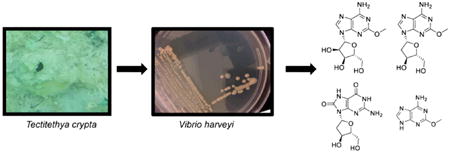
Abstract
Spongosine (1), deoxyspongosine (2), spongothymidine (Ara T) (3), and spongouridine (Ara U) were isolated from the Caribbean sponge Tectitethya crypta and given the general name “spongonucleosides”. Spongosine, a methoxyadenosine derivative, has demonstrated a diverse bioactivity profile including anti-inflammatory activity and analgesic and vasodilation properties. Investigations into unusual nucleoside production by T. crypta-associated microorganisms using mass spectrometric techniques have identified a spongosine-producing strain of Vibrio harveyi and several structurally related compounds from multiple strains.
Graphical abstract
In an attempt to isolate sterols from a marine sponge in the early 1950s, a crystalline material was separated from sponge tissue by refluxing in acetone.1 This material was recrystallized and subjected to a variety of chemical and spectroscopic analyses and was ultimately determined to consist of three compounds, two of which were pyrimidine bases linked to the sugar arabinose [1-β-d-arabinofuranosylthymine (spongothymidine or Ara T) and 1-β-d-arabinofuranosyluracil (spongouridine or Ara U)] and a third purine compound containing ribose [9-β-d-ribofuranosyl-2-methoxyadenosine (spongosine)].1,2A subsequent investigation of Tectitethya crypta from Western Australia yielded another nucleoside analogue, 2′-deoxyspongosine.3 While the arabinonucleosides are exceptional in their utility to cancer chemotherapy, spongosine has been investigated for its vasodilation4 and analgesic activities,5 whereas 2′-deoxyspongosine has shown moderate antitumor activity.6
There is growing recognition that bacteria and sponges engage in significant relationships;7–9 a sponge's biomass may contain up to 35% bacterial cells,7 and over 25 bacterial phyla have been isolated from within marine sponges alone.9 Sponges have been extraordinarily rich sources of structurally diverse and highly bioactive natural products, such as the parent structure halichondrin A, which has given rise to the anticancer agent eribulin.10 On the basis of biosynthetic precedence, it has been speculated that many sponge-derived metabolites actually derive from metabolic processes of their associated microflora.11 However, relatively few unequivocal examples exist in which the biosynthesis of a natural product has been demonstrated from a marine sponge-derived microbe. For example, several diketopiperzines have been isolated from a cultivated Micrococcus species associated with specimens of the sponge Tedania ignis, the originally described source of these compounds.12 Additionally, localization experiments and culture-independent metagenomic methods have been utilized to strongly suggest that associated microbes are the true producers of many described secondary metabolites isolated from marine invertebrates. The examination of the modular biosynthetic pathways present in the associated microbial genomes of a sponge has led to the putative microbial producer of onnamide.13 Metagenomic studies of uncultured invertebrate-associated microbes have led to the discovery of novel biosynthetic pathways and previously undescribed structural classes.14,15 Swinholide A was located in a bacterial cell population separated by differential centrifugation from the sponge cell population.16 Interestingly, this same molecule was isolated from a collection of cyanobacteria, calling into question the true biosynthetic source.17 Gene-based imaging methods such as catalyzed reporter deposition–fluorescence in situ hybridization (CARD-FISH) have been used to co-localize a microbial population and a putative biosynthetic gene within invertebrate tissue, such as in the case of the barbamide homologue dysB1 within a population of cyanobacteria cells in the sponge Dysidea herbacea.11
Herein, we have screened extracts of bacterial isolates from the marine sponge T. crypta using LC-MS/MS-based molecular networking18 and have identified a sponge-associated Vibrio harveyi strain that produces spongosine (1), deoxyspongosine (2), 8-oxo-2′-deoxyguanosine (4), and methoxyadenine (6). These results support a microbial source for the unusual nucleosides originally found in this marine invertebrate, and fermentation of this strain may abrogate the need for sponge collection to acquire these valuable metabolites.
Results and Discussion
Identification of Spongonucleosides in T. crypta
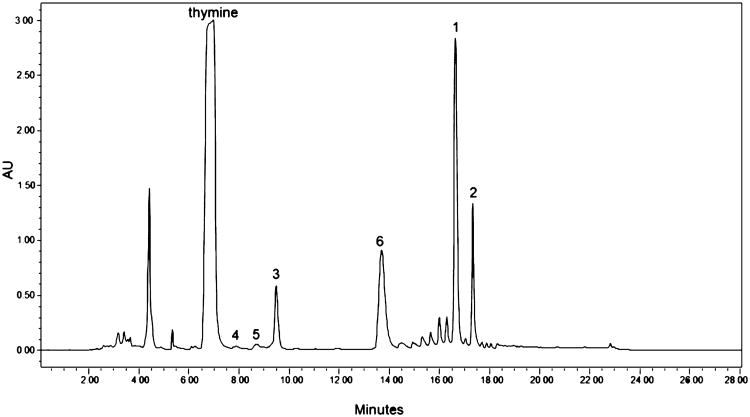
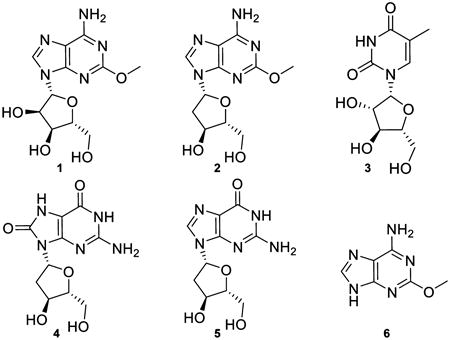
Five nucleosides [spongosine (1), 2′-deoxyspongosine (2), Ara-T (3), 8-oxo-2′-deoxyguanosine (4), and 2′deoxyguanosine (5)] and one modified purine [(2-methoxyadenine (6)] were isolated from field-collected specimens of T. crypta (Figure 1). UV absorbance maxima ranging from 250 to 270 nm were consistent with heterocyclic aromatic compounds. Spongosine was the most abundant nucleoside isolated from T. crypta, accounting for ca. 0.15% of the sponge dry weight. Spectroscopic data for this metabolite matched reported values (Supporting Information).1–3
Figure 1.
Isolation of nucleosides from Tectitethya crypta. HPLC chromatogram monitored at 269 nm. HPLC peaks characterized by MS and NMR analysis and identified as thymine, (1) spongosine, (2) 2′-deoxyspongosine, (3) Ara-T, (4) 8-oxo-2-deoxyguanosine, (5) 2′-deoxyguanosine, and (6) methoxyadenine.
Sponge and Associated-Microbe MS/MS Networking
MS/MS-based molecular networking has emerged as an extremely useful tool for dereplication in natural products research as well as in the search for analogues of known compounds.18,19 In the current report, we used MS/MS-based molecular networking as a method to screen microorganisms isolated from T. crypta for the production of known compounds as well as new analogues. This application of the technique allowed for great insight into the expressed nucleoside chemistry of the sponge holobiont.
Due to their structural similarities, the above identified spongonucleosides from T. crypta clustered together in the MS/MS-based molecular network. Spongonucleoside MS/MS spectra have been uploaded and annotated at gnps.ucsd.edu. Several pure bacterial strains were isolated from T. crypta endoderm, and those that could be cultivated in liquid media were given an alphabetical designation (strains A, C, G, H, I, and J). Subsequently, the 3:1 H2O/MeOH fractions of these sponge-associated microbial MeOH extracts were analyzed by LC-MS/MS in order to determine if unusual nucleosides were produced by any of these strains. Interestingly, consensus nodes were observed between the sponge and a strain of Vibrio harveyi (strain A) for spongosine, 2′-deoxyspongosine, 8-oxo-2′-deoxyguanosine, and a node with m/z 280.0928 for which HRMS analysis gave a molecular formula of C12H13N3O5 (Figure 2). While we have not yet been able to isolate this latter compound from extracts of sponge tissue or bacteria, it seems highly likely that it is a related nucleoside. Two additional metabolites from V. harveyi clustered most closely with the m/z 280.0928 node. HRMS indicated a molecular formula of C12H15N3O3 with an m/z 250.1172 and a formula of C12H13N3O6 with an m/z 296.0860 (Figure 2). Several additional metabolites from V. harveyi as well as a Micrococcus sp. (strain H) and Dietzia sp. (strain I) isolated from the sponge clustered with the nucleoside network and appeared to be related by HRMS analysis: C10H14N2O2, m/z 195.1128 was found in V. harveyi, a Micrococcus sp., and Dietzia sp.; C10H12N2O2, m/z 193.0984, C10H16N2O2, m/z 197.1276, and C9H12N2O2, m/z 181.0971 were found in V. harveyi. The Micrococcus sp. and Dietzia sp. shared a metabolite with m/z 251.1517 and formula C12H18N4O2 (Figure 2) that also clustered with the nucleosides.
Figure 2.
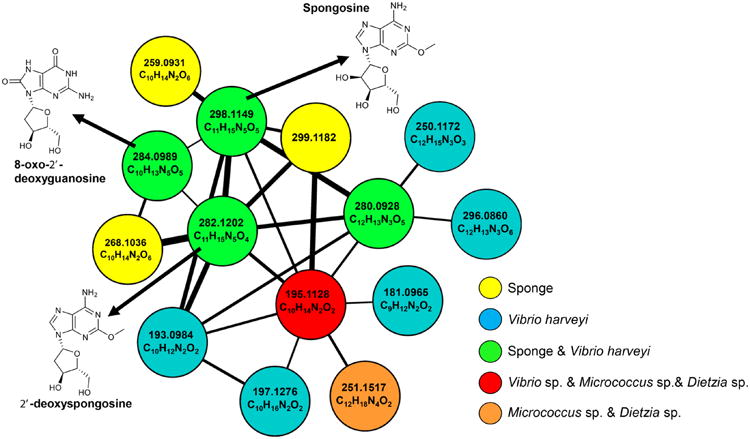
MS/MS-based molecular network. The network was constructed using pure nucleosides isolated from Tectitethya crypta, the T. crypta HPLC prefraction, and prefractions from six bacterial strains (A, C, G, H, I, and J). The cosine level was adjusted to 0.7. Nodes display accurate mass measurements and molecular formulas. Green nodes are metabolites present in sponge tissue and strain A (Vibrio harveyi strain). Strains C, G, and J did not have metabolites that networked with the nucleoside cluster. The m/z 299 is an isotopomer of spongosine (1).
Most intriguing, the sponge and V. harveyi extracts shared a consensus node for spongosine (1) at m/z 298, and an inspection of their MS/MS fragmentation patterns revealed that they were identical (Figure 3A and B). This was also observed for 2′-deoxyspongosine and 8-oxo-deoxyguanosine, thus indicating that at least these three compounds are metabolic products of V. harveyi. Expanding the T. crypta–V. harveyi MS/MS-based network by adjusting the cosine value to 0.6 showed the addition of several nodes from both the sponge and the V. harveyi strain and an additional node at m/z 166. This compound was determined to be 2-methoxyadenine, the aglycone of spongosine, and represented a fourth V. harveyi metabolite (Figure S1).
Figure 3.
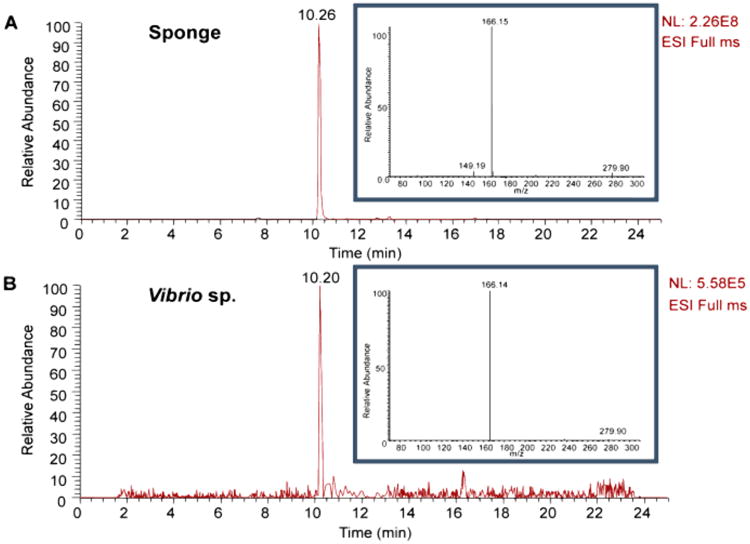
Extracted ion m/z 298 for spongosine (1) from extracts of (A) Tectitethya crypta and (B) Vibrio harveyi shows identical retention time and MS/MS spectra to m/z 166 as the base peak (inset).
While we have demonstrated spongosine and other unusual nucleoside production from this V. harveyi strain, we cannot unequivocally exclude that T. crypta itself is capable of spongosine biosynthesis. Separate analysis of sponge and bacteria cells might provide insight into this question. We cannot discount the possibility of additional spongosine production from microbes other than Vibrio, and thus we are continuing to screen other sponge-derived microbial strains for the presence of unique nucleosides.
Time Course of Spongosine Production
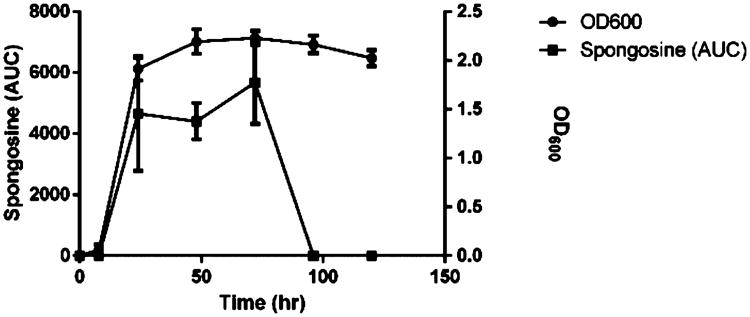
Cultures of V. harveyi were inoculated into 1.5 L flasks, and subsamples were taken every 12 to 24 h over a 5-day period for LC-MS/MS analysis. Spongosine production linearly increased over the first few days of the culture, peaking at 72 h, a time point coinciding with maximal growth (Figure 4). Intriguingly, spongosine could not be detected at time points after 72 h and thus appears to be metabolized to unknown and uncharacterized compounds in late stationary phase.
Figure 4.
Time course of spongosine production. Growth and relative spongosine production of Vibrio harveyi plotted versus time (n = 3). Relative production of spongosine was calculated as the area under the curve (AUC) of the extracted spongosine ion (m/z 298). Vidarabine (Ara A) was used as an internal standard.
Natural products have long been a source of structurally intriguing and bioactive small molecules, and there are several examples of clinical therapeutics and clinical trial agents that are marine organism-derived or inspired.20 A traditional problem in marine natural products research is providing a robust and reliable supply of compound for clinical testing. Total synthesis can provide one solution to this problem; however, another is through cultivation of the source microorganism.21 While the production of spongosine (1) in this Vibrio strain appears to be relatively low at the present time (ca. 72 μg/L), optimization of the culture, extraction, and isolation conditions could likely improve this yield.
Anti-inflammatory Activity of Spongonucleosides
All of the nucleosides isolated from T. crypta, including the HPLC prefraction as well as vidarabine, were evaluated for their ability to inhibit the LPS-induced production of nitric oxide in RAW cells. Interestingly, all of these nucleosides showed appreciable anti-inflammatory activity compared to the LPS control (Figure 5). Spongosine showed the greatest anti-inflammatory effects with a ∼60% reduction in nitric oxide production at 3 μg/mL. This appeared to be a true anti-inflammatory effect, as there was no significant reduction in cell viability in the MTT viability assay (data not shown). The remaining spongonucleosides also showed anti-inflammatory activity; however, none were as potent as spongosine (Figure S2A–E).
Figure 5.
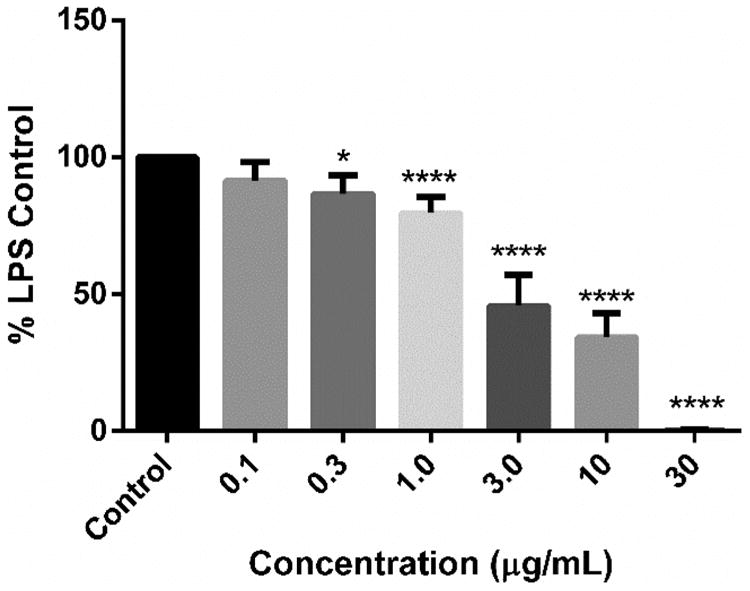
Anti-inflammatory activity of spongosine. Purified spongosine isolated from Tectitethya crypta was tested for its ability to inhibit the LPS-induced inflammation response in RAW cells. Bars are means (n = 6 treatments) ± standard error. Significant difference of treatments from the LPS control were determined using ANOVA followed by Dunnett's multiple comparison procedure *p < 0.05; ****p < 0.0001. Compounds did not significantly alter cell viability (measured as absorbance at 570 nm) in the MTT assay compared to control (p > 0.05) (data not shown).
In this respect, spongosine has significant pharmaceutical potential. It has patented analgesic properties and has been shown to reduce pain in mammals without significant side effects.5 Thus, its isolation from the fermentation of a microbial source could constitute a cost-effective means of production.
MLSA Analysis and Identification of the Spongosine-Producing Microbe
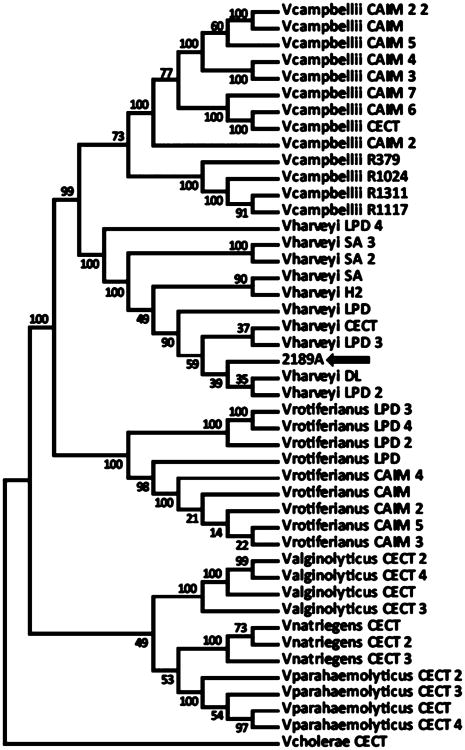
Having observed production of spongosine and other unusual nucleosides from strain A, genomic DNA was isolated from this strain and sequenced to identify the organism and examine the genetic architecture for spongosine biosynthesis. A single 16S rRNA sequence was identified in the genome sequence of strain A. BLAST searching of this sequence identified this strain most closely with V. harveyi (99% identity over 1350 nt). A phylogenetic tree was constructed with this 16S rRNA sequence data from the spongosine-producing strain as well as six core Vibrio species: V. alginolyticus, V. campbellii, V. harveyi, V. natrigens, V. parahemolyticus, and V. rotiferianus. The tree was rooted with V. cholera; however, it had poor branch support, poor node support, and fairly indiscriminate clade separation (data not shown). To improve the species-level identification of this strain, a multilocus sequence analysis (MLSA) was performed, using six well-conserved loci: pyrH, gyrB, rctB, recA, rpoD, and toxR.22 The phylogenetic tree using these data contained far superior clade separation and node support, indicative of the enhanced information provided by MLSA. This analysis indicated that strain A was indeed V. harveyi, and its close yet distinct relationship to other members of the V. harveyi clade suggests that it may be a previously undescribed strain of this species (Figure 6).
Figure 6.
Multilocus sequence analysis using pyrH, gyrB, rctB, recA, rpoD, and toxR genes from Vibrio strains. Black arrow shows position of strain A firmly in the Vibrio harveyi clade. Sequences were compiled from GenBank and http://www.taxvibrio.lncc.br/.
Genome Analysis of Potential Spongosine Biosynthetic Pathway
The size of the assembled V. harveyi genome was 5.9 Mb with a 45% GC content and was composed of 342 contigs, and there were 3537 open reading frames greater than 600 bp in length. The maximum contig length was 426 671 base pairs. AntiSMASH analysis annotated six potential secondary metabolite pathways. Three were bacteriocins,23 and one encoded the osmolyte ectoine.24 BLAST searching of a deduced siderophore pathway showed high identity to the vibrioferrin pathway.25 Finally, a modular NRPS pathway was predicted to encode for a second siderophore, enterobactin.26
The genome was assessed for genes associated with de novo nucleoside biosynthesis and nucleoside salvage pathways27,28 as well as for genes catalyzing oxidation and methylation such as p450, oxidoreductases, and O-methyltransferases. Six oxidoreductases and seven methyltransferases were identified, two of which were predicted to be O-methyltransferases by genome mining and BLAST search. Two gene clusters were identified that had an FAD-dependent oxidoreductase and O-methyltransferase in close proximity. The first cluster appears to be involved in lysophospholipid biosynthesis, whereas the second cluster included genes encoding for an adenylosuccinate synthetase, an oxidoreductase, an O-methyltransferase, and a phosphatase of the haloacid dehalogenase superfamily (HAD). Thus, the latter gene cluster is consistent with the modifications predicted for the biosynthesis of spongosine (1), and is provisionally identified as the spongosine biosynthetic gene cluster (spg).
Spongosine production in V. harveyi was found to reach a maximum in stationary phase, suggesting that the compound may be produced through a nucleoside salvage pathway so as to conserve energy expenditure.29 The known pathway of AMP salvage is via reaction of adenine and phosphoribosyl pyrophosphate by adenine phosphoribosyltransferase. Alternatively, adenylosuccinate generated by adenylosuccinate synthetase could be modified by adenylosuccinate lyase acting in trans to generate AMP; genes encoding these latter enzymes are present in the putative spg pathway in V. harveyi. The conceivable conversion of AMP to spongosine would involve a hydroxylation of C-2 via an oxidoreductase enzyme followed by methylation with S-adenosyl methionine (SAM) and involving an O-methyltransferase. A final step in the production of spongosine (1) would involve a phosphatase-catalyzed dephosphorylation. The spg cluster in the genome of this sponge-derived V. harveyi contains all of these biochemical components. Current efforts are focused on heterologous expression of the cluster or creation of knockout mutants to unequivocally demonstrate its function in spongosine biosynthesis.
Summarizing, we have shown that a source of spongosine (1), 2′-deoxyspongosine (2), and 8-oxo-2′-deoxyguanosine (4) in the sponge T. crypta is a strain of an associated bacterium identified as V. harveyi. While the 16S rRNA analysis did not provide the support necessary to clade the spongosine-producing strain within V. harveyi, MLSA showed strong support for this assignment. It should be noted that we were not able to identify a microbial producer of any of the arabinonucleosides; this remains an area of high interest and continuing investigation. Finally, genome mining and infomatic analysis allowed identification of the putative biosynthetic gene cluster encoding spongosine (spg) biosynthesis.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a JASCO P-2000 polarimeter. LC-MS/MS analysis was carried out using a ThermoFinnigan LCQ AdvantageMax mass detector equipped with an electrospray ionization (ESI) source. HRESIMS spectra were obtained using an Agilent 1290 Infinity system with an Agilent 6530 Accurate Mass Q-TOF LC/MS, and 1H NMR spectra were obtained using a Varian Unity 500 MHz spectrometer. Semipreparative HPLC was carried out using a Waters 515 pump system equipped with a Waters 996 PDA.
Sponge Collection and Extraction
Specimens of Tectitethya crypta were collected by hand from sponge flats surrounding Long Key and Big Pine Key in Florida under State of Florida Scientific Collection Permit in April 2013. The body of the sponge was olive-colored, hemispherical, and covered by a thin layer of sediment with flattened tubercules. The osculum was visible above the sediment layer (Figure 7A). Examination of sponge siliceous spicules showed strongyles and asters consistent with those of T. crypta.30 Specimens were stored in 1:1 2-propanol/seawater for subsequent extraction. Wet sponge biomass (300 g) was frozen and lyophilized to dryness. Dry biomass was extracted three times with MeOH, and the MeOH extract was dried using rotary evaporation. This extract was partitioned between n-butanol and H2O. The n-butanol extract was dried, and the residue was partitioned over a C18 SPE eluting with 3:1 H2O/MeOH.
Figure 7. Tectitethya crypta.

collection and isolation of associated microbes. (A) Underwater photo of T. crypta. Arrow points to visible oscula. (B) Sponges were dissected and inner tissue was streaked onto agar to cultivate associated microbes. (C) Isolation of strain A on marine agar. Strain A was identified as Vibrio harveyi (see Figure 6).
Isolation of Spongonucleosides
The 3:1 H2O/MeOH fraction (HPLC prefraction) was subjected to semipreparative HPLC at a constant flow rate of 3 mL/min. A 10 min hold of 95%/5% solvent A (H2O + 2.5 mM ammonium acetate) and solvent B (CH3CN) was followed by a linear gradient over 10 min to 60% A and 40% B. Initial conditions were restored from 21 to 28 min. Compounds were separated using a Luna 5 μm Phenyl Hexyl column (250 × 10 mm) (Phenomenex, Inc.).
Isolation, Cultivation, and Extraction of Associated Microorganisms
Before preservation of collected sponge tissue, marine agar and SWBG-11 agar plates were streaked with freshly and aseptically exposed sponge endoderm (Figure 7B). Plates were incubated at 28 °C until colonies appeared. Pure colonies obtained by repetitive streaking on agar (Figure 7C) were subsequently inoculated into 100 mL of sterilized LB broth supplemented with 2% NaCl and incubated in a shaker (120 rpm) at 28 °C. Culture flasks were visually observed for growth, and the cultures were grown until OD600 measurements reached a value of 2. Six isolates were cultivated (strains designated A, C, G, H, I, and J) and then harvested by centrifugation. The pelleted biomass was subjected to an identical extraction procedure to that for the sponge tissue, resulting in a 3:1 H2O/MeOH fraction for LC-MS analysis.
LC-MS Analysis and MS/MS-Based Molecular Networking
The 3:1 H2O/MeOH fraction and HPLC fractions from the sponge, as well as the 3:1 H2O/MeOH fractions from the isolated bacteria, were analyzed using LC-MS/MS scanning in positive mode. Solvents used were H2O + 0.1% formic acid (solvent A) and CH3CN (solvent B), and a constant flow rate of 0.7 mL/min was employed. An initial 5 min hold of solvent A at 100% was followed by a linear gradient to 50% A/50% B over 10 min. This was followed by a linear gradient to 25% A/75% B over 5 min, and initial conditions were restored from min 21–25. Compounds were separated using a Kinetex 5 μm C18 column (150 × 4.6 mm) (Phenomenex, Inc.). The polarity of the MS system was set to positive with a mass range of 100–2000. The data type was set to centroid with data-dependent acquisition and dynamic exclusion enabled. The exclusion duration was 3 min. Subsequently, an MS-based metabolomic study was initiated using MS/MS-based molecular networks.18 These data were used for the generation of a sponge and bacteria metabolite molecular network using MS/MS fragmentation patterns as molecular fingerprints. Similarities of fragment ions, relative intensities, and m/z differences between paired spectra were compared, and similarity was determined using a modified cosine calculation as a similarity score (30). Networks were generated using MATLAB scripts, and control spectra from solvent and LB media were subtracted. Networks were visualized using the Cytoscape, where parent masses are represented by nodes (circles) and cosine similarities visualized as edges (lines). The thickness of edges is indicative of similarity, with thicker edges indicating greater similarity.
Time Course of Spongosine Production
Three 1.5 L replicate cultures of strain A were cultured in LB broth + 2% NaCl. A 50 mL aliquot of culture fluid was subsampled at specific time points over a 5-day period. Subsamples were centrifuged, the pelleted biomass was extracted twice in MeOH, and the spongosine production was analyzed by HRESIMS using the LC-MS/MS method described above. Relative spongosine concentration was measured by determining the area under the curve of the spongosine-extracted ion peak compared to that of vidarabine, which was used as an internal standard (25 μg/mL). Spongosine production and bacterial growth (OD600) were plotted using Prism software (GraphPad Software Inc.).
Anti-inflammatory Assay
The anti-inflammatory potential of spongonucleosides isolated from T. crypta and synthetic vidarabine (Ara A) (Alfa Aesar) was evaluated with a nitric oxide assay following previously published methods.31 Murine macrophages (cell line RAW 264.7) were incubated with spongonucleosides for 1 h. Following incubation, an inflammatory agent [bacterial lipopolysaccharide (LPS) 3 μg/mL] was added, and cells were incubated for 24 h. Inflammation was assessed as nitric oxide production and colorimetrically measured with a Griess stain for nitrite, the primary breakdown product of NO. Cells were treated with spongonucleosides at 0.1, 0.3, 1.0, 3.0, 10, and 30 μg/mL (n = 6 wells treated per concentration), and treated cells were compared to LPS controls to determine the percent reduction in nitrite production. An MTT viability assay32 was subsequently conducted with RAW 264.7 cells to determine if the reduced nitrite production was a result of reduced cell viability. Graphs were constructed using Prism software (Graphpad Software Inc.).
DNA Isolation, Genome Sequencing, and MLSA Analysis
Genomic DNA was isolated from strain A (Qiagen Genomic Tip kit, Qiagen), sequenced (MiSeq, Illumina Systems), error corrected using BayesHammer,33 and assembled using SPAdes 3.0 in standard multicell mode.34 Secondary metabolite potential was analyzed using antiSMASH.35 Phylogenetic analysis was carried out for strain A comparing six well-conserved loci: pyrH, gyrB, rctB, recA, rpoD, and toxR. Sequences were aligned using Clustal W and a neighbor-joining tree constructed using the Jukes–Cantor method.36 A bootstrap consensus tree was inferred from 1000 replicates. Sequence alignments and phylogenetic trees were constructed in MEGA v. 5.2.
Supplementary Material
Acknowledgments
We thank the WLG Foundation and NIH CA100851 and GM 107550A for financial support. A.K. was supported in part by the Russian Science Foundation (grant 14-50-00069). We thank the State of Florida for scientific collection permits, the Newfound Harbor Marine Institute and Keys Marine Lab for access to boats and laboratory facilities in the Florida Keys, and the Mevers family for use of their car.
Dedication: Dedicated to Dr. William Fenical of Scripps Institution of Oceanography, University of California–San Diego, for his pioneering work on bioactive natural products.
Footnotes
Supporting Information: An additional MS/MS-based molecular network, MS/MS spectra, 1H NMR spectra, spongonucleoside spectroscopic data, bioactivity data, and photographs of T. crypta spicule preparations are included. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes: The authors declare no competing financial interest.
References
- 1.Bergmann W, Feeney RJ. J Am Chem Soc. 1950;70:2809–2810. [Google Scholar]
- 2.Bergmann W, Feeney RJ. J Org Chem. 1950;16:981–987. [Google Scholar]
- 3.Searle PA, Molinski TF. J Nat Prod. 1994;57:1452–1454. [Google Scholar]
- 4.Bhakuni DS, Rawat DS. Bioactive Marine Natural Products. Springer; New York: 2005. [Google Scholar]
- 5.Richardson P. U.S. Patent 0010475A1. 2007
- 6.Christensen LF, Broom AD, Robins MJ, Bloch A. J Med Chem. 1972;15:735–739. doi: 10.1021/jm00277a010. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MW, Radax R, Steger D, Wagner M. Microbiol Mol Biol R. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piel J. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 9.Webster NS, Taylor MW. Environ Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 10.Hirata Y, Uemura D. Pure Appl Chem. 1986;58:701–710. [Google Scholar]
- 11.Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, Dorrestein PC, Gerwick WG. Proc Natl Acad Sci USA. 2007;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stierle AC, Cardellina JH, Singleton FL. Experientia. 1988;44:1021. doi: 10.1007/BF01939910. [DOI] [PubMed] [Google Scholar]
- 13.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Proc Natl Acad Sci USA. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MC, Piel J. Chem Biol. 2013;20:636–647. doi: 10.1016/j.chembiol.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. J Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 16.Bewley CA, Holland ND, Faulkner DJ. Experientia. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]
- 17.Andrianasolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail K, Leal RM, Mooberry SL, Gerwick WH. Org Lett. 2005;7:1375–1378. doi: 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 18.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raajimakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Proc Natl Acad Sci USA. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC. J Nat Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerwick WH, Moore BS. Chem Biol. 2012;19:85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montaser R, Luesch H. Future Med Chem. 2012;3:1475–1489. doi: 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson FL, Gomez-Gil B, Vasconcelos AT, Sawabe T. Appl Environ Microbiol. 2007;73:4279–4285. doi: 10.1128/AEM.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nes IF, Diep DB, Håvarstein LS, Brurberg MB, Eijsink V, Holo H. Antonie van Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 24.Ono H, Sawada K, Khunajakr N, Tao T, Yamamoto M, Hiramoto M, Shinmyo A, Takano M, Murooka Y. J Bacteriol. 1999;181:91–99. doi: 10.1128/jb.181.1.91-99.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanabe T, Funahashi T, Nakao H, Miyoshi S, Shinoda S, Yamamoto S. J Bacteriol. 2003;185:6938–6949. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehring AM, Bradley KA, Walsh CT. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 27.Nyhan WL. eLS. 2005 doi: 10.1038/npg.els.003909. [DOI] [Google Scholar]
- 28.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5th. W. H. Freeman and Company; New York: 2002. p. 1515. [Google Scholar]
- 29.Proksch P. Toxicon. 1994;32:639–655. doi: 10.1016/0041-0101(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 30.Sará M. In: Systema Porifera: A Guide to the Classification of Sponges. Hooper JNA, Van Soest RWM, editors. Kluwer Academic/Plenum Publishers; New York: 2002. pp. 245–265. [Google Scholar]
- 31.Villa FA, Lieske K, Gerwick L. Eur J Pharmacol. 2010;629:140–146. doi: 10.1016/j.ejphar.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 33.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelski A, Pyshkin A, Sirotkin A, Sirotkin Ya, Stepanauskas R, Clingenpeel S, Woyke T, Mc Lean J, Lasken R, Tesler G, Alekseyev M, Pevzner P. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolenko S, Korobeynikov A, Alekseyev M. BMC Bioinf. 2014;14:S7. doi: 10.1186/1471-2164-14-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. Nucleic Acids Res. 2011;39:1–8. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jukes TH, Cantor CR. In: Mammalian Protein Metabolism. Munro HN, editor. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.