Abstract
In vitro liver models provide essential information for evaluating drug metabolism, metabolite formation, and hepatotoxicity. Interfacing liver models with other organ models could provide insights into the desirable as well as unintended systemic side effects of therapeutic agents and their metabolites. Such information is invaluable for drug screening processes particularly in the context of secondary organ toxicity. While interfacing of liver models with other organ models has been achieved, platforms that effectively provide human-relevant precise information are needed. In this concise review, we discuss the current state-of-the-art of liver-based multiorgan cell culture platforms primarily from a drug and metabolite perspective, and highlight the importance of media-to-cell ratio in interfacing liver models with other organ models. In addition, we briefly discuss issues related to development of optimal liver models that include recent advances in hepatic cell lines, stem cells, and challenges associated with primary hepatocyte-based liver models. Liver-based multiorgan models that achieve physiologically relevant coupling of different organ models can have a broad impact in evaluating drug efficacy and toxicity, as well as mechanistic investigation of human-relevant disease conditions.
Introduction
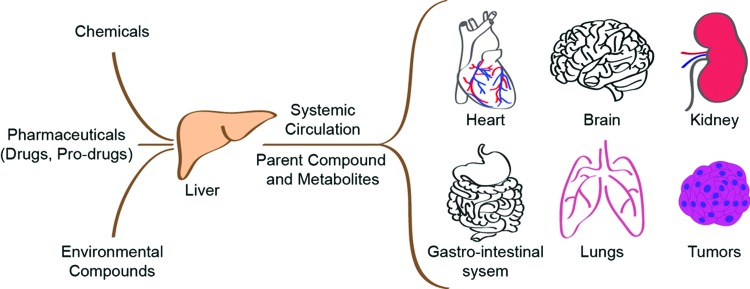
Liver is the principal organ responsible for metabolizing drugs and the primary site for drug-induced toxicity. It accounts for more than 50% of cases of acute liver failure and remains a major factor responsible for withdrawal or restriction of approved drugs.1,2 Apart from hepatic toxicity, liver-generated metabolites are transported through systemic circulation to other tissues resulting in interaction with other organs, including kidney, heart, brain, gut, and lungs, and targets such as tumors (Fig. 1). The drug–metabolite interaction of liver with other organ(s) can be exploited for the desired therapeutic action of the drug as exemplified in the case of chemotherapy prodrug tegafur, where liver biotransformation generates toxic metabolite 5-fluorouracil (5-FU), which is responsible for the antitumor efficacy of the drug.3,4 The interaction can also be responsible for unintended toxic side effects such as renal toxicity observed in the case of the chemotherapeutic prodrug, ifosfamide (IFO), where liver generates nephrotoxic metabolite chloroacetaldehyde (CAA), in addition to the desired antitumor metabolite 4-hydroxy-IFO.5 Thus, liver interaction with other organs needs to be carefully considered and evaluated in the context of drug metabolism and toxicity.
FIG. 1.
Liver is the central metabolizing organ in the human body. Pharmaceuticals, chemicals, and environmental compounds interact with liver and other organs in the body directly or in metabolized form. Color images available online at www.liebertpub.com/teb
Due to high rates of drug hepatotoxicity, liver-based stand-alone systems are widely used in the pharmaceutical industry to identify hepatotoxicity of potential drug candidates.6–8 To capture the full range of interorgan interactions and identify potential toxicity of drugs and metabolites, animal models are widely used during the drug development phase. However, animal models are limited due to potential difference in drug metabolism from humans.9 The situation becomes even more complex when different animal species give conflicting drug toxicity data, thereby further complicating extrapolation of animal data to humans. Development of in vitro systems composed of human cells, and conducive to capturing inter-organ interaction, can lead to better understanding and identification of toxic drug candidates before initiating costly human clinical trials. Furthermore, these systems also offer the opportunity of evaluating efficacy and toxicity of drugs under human-relevant disease conditions.
Importance of Human Relevance in Drug Metabolism and Toxicity Platforms
A critical lack in current drug screening and development pipeline is the ability to obtain human-relevant information at the early stages. In a comprehensive study compiled from 150 compounds tested by 12 pharmaceutical companies, Olson et al.9 compared drug toxicities in humans to various animal species, including dog, primate, rat, mouse, and guinea pig. Their analysis indicated an overall concordance rate between human and animal toxicity to be 71%, which further reduced to 63% and 43% for nonrodent and rodent alone studies, respectively. Considering analysis pertinent to the liver, the target organ toxicity data indicated that the concordance rate between human and animal was around 50% for liver toxicity. A notable example of poor hepatic concordance is the drug fialuridine, a nucleoside analogue, where hepatic failure was detected during clinical trials, whereas animal studies with monkeys, dogs, and rats failed to show any appreciable hepatic toxicity, despite long-term treatment.10
A clear case of species variability for the detection of drug metabolism and metabolite toxicity is efavirenz, a retroviral drug. Animal testing conducted in rats alone produced a toxic metabolite causing renal tubular epithelial cell necrosis. However, the toxic metabolite is not observed in primates (monkeys) or humans due to the absence of the metabolic component (Glutathione-S-transferase) that leads to its formation.11,12 While the toxic metabolite is observed in some animal models, the lack of the specific CYP enzyme in human models highlights the importance of developing comprehensive in vitro models that can demonstrate drug metabolism in a human-relevant way.
Design Considerations for Liver Models that Effectively Capture Metabolite Formation and Interaction with Other Organs
Advancement in design and development of liver models has enabled in vitro examination of the drug metabolism function of liver, both in physiological and pathophysiological states. Central to these models is choosing the appropriate cell type(s) and providing the right microenvironment to mimic both healthy and pathological conditions. Multiorgan systems that interface a liver model with other organ model(s) need addressing several design considerations, including realization of drug–metabolite interaction across different organs, compatibility of culture condition for different organ models, overall complexity, and throughput of the system. The latter is especially important if such a system has to be used at the screening stage of the drug development process. In this section, we briefly evaluate the current state-of-the-art of hepatic cells, challenges associated with long-term maintenance of optimal liver models and considerations for scaling in vitro liver designs to effectively interface with other organ models.
Progress in hepatic cells for use in liver platforms
Since hepatocytes are the principal cells responsible for drug metabolizing function of liver, they form an integral part of any liver cell-based model. Several hepatic cell choices are available for these models, including primary human hepatocytes (PHH), cell lines (HepaRG, HepG2), and recently hepatocyte-like cells (HLCs) derived from human pluripotent stem cell sources. While the choice of cells has several wanted features, there are disadvantages to each choice that must be considered. In comparison to other cell choices, PHH exhibit the most optimal drug metabolizing enzymatic profile closest to the human liver. This is seen in expression levels of key phase I enzymes, CYP450s, including CYP3A4, 1A1, and 2B6, as well as phase II enzymes, sinusoidal transporters, and nuclear receptors, which are key to drug metabolism.7,13–15 Cell lines often have a more stable genetic expression profile than PHH, but they often exhibit CYP450s at much lower levels.14,16 In this regard, HepaRG cells have higher expression of key CYP450s than HepG2 cells, whose expression is usually low or undetectable.13,15,16 Specifically, the most important and abundant isoform in the human liver, CYP3A4, is not detected at all in HepG2, and several key enzymes, including CYP1A2, CYP2A6, and CYP2B6, are expressed at extremely low levels.14,16 HepaRG expresses most CYP450s at a higher level than HepG2 cell lines, but still do not match the expression of PHH.15,16 Thus, while PHH are the best choice, HepaRG cells offer an attractive alternative for conducting xenobiotic metabolism and toxicology experiments, as well as chemical carcinogenesis studies.14,17,18
One of the primary challenges associated with PHH is the lack of renewable source of cells. In vitro, hepatocytes lose proliferation ability, which makes their availability dependent on frequent donor source. In the absence of a large batch of cryopreserved PHH from a single donor, donor-to-donor variation can introduce variability in the CYP450 expression profiles. Recent studies have attempted to address the non-proliferation ability of PHH with some success. Shan et al.19 conducted a screen of small molecules to identify inducers of PHH proliferation, whereas Levy et al.20 described an oncostatin M-dependent approach for expanding PHH. The latter approach is especially promising, where the authors reported up to 40 population doublings of PHH.
An alternate strategy for addressing cell source limitation of PHH is presented by the recent developments in the use of renewable cells that are derived from embryonic stem cells (ESCs) or the adult-induced pluripotent stem cells (iPSCs). Duan et al.21 generated highly enriched albumin-positive HLCs from hESCs that expressed phase I and II drug-metabolizing enzymes and phase III transporters, some of them comparable to PHH. However, the expression of asialoglycoprotein receptor indicated that significant proportion of the cells in the HLC population did not mature. In another study, Ogawa et al.22 combined 3D cell aggregation with cAMP signaling to enhance maturation of HLCs from hESCs as observed by the increased expression of adult phase I enzymes, such as CYP3A4, 1A2, and 2B6, along with phase II enzyme UGT1A1. The authors demonstrated wider applicability of their maturation approach whereby HLCs derived from iPSCs exhibited reduced expression of fetal markers and enhanced expression of some of the adult markers, including CYP3A4. In earlier reports,23,24 HLCs derived from iPSCs expressed a number of hepatic markers, including albumin, however, the profile was closer to a fetal phenotype as shown by the expression of α-fetoprotein (AFP) and highly reduced activity of adult phase I enzymes such as CYP 3A4. Shan et al.19 identified two molecules within their small-molecule screen that promoted maturation of HLCs derived from iPSCs as shown by the increased activity of adult phase I enzymes CYP 3A4 and 2A6 and reduction in secretion of AFP. As opposed to using pluripotent stem cells, Du et al.25 applied lineage reprogramming to generate HLCs directly from human embryonic fibroblasts without passing through an intermediate state. The basal metabolic activities of several enzymes, including CYP3A4, 1A2, 2B6, 2C9, and 2C19, were comparable between HLCs and freshly isolated PHH.
In contrast to the embryonic source, adult iPSC-derived hepatocytes offer a unique opportunity to revolutionize pharmacological and toxicological assessment because of their potential to test cells from normal and diseased state, as well as from genetically diverse adult humans. Within the human population, CYP2D6, 2C9, and 2C19 polymorphism accounts for the most frequent variation in phase I metabolism of drugs. Additionally, extensive polymorphism also occurs in other CYP genes, such as CYP1A1, 2A6, 2A13, 2C8, 3A4, and 3A5.26 Hepatocytes generated from iPS cells derived from such genetically diverse donors can potentially capture interindividual variation in drug metabolism. Although such iPS cell libraries are currently lacking, in a recent study, Takayama et al.27 utilized PHHs from multiple donors to reprogram them into iPSCs that were subsequently differentiated into HLCs. Their results indicated that CYP450 metabolism capacity and drug responsiveness of HLCs were highly correlated to PHHs. Specifically, the authors were able to reproduce in HLCs the interindividual differences in CYP2D6 metabolism capacity and drug responsiveness (as assessed by PHH response) due to polymorphism in CYP2D6. However, the CYP induction capacities of HLCs were weakly correlated with PHHs. Extension of this approach, where iPSCs are derived from more accessible tissue/blood cell source, rather than PHH, holds great promise for drug toxicity and efficacy, and disease-modeling studies in the context of personalized medicine. Despite the progress, HLCs derived from pluripotent stem cells (especially iPSCs) exhibit immature phenotype–more closely resembling fetal rather than adult hepatocytes.28 Future advancement depends on the significant development in protocols and their validation to achieve efficient differentiation and maturation that yields homogeneous cell population exhibiting full range of adult PHH phenotype, including CYP induction.
Challenges associated with long-term maintenance of primary hepatocyte-based liver model
Given the importance of primary hepatocytes in creating an optimal liver model, it will be useful to briefly discuss the sophisticated conditions required for long-term maintenance of these cells in culture. Primary hepatocytes, from human and animal sources, share similar challenges with regard to long-term maintenance in culture and, thus, our discussion also references studies that employed primary hepatocytes from animal source. Typically, monocultures of primary hepatocytes can be maintained for several weeks by providing specialized condition such as overlaying extracellular matrix (ECM) on top of the cells (sandwich configuration) as demonstrated for collagen gel and Matrigel®.29–31 The sandwich configuration is reminiscent of the in vivo ECM topology, where hepatocytes are bounded by the ECM of the space of Disse. Inclusion of other native liver tissue matrix components such as glycosaminoglycans in the ECM microenvironment of hepatocytes have been shown to improve their function,32 which further highlights the important role ECM plays in maintaining hepatocytes ex vivo. Other methodologies that permit long-term monoculture of hepatocytes include promoting 3D spheroid formation by culturing them on a nonadherent surface33 or embedding them in scaffolds based on a variety of materials, including alginate,34 peptide hydrogel (PuraMatrix),35 and poly(lactic-co-glycolic acid).36
An alternate approach that facilitates long-term maintenance is coculturing hepatocytes with other cell types, including fibroblast,37 endothelial cells,38,39 or other nonparenchymal cells of the liver.40–42 The latter option offers additional advantage whereby inclusion of nonparenchymal cells such as stellate, sinusoidal endothelial cells, and Kupffers can promote the creation of an in vivo-like microenvironment experienced by hepatocytes. This is particularly useful for examining metabolic function and drug toxicity under adverse conditions such as exposure to lipopolysaccharide, where nonparenchymal cells, especially Kupffers (resident macrophages of liver), may play a pivotal role in modulating CYP 450 function and drug toxicity.43 Other important parameters that can affect hepatocyte performance include shear stress, oxygen availability, and flow-induced change in the chemical environment. Primary hepatocytes are sensitive to shear, with a shear stress >5 dyne/cm2 shown to be detrimental to their function.44 They are highly metabolically active cells and may require separate provision for oxygenation in the flow systems, where media perfusion is the only source of oxygen, which can get depleted due to the constraints imposed on flow rate by the shear stress.44 Lastly, continuous flow, where media gets constantly replenished, can positively impact performance of hepatocytes as demonstrated in a recent study, where induced CYP1A1/2 activity under flow was higher than in static condition.45 These are important issues that need to be considered while designing primary hepatocyte-based liver models that can be interfaced with other organ models in static and flow environment.
Scaling and throughput consideration for systems that facilitates interfacing of liver models with other organ models
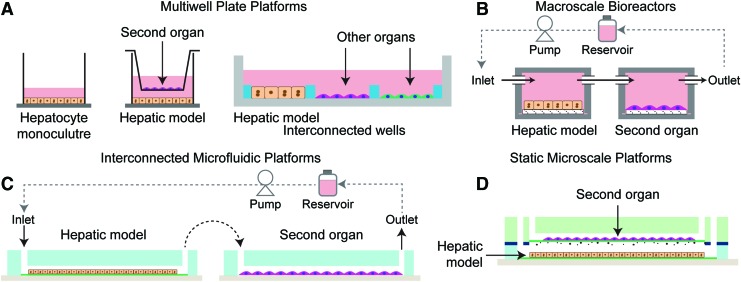
An important parameter to consider, in the design of in vitro systems that incorporate liver and a second organ model is the impact of media-to-cell ratio on drug metabolism, metabolite formation, and metabolite stability.46,47 Multiwell plates are convenient and widely used cell culture platforms that provide an environment resulting in a typical culture media of ∼1 nL/hepatocyte in the case of monolayer culture of hepatocytes. Cell culture platforms can be broadly divided and evaluated based on the amount of media per hepatocyte (Fig. 2). Considering that hepatocytes are the major metabolic components and secrete the metabolized product into the media, the media-to-cell ratio is a significant parameter leading to metabolite accumulation in the system at different concentrations. This could be particularly important in cases where the metabolite is short-lived, or the therapeutic/toxic molecule is a higher-order metabolite. Broadly, cell culture systems can be classified as a multiwell plate (monoculture, transwell, and interconnected culture), macroscale bioreactor, and microscale (microfluidic and static microfabricated) platforms (Fig. 3). In the multiwell plate format, the addition of transwell (culturing a different cell-type representing a different organ model) or partial seeding in an interconnected well to accommodate other organ models (Fig. 3A), leads to an increase in media per hepatocyte and, thus, dilute the metabolite generated by hepatocytes (liver model). In both microfluidic and macroscale bioreactor platforms, typically the second organ model is introduced in the daisy chain configuration (in series) with respect to the liver model and metabolite interaction is achieved by flowing the media from the liver model to the desired organ model (Fig. 3B, C). These systems typically require interconnects for integrating different organ modules and a reservoir for recirculating the media. This additional dead volume can further increase media volume/hepatocyte in addition to the volume added by the second organ model. By contrast, significantly lower media-to-cell ratios can be achieved in static microfabricated systems (Fig. 3D), resulting in higher product and metabolite formation and greater resolution of drug metabolism. It is important to note that despite achieving relatively lower media volume/hepatocyte ratio in the static microfabricated system, it remains an order of magnitude higher than in vivo, based on simple scaling consideration (blood volume/hepatocyte).46
FIG. 2.
Comparison of media-to-cell ratio and throughput for in vitro liver and multiorgan systems. Traditional monolayer culture of hepatocytes in multiwell plate provide 1 nL per hepatocyte, which is used as a basis for comparing various multiorgan systems. Color images available online at www.liebertpub.com/teb
FIG. 3.
Broad classification of cell-culture systems used for developing liver-based multiorgan models. (A) Multiwell plate (monoculture, transwell, and interconnected culture), (B) Macroscale bioreactor, (C) Interconnected microfluidic platforms and (D) Static microscale platforms. Color images available online at www.liebertpub.com/teb
Another important consideration for these systems is the relative throughput especially if they intend to be used at the drug screening stage. By far, multiwell plate static configuration offers the highest throughput followed by static microfabricated systems and lastly the modular macroscale bioreactor and microfluidic flow platforms. Despite the relative complexity, many studies rely on modular flow systems for achieving organ–organ interaction as described in the following section below. One advantage of modular flow systems is that they are potentially closer in mimicking the organ–organ interaction in vivo, where interaction is mediated by flow-induced transport of metabolite from one organ to the other. Additionally, they offer greater flexibility in integrating multiple (>2) organ models in desired configurations and are likely more conducive to incorporating highly sophisticated 3D tissue models, which are currently in development. Nevertheless, relatively low throughput remains an important issue with the modular flow systems. Next section discusses examples of various multiorgan model systems that employ liver as one of the organ models and broadly falls into one of the categories discussed above.
Current In Vitro Systems for Examining Liver Interaction with Other Organs
A lot of progress has been made toward creating model systems that interface a liver model with other organ models. Table 1 summarizes the various model systems that have been designed to interrogate liver interaction with other organ(s). The list includes organ models based on animal cells in addition to those based on human cells as they share some of the same technological challenges. We discuss below some of the key insights provided by these models.
Table 1.
Systems Conducive to Interfacing Liver Models to Other Organ Models
| Organ systems | Drug (metabolite) | Observations |
|---|---|---|
| Multiwell transwell coculture | ||
| Liver–gastrointestinal (PHH)—(Caco-2)48 | Bretrylium, Chlorothiazide, Diazepam, Theophylline, Acyclovir, Terbutaline, Lorazepam, Oxazepam, Ganciclovir, Caffeine, Pirenzepine, Metoclopramide, Cyclosporine, Sildenafil, Timolol, Midazolam, Nifedipine, Imipramine, Chlorpromazine, Fluoxetine, Desipramine, Propranolol, Nicardipine, and Verapamil | Reasonable correlation (r2 = 0.86) between the model prediction and the oral bioavailability |
| Liver–gastrointestinal (PHH)-(Caco-2)49 | PEG-4000, Doxorubicin, Mannitol, Acyclovir, Nadolol, Terbutaline, Ranitidine, Furosemide, Atenolol, Cimetidine, Etoposide, Metolazone, Sulfasalazine, Hydrochlorothiazide, Methylprednisolone, Propranolol, Hydrocortisone, Alprenolol, Metoprolol, Antipyrine, Caffeine, Carbamazepine, Ketoprofen, Naproxen, Theophylline, Verapamil, Corticosterone, and Dexamethasone | Reasonable correlation (r2 = 0.73) between the model prediction and the oral bioavailability |
| Interconnected multiwell plate | ||
| Liver–Kidney–Lung–CNS–Blood vessels–Cancers (primary hepatocytes)—(proximal tubule epithelial cells)—(small airway epithelial cells)—(astrocytes)—(aortic endothelial cells)—(MCF-7)50 | Tamoxifen | ↓ Viability of MCF-7 cells |
| Relative cytotoxicity to other organs | ||
| Liver–Kidney–Lung (PHH)–(renal proximal tubule cells)—(small airway epithelial cells)52 | Aflatoxin B1 ((AFB1)-8,9-epoxide) | ↓ Viability of hepatocytes |
| Relative cytotoxicity to other organs | ||
| Liver–fibroblasts (PHH)—(3T3)53 | Aflatoxin B1 ((AFB1)-8,9-epoxide) cyclophosphamide (4-hydroxycyclophosphamide and phosphoramide mustard) tamoxifen | ↓ Viability of 3T3 cells |
| ↓ Viability of hepatocytes | ||
| Macroscale bioreactor platform | ||
| Liver–Vascular (HepG2)—(HUVEC)55 | ↑Albumin production | |
| ↑Urea production | ||
| ↑Viability | ||
| ↑Nitric oxide production | ||
| Liver–fat–vascular (HepG2)—(omental adipose tissue)—(HUVEC)56 | Network of interactions between various systems in fasting, postabsorptive states and postprandial states in type 1 and type 2 diabetes. | |
| Liver–vascular (HepG2/C3a)—(HUVEC)57 | Numeric relationship between cells in cell number scaling mode and metabolic and surface scaling models. | |
| Liver–gastrointestinal (Liver slices from male Wistar rats)—(Jejunum/Ileum slices from male Wistar rats)58 | 7-ethoxyxoumarin | Short-term culture in connected system |
| 7-hydroxycoumarin | ↓CYP7A1 Expression | |
| Lidocaine (MEG-X) | ||
| Liver–gastrointestinal (HepG2)—(Caco-2)59 | benzo[a]pyrene | ↑Permeability of Caco-2 in flow model |
| Microfluidic interconnected system | ||
| Liver–lung–fat (HepG2/C3A)—(L2)—(3T3-L1)60 | Naphthalene (Naphthoquinone) | Reduced naphthalene and naphthoquinone-induced glutathione (GSH) depletion |
| Liver–bone marrow–tumor-resistant tumor) (HepG2/C3A)—(MEG-01)—(MES-SA)—(MES-SA/DX-5)61 | Doxorubicin (Doxorubicinol), Nicardipine, Cyclosporine A | Synergistic effect of drugs on cell proliferation |
| Liver–GI–Fat–Kidney–Bone Marrow (HepG2/C3A)—(Caco-2/HT29-MTX)–Empty compartments62 | Acetaminophen | Dose-dependent hepatotoxicity |
| Liver–lung–kidney–fat (HepG2/C3A)—(A549)—(HK-2)—(Primary human preadipocyte, HPA)63 | ↑A549 PROD enzyme activity | |
| ↑HPA adiponectin secretion | ||
| ↓C3A albumin secretion | ||
| ↓HK-2 GGT enzyme activity | ||
| Liver–colon tumor–bone marrow (HepG2/C3A)—(HCT-116)—(Kasumi-1)64 | 5-Fluorouracil | ↓ Viability of the cells |
| Liver–GI–intestine–cancer (HepG2)—(Caco-2)—(MCF-7)65 | Cyclophosphamide (phosphoramide mustard), Tegafur (5-Fluorouracil) | ↓MCF-7 viability |
| Liver–tumor (HepG2/C3A)—(GBM-M059K)66 | Temozolomide (MTIC), Ifosfamide (IPM) | ↓GBM Viability |
| Liver–skin (HepaRG, primary human hepatic stellate cells)—(Skin Biopsy)67 | Troglitazone | Long-term tissue culture |
| Liver–kidney (HepG2/C3A and HepaRG)—(MDCK)15 | Ifosfamide (Chloroacetaldehyde) | ↑CYP3A4, CYP3A5, CYP2B6 |
| ↓MDCK Proliferation | ||
| Liver–kidney (HepaRG)—(MDCK)68 | Ifosfamide (Chloroacetaldehyde) | Pharmacokinetic model coupling ROS and glutathione (GSH) |
| Static microscale platform | ||
| Liver–cancer46(hepatocytes)-(MCF-7) | Tegafur–uracil (5-Fluorouracil) | ↓MCF-7 viability |
↓, decrease; ↑, increase; ↔, no change; GMB, glioblastoma multiforme; PHH, primary human hepatocytes; ROS, reactive oxygen species.
Multiwell plate systems (transwell coculture)
Cheng and coworkers48 utilized a transwell coculture model composed of Caco-2 cells (GI tract, transwell) with PHH (liver, well) to evaluate intestinal absorption and first-pass metabolism stages of oral bioavailability. The model was tested on 24 commonly prescribed drugs, comparing their results to reported oral bioavailabilities, showing a reasonable correlation as a predictive model. This model effectively captured the Caco-2 permeability along with hepatocyte clearance of the drugs and could provide predictive information estimating the range of oral availability in humans. In further works, this model was improved with the addition of an elimination phase to the intestinal absorption and metabolism phases.49
Interconnected multiwell plate system
Albert Li and coworkers have developed an interconnected multiwell system, which allows tissue slices and cells from different organs to be cultured within their own well, but share media with other wells, as a tool for evaluating human-specific xenobiotic metabolism. The Integrated Discrete Multiple Organ Coculture (IdMOC) is created on a 96-well plate with very shallow wells so that media covers the top of the cultured cells in each well, while enabling interaction between different organ systems through secreted molecules, without requirement of any media flow.50,51 This allows for evaluation of each cell type for specific toxicity after exposure to any toxic compounds. Their experiments tested the toxicity of tamoxifen not only to breast cancer cells (MCF-7), but also to five other organs (liver, kidney, lung, central nervous system, and blood vessels). This system produced results that can be used for the quantitative evaluation of a drug's anticancer effect versus its toxicity toward healthy organs.50 In later work, they have demonstrated aflatoxin B1's hepatocyte-specific toxicity and compared the findings using metabolism comparative cytotoxicity assay (MCCA) and cytotoxic metabolic pathway identification assay (CMPIA) methods.52 This proof-of-concept study suggests these methods (MCCA and CMPIA) with cryopreserved human hepatocytes are potentially useful for the evaluation of the relationship between human xenobiotic metabolism and toxicity. Recently, they demonstrated the diverse toxicity testing capability of IdMOCs by testing PHH and mouse 3T3 fibroblasts (metabolically incompetent cell line) with Tamoxifen (direct toxicant), and two prodrugs—aflatoxin B1, and cyclophosphamide. The system was able to differentiate the metabolism of the drugs and the distinct patterns of cytotoxic profile based on the metabolizing capability of the cell type.53 A key advantage of the IdMOC platform is the similarity with standard multiwell plate cultures, allowing for the use of various cell types or organ slices to demonstrate multiple organ interactions. The IdMOC system can also be used to create specific organ toxicity models by culturing different cells from the same organ in nearby wells and allow for paracrine signaling.54
Macroscale bioreactor platforms
Guzzardi et al.55 developed a multicompartment bioreactor with liver (HepG2) and endothelial (HUVEC) cells, enabling crosstalk by soluble molecules. The culture setup was based on ratios of cells found in the human body, and evaluated the coculture effects on each cell based on glucose consumption, and albumin and urea secretion. In a continuation, Iori et al.56 utilized the modular bioreactor system to demonstrate crosstalk between liver, endothelial, and fat cells. This system consisted of two low-shear flow compartments (liver and fat), one high-shear flow compartment (endothelial), a mixing chamber and a pump, all connected in series. Utilizing this system, the authors simulated multiple physiological and pathological conditions–namely fasting, postabsorptive states, and postprandial states in type 1 and type 2 diabetes demonstrating the fractional variation of metabolite concentrations. The system is a clear demonstration of an in vitro model of endogenous metabolism analyzing the network of interactions between different cell types and organ models. Ucciferri et al.57 took a different approach, demonstrating the importance of allometric scaling in creating a physiologically accurate model of human organ crosstalk that is metabolically relevant. In this work, two models were tested: cell number scaling model (CNSM) and metabolic and surface scaling model (MSSM). While CNSM considers the mass ratios independent of the cell function, the MSSM considered the whole-body metabolism and nutrient distribution. The experimental setup consisted of human hepatoma-derived cell line HepG2/C3A (liver) and HUVEC (endothelial) cells connected in series, with varying endothelium-to-liver ratios. The results demonstrate the importance of design and scaling of organs in a multiorgan system to accurately represent physiological conditions, with an emphasis on maintenance of homeostatic balance within the culture.
In a different approach, Van Midwoud et al. created a gut–liver coculture system consisting of precision-cut intestinal slices (jejunum and ileum) and rat liver slices (from male Wistar rats) connected in series representing intestinal absorption and liver metabolism, respectively.58 Within each chamber, the precision-cut slices were suspended between two membranes allowing perfusion of media around the tissues and transport to the next chamber. Experiments with chenodeoxycholic acid (a primary bile acid) demonstrated the upregulation of fibroblast growth factor 15, resulting in a downregulation of CYP 7A1, demonstrating the retention of function within the platform and interorgan communication. While tissue slices offer the advantage of preserving the composition of the organ along with vasculature, a major drawback with tissue culture slices is the short life-span of the isolated slices (8 h for intestine and 24 h for liver slices) and deteriorating responses within 24 h.
Ouattara et al.59 utilized both static and perfused models to evaluate effects of drugs on the GI tract and their interactions with a liver model. The perfusion system consisted of transwell culture-type systems that were connected in a physiological manner. They studied the adsorption and metabolism of benzo[a]pyrene on Caco-2 cells (GI tract), with supplemental CYP 450s contributed by HepG2 cells (liver). The perfused system consisted of two connected but distinct compartments plated with Caco-2 cells and HepG2 cells, respectively. Their results indicate effective capture of intestinal permeability in dynamic flow, and higher metabolism in static coculture.
Microfluidic interconnected systems
One of the microfluidic interconnected system designs has been by Shuler and coworkers, which includes a four-chamber interconnected microscale cell culture analog (μCCA) utilizing microfluidic channels to connect media between each chamber. In the initial work, the μCCA consisted of four different organ mimics, namely liver, lung, and fat cells with the media connecting the organs as an effective circulatory system mimic.60 The authors evaluated HepG2/C3A cells' ability to process naphthalene into its metabolites, which are known to cause undesirable effects on lung cells (L2) and can accumulate in fat cells (3T3-L1). The addition of the fat compartment is interesting due to the resulting protection of L2 cells from the toxic metabolite—by sequestering the metabolite from the media. The use of interconnected systems, with liver (metabolism), lung (target organ), and adipose (bioaccumulation) aspects provides better correlations with conditions in the human body. As another example of the applicability of the system, Tatosian and Shuler61 demonstrated the use of μCCA to evaluate the difference of doxorubicin's effect on regular uterine cancer and multidrug-resistant uterine cancer. In this format, the other two chambers in μCCA represented liver (hepatocytes) and immune system (bone marrow). The premise of the study was to evaluate the effectiveness of a combined treatment of doxorubicin and modulators cyclosporine and nicardipine, to doxorubicin alone. The results indicated that the combined treatment was beneficial in that it showed a significant decrease in cancer cell viability compared to both the control and doxorubicin alone. In continuation, Mahler et al.62 demonstrated the use of μCCA to investigate the ADME (absorption, distribution, metabolism, and excretion) properties of drugs. The device included an upstream chamber (GI tract comprising of Caco-2 cells), which delivered the absorbed metabolites to the μCCA with liver (HepG2/C3A cells), bone marrow, kidney, and fat compartments. The authors evaluated the uptake of acetaminophen and its conversion to NAPQI (toxic metabolite), in the output media reservoir. Their results indicated decreased NAPQI levels in the presence of an upstream GI compartment leading to increased liver viability, making this a useful diagnostic tool for orally administered drugs. Zhang et al.63 developed a 3D microfluidic cell culture system, which incorporated an upstream lung (A549) compartment interfacing with liver (HepG2/C3A), kidney (HK-2), and fat (HPA) downstream in parallel. The device evaluated the crosstalk in between cell culture compartments, and how TGF-β1 affected A549 cells and the other downstream compartments. A recent advance in the μCCA incorporates a 3D layered model, which uses gravity to induce flow, eliminating the need of pumping mechanisms.64 The new format incorporates liver (HepG2/C3A), tumor (HCT-116) and bone marrow (Kasumi-1) cells in parallel chambers connected by common reservoirs. The authors observed increased sensitivity to low doses of 5-fluorouracil (with and without uracil) in dynamic flow in comparison with a static system.
Imura et al.65 have developed a microfluidic system with four compartments comprising of GI tract, intestine, liver, and a target cell (cancer) compartment as a physiologically based pharmacokinetic (PBPK) model for prodrugs. They demonstrated the digestion, intestinal absorption, hepatic metabolism, and bioactivity of known anticancer prodrugs, tegafur and cyclophosphamide. The design included a GI tract complete with a stomach, duodenum, colon (Caco-2), liver (HepG2), and target cancer cells (MCF-7). The results showed a clear difference between cyclophosphamide and tegafur: Cyclophosphamide retained efficacy during digestion while tegafur is degraded before it can be metabolized in the liver, losing its effect on cancer cells. Ma et al.66 developed a 3D microscale perfusion-based two-chamber device to evaluate the role of drug metabolism on the efficacy of anticancer drugs. The device consists of two compartments (connected in series) within which polylactic acid scaffolds containing HepG2/C3A cells (liver) and glioblastoma multiforme (brain tumor cells) were introduced representing liver and cancer cells, respectively. Both drugs (temozolomide and IFO) required a metabolic breakdown to form active therapeutic agents. Their observations reveal that cells in a 3D structure were much more resistant to drugs than those in a 2D monolayer (which is the current standard for testing). They demonstrated that temozolomide is much less toxic to glioblastoma multiforme cells in the presence of liver cells, while IFO is more toxic, which helps to specify drug doses and schedules.
Wagner et al.67 used a Multi-Organ-Chip (MOC) with two compartments to model the 3D long-term stability of liver cells (HepaRG and hepatic stellate cells) and fresh skin biopsies. The model showed steady albumin levels due to adequate crosstalk between the liver cells and skin biopsies, and the ability to maintain a stable culture for 28 days. The study tested repeated doses of troglitazone, a drug with high liver toxicity. The MOC could provide a simplistic model for analyzing the effects of new cosmetics on the liver and the skin.
Leclerc and coworkers developed a 3D microfluidic biochip system with two compartments cultured with liver cells (HepG2/C3A and HepaRG) and kidney cells (MDCK), respectively.15 They used this model to study the toxic effect of the anticancer drug IFO, whose metabolite CAA causes kidney cell toxicity. When in coculture, MDCK cell viability was reduced due to the metabolism of IFO by the hepatocytes. They also demonstrated that HepaRG cells are more suitable than HepG2/C3A cells for metabolizing drugs. In a continuation work,68 they have integrated the in vitro findings with a pharmacokinetic model to describe the production of reactive oxygen species induced by CAA in renal cells.
Static microfabricated platforms
Bale et al.46 demonstrated the importance of media-to-cell ratio by utilizing the microscale features of a static coculture system in evaluating drug metabolism and metabolite toxicity/efficacy on the target organ. Using a simple microfabricated two-chamber platform, separated with a tissue culture membrane seeded with hepatocytes and MCF-7 cells, they demonstrated the ability of primary hepatocytes to metabolize the prodrug tegafur, and the subsequent metabolite's (5-fluorouracil) effect on MCF-7 cells. The toxic metabolite, 5-FU, produced in the hepatocyte compartment (lower chamber) diffused through the membrane to induce toxicity in breast cancer cells (in the upper chamber). A clear advantage of the system is the utilization of static culture conditions leading to low media-to-cell ratio, which significantly increases the product formation and its accumulation.
Summary
Drug development requires comprehensive evaluation of drug–metabolite interaction between liver and other organs to identify any potential toxicity in humans before clinical trials. Systems that provide human-relevant information at the preclinical stage of drug development can complement the animal models that have limited concordance with humans. Integration of microfabrication and microfluidics technology with cell culture has not only spurred development of highly sophisticated liver models that increasingly mimic liver function in vitro, but also facilitate interfacing with other organ models. In fact, there is tremendous interest in developing a “human-on-a-chip” system, where the goal is to integrate multiple organ models, including liver, containing human cells. However, despite the progress in development of individual organ models, and a few proof-of-concept demonstrations of organ–organ interaction, there are significant challenges that need to be overcome, including realization of culture conditions that promote maintenance of multiple organ models in a coupled configuration and concomitantly achieve physiologically relevant coupling. The successful development of multiorgan human cell-based systems can have a broad impact in evaluating drug efficacy and toxicity, as well as mechanistic investigation of human-relevant disease conditions.
Future Outlook
There is impetus within both academic and commercial sectors for developing advanced human-on-a-chip systems. Initiatives in the USA by Defense Advanced Research Projects Agency (DARPA, www.darpa.mil/program/microphysiological-systems) and the National Institute of Health (NIH, www.ncats.nih.gov/tissuechip)69,70 towards a human-on-a-chip platform aim to “develop and use new tools, standards, and approaches to efficiently develop therapies and to more effectively evaluate product safety, efficacy, and quality.” Other programs in the USA include the Department of Defense (through the Defense Threat Reduction Agency) sponsored effort to develop a body-on-a-chip system using bioprinting of organs approach. Initiatives in Europe, SEURAT (www.seurat-1.eu/) aim toward the replacement of in vivo repeated dose systemic toxicity testing. In addition, several commercial companies are developing platforms to address the lack of human-relevant efficacy models and thorough testing during drug screening process, including Hμrel, Kirkstall, Organovo, Insphero, Emulatebio, and Mimetas, to name a few.
Besides continuing advancement in models of liver and other organs, one critical factor that needs to be addressed for developing human-on-a-chip system is the physiologically relevant coupling of different organ models. This includes taking into account relative sizes of different organs, and the accompanying transport (flow rate/residence time)-mediated interaction between different organs, as well as development of common media formulations that support maintenance of multiple organ models while also promoting achievement of in vivo-like function for different models. Even though various approaches have been proposed to address some aspects of the first two issues, including allometric57 and functional scaling,71 PBPK/pharmacodynamic modeling,64,72 and “metabolically supported functional scaling,”73 several technical and fundamental challenges remain. While optimal organ function in vitro may be achieved by utilization of primary parenchymal cells, their use typically requires specialized media (optimized in single organ setting) formulation for achieving ideal function and can also impose severe constraints on the transport parameters (flow rate/residence time) due to shear stress and oxygen requirement. Within the coupled system, this complicates common media development as well as matching of transport parameters vis-à-vis relative size of organ model without compromising the function. Approaches that comprehensively tackle these challenges will likely provide a framework for accelerating the development of advanced human-on-a-chip systems. While a human-on-a-chip system is the ideal goal, drug screening processes could benefit from a minimal organ interaction system comprising of a smaller combination of critical organ systems involved in absorption, metabolism, and excretion (gut, liver, and kidney).
Acknowledgments
This work was supported by grants from the National Institutes of Health NIH-5UH2TR000503, and HMS Shore Fellowship for R.J.
Disclosure Statement
No competing financial interests exist.
References
- 1.Lee W.M. Drug-induced hepatotoxicity. N Engl J Med 349, 474, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Schuster D., Laggner C., and Langer T. Why drugs fail—a study on side effects in new chemical entities. Curr Pharm Des 11, 3545, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Ikeda K., Yoshisue K., Matsushima E., Nagayama S., Kobayashi K., Tyson C.A., Chiba K., and Kawaguchi Y. Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res 6, 4409, 2000 [PubMed] [Google Scholar]
- 4.Longley D.B., Harkin D.P., and Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3, 330, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Bruggemann S.K., Kisro J., and Wagner T. Ifosfamide cytotoxicity on human tumor and renal cells: role of chloroacetaldehyde in comparison to 4-hydroxyifosfamide. Cancer Res 57, 2676, 1997 [PubMed] [Google Scholar]
- 6.Soldatow V.Y., LeCluyse E.L., Griffith L.G., and Rusyn I. In vitro models for liver toxicity testing. Toxicol Res 2, 23, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bale S.S., Vernetti L., Senutovitch N., Jindal R., Hegde M., Gough A., McCarty W.J., Bakan A., Bhushan A., Shun T.Y., Golberg I., Debiasio R., Usta B.O., Taylor D.L., and Yarmush M.L. In vitro platforms for evaluating liver toxicity. Exp Biol Med (Maywood) 239, 1180, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godoy P., Hewitt N., Albrecht U., Andersen M., Ansari N., Bhattacharya S.,et al. , Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87, 1315, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van Deun K., Smith P., Berger B., and Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol 32, 56, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Greaves P., Williams A., and Eve M. First dose of potential new medicines to humans: how animals help. Nat Rev Drug Discov 3, 226, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Mutlib A.E., Gerson R.J., Meunier P.C., Haley P.J., Chen H., Gan L.S., Davies M.H., Gemzik B., Christ D.D., Krahn D.F., Markwalder J.A., Seitz S.P., Robertson R.T., and Miwa G.T. The species-dependent metabolism of efavirenz produces a nephrotoxic glutathione conjugate in rats. Toxicol Appl Pharmacol 169, 102, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Ogburn E.T., Jones D.R., Masters A.R., Xu C., Guo Y., and Desta Z. Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metab Dispos 38, 1218, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkening S., Stahl F., and Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos 31, 1035, 2003 [DOI] [PubMed] [Google Scholar]
- 14.Guo L., Dial S., Shi L., Branham W., Liu J., Fang J.L., Green B., Deng H., Kaput J., and Ning B. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 39, 528, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choucha-Snouber L., Aninat C., Grsicom L., Madalinski G., Brochot C., Poleni P.E., Razan F., Guillouzo C.G., Legallais C., Corlu A., and Leclerc E. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol Bioeng 110, 597, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Antherieu S., Chesne C., Li R., Camus S., Lahoz A., Picazo L., Turpeinen M., Tolonen A., Uusitalo J., Guguen-Guillouzo C., and Guillouzo A. Stable expression, activity, and inducibility of cytochromes P450 in differentiated HepaRG cells. Drug Metab Dispos 38, 516, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Aninat C., Piton A., Glaise D., Le Charpentier T., Langouet S., Morel F., Guguen-Guillouzo C., and Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34, 75, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hart S.N., Li Y., Nakamoto K., Subileau E.A., Steen D., and Zhong X.B. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos 38, 988, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan J., Schwartz R.E., Ross N.T., Logan D.J., Thomas D., Duncan S.A., North T.E., Goessling W., Carpenter A.E., and Bhatia S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol 9, 514, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy G., Bomze D., Heinz S., Ramachandran S.D., Noerenberg A., Cohen M., Shibolet O., Sklan E., Braspenning J., and Nahmias Y. Long-term culture and expansion of primary human hepatocytes. Nat Biotechnol 33, 1264, 2015 [DOI] [PubMed] [Google Scholar]
- 21.Duan Y., Ma X., Zou W., Wang C., Bahbahan I.S., Ahuja T.P., Tolstikov V., and Zern M.A. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells 28, 674, 2010 [DOI] [PubMed] [Google Scholar]
- 22.Ogawa S., Surapisitchat J., Virtanen C., Ogawa M., Niapour M., Sugamori K.S., Wang S., Tamblyn L., Guillemette C., Hoffmann E., Zhao B., Strom S., Laposa R.R., Tyndale R.F., Grant D.M., and Keller G. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 140, 3285, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., and Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan G.J., Hay D.C., Park I.H., Fletcher J., Hannoun Z., Payne C.M., Dalgetty D., Black J.R., Ross J.A., Samuel K., Wang G., Daley G.Q., Lee J.H., Church G.M., Forbes S.J., Iredale J.P., and Wilmut I. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 51, 329, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T., Zhao D., Sun Y., Shu J., Guo Q., Yin M., Sun D., Lu S., Shi Y., and Deng H. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14, 394, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Zhou S.F., Liu J.P., and Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev 41, 89, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Takayama K., Morisaki Y., Kuno S., Nagamoto Y., Harada K., Furukawa N., Ohtaka M., Nishimura K., Imagawa K., Sakurai F., Tachibana M., Sumazaki R., Noguchi E., Nakanishi M., Hirata K., Kawabata K., and Mizuguchi H. Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proc Natl Acad Sci U S A 111, 16772, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz R.E., Fleming H.E., Khetani S.R., and Bhatia S.N. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol Adv 32, 504, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn J.C., Yarmush M.L., Koebe H.G., and Tompkins R.G. Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. FASEB J 3, 174, 1989 [DOI] [PubMed] [Google Scholar]
- 30.Moghe P.V., Berthiaume F., Ezzell R.M., Toner M., Tompkins R.G., and Yarmush M.L. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials 17, 373, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Ryan C.M., Carter E.A., Jenkins R.L., Sterling L.M., Yarmush M.L., Malt R.A., and Tompkins R.G. Isolation and long-term culture of human hepatocytes. Surgery 113, 48, 1993 [PubMed] [Google Scholar]
- 32.Lin K.H., Hino H., Maeda S., Inagaki H., Airat J.V., and Saito T. Albumin synthesis by rat hepatocytes cultured on collagen gels is sustained specifically by heparin. Exp Cell Res 219, 717, 1995 [DOI] [PubMed] [Google Scholar]
- 33.Tong J.Z., Sarrazin S., Cassio D., Gauthier F., and Alvarez F. Application of spheroid culture to human hepatocytes and maintenance of their differentiation. Biol Cell 81, 77, 1994 [DOI] [PubMed] [Google Scholar]
- 34.Glicklis R., Shapiro L., Agbaria R., Merchuk J.C., and Cohen S. Hepatocyte behavior within three-dimensional porous alginate scaffolds. Biotechnol Bioeng 67, 344, 2000 [DOI] [PubMed] [Google Scholar]
- 35.Wang S., Nagrath D., Chen P.C., Berthiaume F., and Yarmush M.L. Three-dimensional primary hepatocyte culture in synthetic self-assembling peptide hydrogel. Tissue Eng Part A 14, 227, 2008 [DOI] [PubMed] [Google Scholar]
- 36.Hasirci V., Berthiaume F., Bondre S.P., Gresser J.D., Trantolo D.J., Toner M., and Wise D.L. Expression of liver-specific functions by rat hepatocytes seeded in treated poly(lactic-co-glycolic) acid biodegradable foams. Tissue Eng 7, 385, 2001 [DOI] [PubMed] [Google Scholar]
- 37.Khetani S.R., and Bhatia S.N. Microscale culture of human liver cells for drug development. Nat Biotechnol 26, 120, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Jindal R., Nahmias Y., Tilles A.W., Berthiaume F., and Yarmush M.L. Amino acid-mediated heterotypic interaction governs performance of a hepatic tissue model. FASEB J 23, 2288, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jindal R., Patel S.J., and Yarmush M.L. Tissue-engineered model for real-time monitoring of liver inflammation. Tissue Eng Part C Methods 17, 113, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bale S.S., Golberg I., Jindal R., McCarty W.J., Luitje M., Hegde M., Bhushan A., Usta O.B., and Yarmush M.L. Long-term coculture strategies for primary hepatocytes and liver sinusoidal endothelial cells. Tissue Eng Part C Methods 21, 413, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostadinova R., Boess F., Applegate D., Suter L., Weiser T., Singer T., Naughton B., and Roth A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol 268, 1, 2013 [DOI] [PubMed] [Google Scholar]
- 42.Esch M.B., Prot J.-M., Wang Y.I., Miller P., Llamas-Vidales J.R., Naughton B.A., Applegate D.R., and Shuler M.L. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip 15, 2269, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milosevic N., Schawalder H., and Maier P. Kupffer cell-mediated differential down-regulation of cytochrome P450 metabolism in rat hepatocytes. Eur J Pharmacol 368, 75, 1999 [DOI] [PubMed] [Google Scholar]
- 44.Tilles A.W., Baskaran H., Roy P., Yarmush M.L., and Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol Bioeng 73, 379, 2001 [DOI] [PubMed] [Google Scholar]
- 45.Hegde M., Jindal R., Bhushan A., Bale S.S., McCarty W.J., Golberg I., Usta O.B., and Yarmush M.L. Dynamic interplay of flow and collagen stabilizes primary hepatocytes culture in a microfluidic platform. Lab Chip 14, 2033, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bale S.S., Sridharan G.V., Golberg I., Prodanov L., McCarty W.J., Usta O.B., Jindal R., and Yarmush M.L. A novel low-volume two-chamber microfabricated platform for evaluating drug metabolism and toxicity. Technology 03, 155, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehling M., and Tay S. Microfluidic cell culture. Curr Opin Biotechnol 25, 95, 2014 [DOI] [PubMed] [Google Scholar]
- 48.Lau Y.Y., Chen Y.H., Liu T.T., Li C., Cui X., White R.E., and Cheng K.C. Evaluation of a novel in vitro Caco-2 hepatocyte hybrid system for predicting in vivo oral bioavailability. Drug Metab Dispos 32, 937, 2004 [PubMed] [Google Scholar]
- 49.Li C., Liu T., Cui X., Uss A.S., and Cheng K.C. Development of in vitro pharmacokinetic screens using Caco-2, human hepatocyte, and Caco-2/human hepatocyte hybrid systems for the prediction of oral bioavailability in humans. J Biomol Screen 12, 1084, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Li A.P., Bode C., and Sakai Y. A novel in vitro system, the integrated discrete multiple organ cell culture (IdMOC) system, for the evaluation of human drug toxicity: comparative cytotoxicity of tamoxifen towards normal human cells from five major organs and MCF-7 adenocarcinoma breast cancer cells. Chem Biol Interact 150, 129, 2004 [DOI] [PubMed] [Google Scholar]
- 51.Li A.P. The use of the Integrated Discrete Multiple Organ Co-culture (IdMOC) system for the evaluation of multiple organ toxicity. Altern Lab Anim 37, 377, 2009 [DOI] [PubMed] [Google Scholar]
- 52.Li A.P. Metabolism Comparative Cytotoxicity Assay (MCCA) and Cytotoxic Metabolic Pathway Identification Assay (CMPIA) with cryopreserved human hepatocytes for the evaluation of metabolism-based cytotoxicity in vitro: proof-of-concept study with aflatoxin B1. Chem Biol Interact 179, 4, 2009 [DOI] [PubMed] [Google Scholar]
- 53.Li A.P., Uzgare A., and LaForge Y.S. Definition of metabolism-dependent xenobiotic toxicity with co-cultures of human hepatocytes and mouse 3T3 fibroblasts in the novel integrated discrete multiple organ co-culture (IdMOC) experimental system: results with model toxicants aflatoxin B1, cyclophosphamide and tamoxifen. Chem Biol Interact 199, 1, 2012 [DOI] [PubMed] [Google Scholar]
- 54.Li A.P. Evaluation of adverse drug properties with cryopreserved human hepatocytes and the Integrated Discrete Multiple Organ Co-culture (IdMOC(TM)) System. Toxicol Res 31, 137, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzzardi M.A., Vozzi F., and Ahluwalia A.D. Study of the crosstalk between hepatocytes and endothelial cells using a novel multicompartmental bioreactor: a comparison between connected cultures and cocultures. Tissue Eng Part A 15, 3635, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Iori E., Vinci B., Murphy E., Marescotti M.C., Avogaro A., and Ahluwalia A. Glucose and fatty acid metabolism in a 3 tissue in-vitro model challenged with normo- and hyperglycaemia. PLoS One 7, e34704, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ucciferri N., Sbrana T., and Ahluwalia A. Allometric scaling and cell ratios in multi-organ in vitro models of human metabolism. Front Bioeng Biotechnol 2, 74, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Midwoud P.M., Merema M.T., Verpoorte E., and Groothuis G.M. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 10, 2778, 2010 [DOI] [PubMed] [Google Scholar]
- 59.Ouattara D.A., Choi S.-H., Sakai Y., Péry A.R.R., and Brochot C. Kinetic modelling of in vitro cell-based assays to characterize non-specific bindings and ADME processes in a static and a perfused fluidic system. Toxicol Lett 205, 310, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Viravaidya K., and Shuler M.L. Incorporation of 3T3-L1 cells to mimic bioaccumulation in a microscale cell culture analog device for toxicity studies. Biotechnol Prog 20, 590, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Tatosian D.A., and Shuler M.L. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng 103, 187, 2009 [DOI] [PubMed] [Google Scholar]
- 62.Mahler G.J., Esch M.B., Glahn R.P., and Shuler M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng 104, 193, 2009 [DOI] [PubMed] [Google Scholar]
- 63.Zhang C., Zhao Z., Rahim N.A.A., Noort D.v., and Yu H. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip 9, 3185, 2009 [DOI] [PubMed] [Google Scholar]
- 64.Sung J.H., Kam C., and Shuler M.L. A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 10, 446, 2010 [DOI] [PubMed] [Google Scholar]
- 65.Imura Y., Yoshimura E., and Sato K. Micro total bioassay system for oral drugs: evaluation of gastrointestinal degradation, intestinal absorption, hepatic metabolism, and bioactivity. Anal Sci 28, 197, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Ma L., Barker J., Zhou C., Li W., Zhang J., Lin B., Foltz G., Kublbeck J., and Honkakoski P. Towards personalized medicine with a three-dimensional micro-scale perfusion-based two-chamber tissue model system. Biomaterials 33, 4353, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagner I., Materne E.-M., Brincker S., Süßbier U., Frädrich C., Busek M., Sonntag F., Sakharov D.A., Trushkin E.V., Tonevitsky A.G., Lauster R., and Marx U. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 13, 3538, 2013 [DOI] [PubMed] [Google Scholar]
- 68.Leclerc E., Hamon J., and Bois F.Y. Investigation of ifosfamide and chloroacetaldehyde renal toxicity through integration of in vitro liver-kidney microfluidic data and pharmacokinetic-system biology models. J Appl Toxicol 36, 330, 2016 [DOI] [PubMed] [Google Scholar]
- 69.Sutherland M.L., Fabre K.M., and Tagle D.A. The National Institutes of Health microphysiological systems program focuses on a critical challenge in the drug discovery pipeline. Stem Cell Res Ther 4 Suppl 1, I1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabre K.M., Livingston C., and Tagle D.A. Organs-on-chips (microphysiological systems): tools to expedite efficacy and toxicity testing in human tissue. Exp Biol Med (Maywood) 239, 1073, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Wikswo J.P., Curtis E.L., Eagleton Z.E., Evans B.C., Kole A., Hofmeister L.H., and Matloff W.J. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 13, 3496, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viravaidya K., Sin A., and Shuler M.L. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog 20, 316, 2004 [DOI] [PubMed] [Google Scholar]
- 73.Moraes C., Labuz J.M., Leung B.M., Inoue M., Chun T.H., and Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr Biol (Camb) 5, 1149, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]