μm were prepared and stained with H&E for subsequent histopathological analyses. See Supplemental Experimental Procedures for details of antibodies used for staining.
Gene Expression Profiling
The procedure is described in Supplemental Experimental Procedures.
Author Contributions
G.F. conceived the experiments and conducted most of them, coordinated the project, and wrote the paper. J.Y.S. analyzed tumors and scored human and mouse IHC staining. K.D.S. performed the cloning of adenoviral vectors. R.B. performed RNA-seq data analysis. K.M. provided human material and scored human IHC staining. J.P.L. and N.P. gave technical support for intratracheal injections. G.G. helped in the interpretation of RNA-seq data and statistical analysis, and provided some input in manuscript writing. A.B. supervised the whole project.
Acknowledgments
We wish to thank members of the animal facility of the Netherlands Cancer Institute for maintaining the mice; the Animal Pathology department for producing high-quality histopathological preparations; the NKI-AVL core facility Molecular Pathology & Biobanking for supplying NKI-AVL Biobank material and laboratory support; Ivo Huijbers and Colin Pritchard for assisting in generating the mutant ESC lines; and Sarah Best for experiments that were not included in the final manuscript. We wish to thank Paul Krimpenfort for his input during this work and critically reading the manuscript. This study was supported by a WKO grant to A.B. by the Dutch Cancer Society and by a Synergy ERC grant (COMBATCANCER) in which A.B. is one of the principal investigators. K.D.S. is a recipient of a National Health and Medical Research Council of Australia Overseas based Biomedical Training Fellowship (No. 516781). This work also was made possible through Victorian State Government Operational Infrastructure Support and Australian Government IRIISS, and Worldwide Cancer Research grant 14-0433.
Published: October 10, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2016.09.001.
Accession Numbers
Sequencing data relative to tumors from Kras;p53±Eed mice are available under accession number GEO: GSE61190. Sequencing data relative to tumors from Sox2PC mice are available under accession number GEO: GSE78948.
Supplemental Information
References
- Adamson I.Y., Bowden D.H. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 1974;30:35–42. [PubMed] [Google Scholar]
- Bass A.J., Watanabe H., Mermel C.H., Yu S., Perner S., Verhaak R.G., Kim S.Y., Wardwell L., Tamayo P., Gat-Viks I. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- Brcic L., Sherer C.K., Shuai Y., Hornick J.L., Chirieac L.R., Dacic S. Morphologic and clinicopathologic features of lung squamous cell carcinomas expressing Sox2. Am. J. Clin. Pathol. 2012;138:712–718. doi: 10.1309/AJCP05TTWQTWNLTN. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioda M., Peranzoni E., Desantis G., Papalini F., Falisi E., Solito S., Mandruzzato S., Bronte V. Myeloid cell diversification and complexity: an old concept with new turns in oncology. Cancer Metastasis Rev. 2011;30:27–43. doi: 10.1007/s10555-011-9268-1. [DOI] [PubMed] [Google Scholar]
- Condamine T., Gabrilovich D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y., Liao G., Dixon D., Linnoila R.I., Lori A., Randell S.H., Oren M., Jetten A.M. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am. J. Physiol. Cell Physiol. 2004;287:C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Cabral L.J., Stephens R.J., Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp. Mol. Pathol. 1975;22:142–150. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- Ferone G., Thomason H.A., Antonini D., De Rosa L., Hu B., Gemei M., Zhou H., Ambrosio R., Rice D.P., Acampora D. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol. Med. 2012;4:192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G., Mollo M.R., Thomason H.A., Antonini D., Zhou H., Ambrosio R., De Rosa L., Salvatore D., Getsios S., van Bokhoven H. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum. Mol. Genet. 2013;22:531–543. doi: 10.1093/hmg/dds464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipazzi P., Huber V., Rivoltini L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol. Immunother. 2012;61:255–263. doi: 10.1007/s00262-011-1161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funai K., Yokose T., Ishii G., Araki K., Yoshida J., Nishimura M., Nagai K., Nishiwaki Y., Ochiai A. Clinicopathologic characteristics of peripheral squamous cell carcinoma of the lung. Am. J. Surg. Pathol. 2003;27:978–984. doi: 10.1097/00000478-200307000-00013. [DOI] [PubMed] [Google Scholar]
- Giangreco A., Groot K.R., Janes S.M. Lung cancer and lung stem cells: strange bedfellows? Am. J. Respir. Crit. Care Med. 2007;175:547–553. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Sano H., Egashira R., Tabata K., Tanaka T., Nakayama T., Kashima Y., Hori T., Nunomura S., Fukuoka J. Difference of morphology and immunophenotype between central and peripheral squamous cell carcinomas of the lung. Biomed. Res. Int. 2013;2013:157838. doi: 10.1155/2013/157838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K.U., Reynolds S.D., Watkins S., Fuchs E., Stripp B.R. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- Hussenet T., Dali S., Exinger J., Monga B., Jost B., Dembele D., Martinet N., Thibault C., Huelsken J., Brambilla E., du Manoir S. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., Tuveson D.A. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., Ijpenberg A., Song J.Y., van der Valk M., Nawijn M., Zevenhoven J., Berns A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen Z., Zhu F., Hu Y. IkappaB kinase alpha and cancer. J. Interferon Cytokine Res. 2012;32:152–158. doi: 10.1089/jir.2011.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Jiang M., Lu Y., Chen H., Sun J., Wu S., Ku W.Y., Nakagawa H., Kita Y., Natsugoe S. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Futtner C., Rock J.R., Xu X., Whitworth W., Hogan B.L., Onaitis M.W. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkoski S.P., Cleaver T.G., Thompson J.J., Sutton W.P., Haeger S.M., Rodriguez K.J., Lu S.L., Merrick D., Wang X.J. Role of PTEN in basal cell derived lung carcinogenesis. Mol. Carcinog. 2014;53:841–846. doi: 10.1002/mc.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Krimpenfort P., Leung C., van der Korput H.A., Trapman J., Camenisch I., Berns A., Brandner S. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Berrett K.C., Kc U., Clair P.M., Pop S.M., Carr S.R., Witt B.L., Oliver T.G. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep. 2014;8:40–49. doi: 10.1016/j.celrep.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch C., Muthana M., Coffelt S.B., Lewis C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Ramirez A., Page A., Gandarillas A., Zanet J., Pibre S., Vidal M., Tusell L., Genesca A., Whitaker D.A., Melton D.W., Jorcano J.L. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- Rawlins E.L., Okubo T., Xue Y., Brass D.M., Auten R.L., Hasegawa H., Wang F., Hogan B.L. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Asamura H., Watanabe S., Suzuki K., Tsuchiya R. Clinicopathologic features of peripheral squamous cell carcinoma of the lung. Ann. Thorac. Surg. 2004;78:222–227. doi: 10.1016/j.athoracsur.2004.01.029. [DOI] [PubMed] [Google Scholar]
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Serresi M., Gargiulo G., Proost N., Siteur B., Cesaroni M., Koppens M., Xie H., Sutherland K.D., Hulsman D., Citterio E. Polycomb repressive complex 2 is a barrier to KRAS-driven inflammation and epithelial-mesenchymal transition in non-small-cell lung cancer. Cancer Cell. 2016;29:17–31. doi: 10.1016/j.ccell.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Sholl L.M., Long K.B., Hornick J.L. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl. Immunohistochem. Mol. Morphol. 2010;18:55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Sutherland K.D., Berns A. Cell of origin of lung cancer. Mol. Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland K.D., Proost N., Brouns I., Adriaensen D., Song J.Y., Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- Sutherland K.D., Song J.Y., Kwon M.C., Proost N., Zevenhoven J., Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl. Acad. Sci. USA. 2014;111:4952–4957. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H.A., Zhou H., Kouwenhoven E.N., Dotto G.P., Restivo G., Nguyen B.C., Little H., Dixon M.J., van Bokhoven H., Dixon J. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J. Clin. Invest. 2010;120:1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon G., Wong K.K., Maulik G., Brennan C., Feng B., Zhang Y., Khatry D.B., Protopopov A., You M.J., Aguirre A.J. High-resolution genomic profiles of human lung cancer. Proc. Natl. Acad. Sci. USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W.D. Pathology of lung cancer. Clin. Chest Med. 2002;23:65–81. doi: 10.1016/s0272-5231(03)00061-3. viii. [DOI] [PubMed] [Google Scholar]
- Tsuta K., Tanabe Y., Yoshida A., Takahashi F., Maeshima A.M., Asamura H., Tsuda H. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. J. Thorac. Oncol. 2011;6:1190–1199. doi: 10.1097/JTO.0b013e318219ac78. [DOI] [PubMed] [Google Scholar]
- Visvader J.E. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Walker S. Updates in non-small cell lung cancer. Clin. J. Oncol. Nurs. 2008;12:587–596. doi: 10.1188/08.CJON.587-596. [DOI] [PubMed] [Google Scholar]
- Weiss J., Sos M.L., Seidel D., Peifer M., Zander T., Heuckmann J.M., Ullrich R.T., Menon R., Maier S., Soltermann A. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson M.D., Yin X., Hoadley K.A., Liu Y., Hayward M.C., Cabanski C.R., Muldrew K., Miller C.R., Randell S.H., Socinski M.A. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin. Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Pang Y., Wilkerson M.D., Wang D., Hammerman P.S., Liu J.S. Gene-expression data integration to squamous cell lung cancer subtypes reveals drug sensitivity. Br. J. Cancer. 2013;109:1599–1608. doi: 10.1038/bjc.2013.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Jiang Q., Willette-Brown J., Xi S., Zhu F., Burkett S., Back T., Song N.Y., Datla M., Sun Z. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell. 2013;23:527–540. doi: 10.1016/j.ccr.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fillmore C.M., Koyama S., Wu H., Zhao Y., Chen Z., Herter-Sprie G.S., Akbay E.A., Tchaicha J.H., Altabef A. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Gabrilovich D.I. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur. J. Immunol. 2010;40:2969–2975. doi: 10.1002/eji.201040895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Nagaraj S., Collazo M., Gabrilovich D.I. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Collazo M., Shalova I.N., Biswas S.K., Gabrilovich D.I. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousem S.A. Peripheral squamous cell carcinoma of lung: patterns of growth with particular focus on airspace filling. Hum. Pathol. 2009;40:861–867. doi: 10.1016/j.humpath.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl Med. 2016;8:328rv324. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.