Abstract
Aim
To establish a connection between microRNA (miRNAs), circadian rhythm, and colorectal cancer patient survival.
Methods
Genomic and clinical data were extracted from The Cancer Genome Atlas (TCGA) colorectal cancer database, and the expression levels of candidate miRNAs and a set of circadian rhythm-related genes (Per1, Per2, Per3, Bmal1), and genes associated with chemosensitivity (thymidylate synthase, dihydrofolate reductase) were assessed for any correlations among their expression. In addition, survival analyses specific to different colorectal cancer stages were performed to determine if these genes contribute to patient outcomes.
Results
Significant inverse correlation between the expression of Per1 and that of miR-192 and miR-194 was observed. In survival analyses, high miR-192 and miR-194 correlate with better overall survival in Stage II patients, but worse survival in more advanced Stage III/IV patients. The expression of Per1, but Per2 or Bmal1, is marginally associated with patient survival for Stage II patients. Low thymidylate synthase expression correlates with better overall survival in Stage II patients but worse survival in Stage III/IV patients.
Conclusion
This study establishes a foundation based on a large genomic database of colorectal cancer, for further investigation into the importance of regulatory mechanisms of circadian rhythm by miRNAs in colorectal cancer.
Keywords: Circadian Rhythm, colorectal cancer, epigenetic regulation, microRNA
INTRODUCTION
The circadian rhythm is the coordination and fluctuation of various biological processes and responses on a daily cyclic pattern. Circadian rhythm is influenced by external cues such as light cycles, and system-wide is regulated by the suprachiasmatic nucleus in the hypothalamus. Through various endocrine and neuronal signals, the rhythmic functions of other organs are coordinated. In addition to the system-wide regulation directed in the hypothalamus, each cell of the body has its own internal molecular clock that is regulated through the expression of a set of genes including Per1, Per2, Per3, and Bmal1.1 The regulation of the clock genes in the cell can influence the transcription of various other genes such as p53 and cMyc and coordinate important cellular functions such as cell cycle progression and DNA repair.2 The circadian rhythm coordination of each cell can be important to maintain proper cellular growth and function. For example, in the colon, the cycling of expression of these genes can affect colon function as well as influence cell cycle regulation.3 In addition, in epidermal proliferating stem cells, the cycling of these genes helps to regulate DNA synthesis, oxidative phosphorylation, and glycolysis and may help to prevent genomic damage.4
It has been well appreciated for a period of time that circadian rhythm is an important factor to consider during chemotherapeutic treatment. The concept of chronotherapy takes advantage of this understanding to optimize the timing of drug delivery with circadian rhythms to not only improve drug efficacy but also to decrease toxicity to the patient. This can be important to improving patient outcomes. In addition, disruptions of circadian rhythm during chemotherapy are associated with worse patient survival.5
5-Fluoropyrimidine (5-FU, tegafur, capecitabine)-based chemotherapy is an effective treatment option for advanced colorectal cancer.6 The response and survival rate of colorectal cancer has been improved over the years by extensive research efforts.7 It is well established that 5-FU targets a critical enzyme thymidylate synthase (TYMS, TS).8 TS is a folate-dependent enzyme that catalyzes the reductive methylation of 2′-deoxyuridine 5′-monophosphate by 5, 10-methylenetetrahydrofolate to form deoxythymidine monophosphate and dihydrofolate. Because the TS-catalyzed enzymatic reaction provides the sole intracellular de novo source of thymidylate, an essential precursor for DNA biosynthesis, this enzyme has been an important target for cancer chemotherapy for over a half century.9 5-FU also directly incorporates into DNA and RNA to trigger cell death.10 Studies investigating the chronomodulation of 5-FU therapy in advanced colorectal cancer with metastasis have demonstrated that when the timing of drug delivery is adjusted to account for circadian rhythm, patients not only exhibit a greater response, with improved survival but also exhibited less toxicity and side effects.11,12 These differences can be understood by considering the rhythmic fluctuation in different anabolic and catabolic pathways associated with 5-FU metabolism. Due to the activity of enzymes that process 5-FU and can help to clear it from the body peaking at night, 5-FU can better be processed and its toxicity reduced when administered at night.13 In addition, as 5-FU targets DNA synthesis that is reduced at night and TS, which is also less active at night, a night time administration of 5-FU is less toxic to healthy tissues.14,15 Thus, there is a solid link between circadian rhythm and chemotherapy.
With the advancement of sequencing technology and the discovery of noncoding RNAs, the epigenetic regulations mediated by microRNAs (miRNAs) on circadian rhythm have now been identified in many organ systems.16,17 However, the contribution that miRNA regulation of circadian rhythm makes to chemoresistance has not been intensively investigated in cancer. With regards to colorectal cancer and chemoresistance, our laboratory has discovered several miRNAs, which contributes to 5-FU chemoresistance via various mechanisms. More specifically, we and others have discovered that the target protein of 5-FU, TS, is regulated by miR-192 and miR-215.18–20 In addition to TS, miR-192 also regulates dihydrofolate reductase (DHFR), a protein target of methotrexate.19 Targeting DHFR represents another important therapeutic strategy in antifolate-based cancer chemotherapy.21
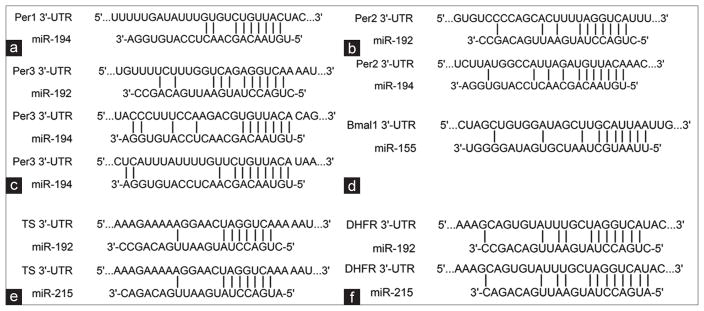
It is interesting to note that some of these miRNAs also regulate the expression of proteins that control circadian rhythm.17 It has been reported that the miR-192/194 cluster regulates the Period gene family (Per1, Per2, and Per3) and the circadian clock.16 The inhibition of these genes by miR-192/194 shortens the length of the circadian period.16 miR-192 also forms a positive feedback loop with p53 to regulate cell cycle and proliferation.19 miR-215 suppressed DTL expression, which is involved in destabilizing p53.20 p53 is directly involved in circadian rhythm controlled by modulating Per2 protein expression.22,23 These studies established a direct link between miRNA, p53, circadian rhythm, and genotoxic stress response.22,23 miR-155 also has been found to target Bmal1. In macrophages, the regulation of Bmal1 by miR-155 plays a role in the regulation of the inflammation response.24 The immune response has an important role in chemoresistance. Binding sites between these miRNA and their mRNA targets are shown in Figure 1.
Figure 1.
The depiction of previously validated microRNA-target mRNA pairs shown is base pairing between microRNA and binding sites in the 3′UTR of the target mRNA. (a) Per1 with miR-19216 (b) Per2 with miR-192 and miR-19416 (c) Per3 with miR-192 and miR-19416 (d) Bmal1 with miR-15524 (e) thymidylate synthase with miR-192 and miR-21528 (f) dihydrofolate reductase with miR-192 and miR-21519,20
Clearly, strong connections have been established between miRNA and circadian rhythm, and circadian rhythm has been associated with chemoresistance. So far, however, there has been no attempt to link epigenetic control mediated by miRNAs with circadian rhythm, chemoresistance, and clinical outcome of colorectal cancer. The rationale for the study is to determine whether some of these miRNAs and the relevant mRNAs targets will have any impact on the clinical outcome of colorectal cancer patient survival. The rationale for the study is illustrated in Figure 2. The study is based on the fact that circadian rhythm, 5-FU target protein TS and its miRNA regulators (miR-192, miR-215) have been demonstrated as key factors for influencing chemosensitivity and survival of the colorectal cancer patients. In addition, these miRNAs also have been reported to regulate circadian rhythm. The availability of The Cancer Genome Atlas (TCGA) colorectal cancer database and the large initiative to explore this database to answer biological and clinical driven questions provides a powerful tool and opportunity to utilize a large patient sample cohort to test the feasibility of the connection and establish a solid base to build on these connections going forward. In this study, we analyzed the expression profiles of miRNAs and circadian rhythm-related genes in TCGA colorectal cancer database, with the hope of gaining some insight into the role miRNA regulation of circadian rhythm may play in patient outcomes in colorectal cancer.
Figure 2.
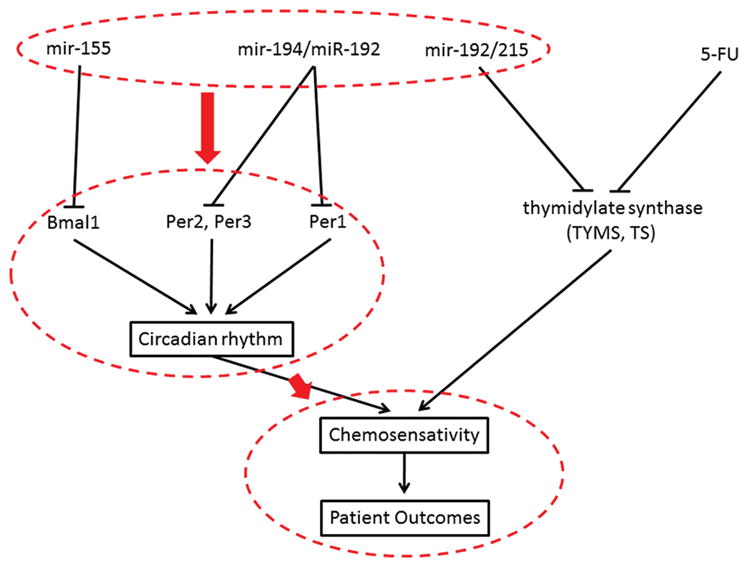
Schematic flow diagram of study rationale to investigate the circadian rhythm control microRNAs with colorectal cancer treatment outcome. Known connections between circadian rhythm and chemosensitivity, as well as validated microRNA-mRNA targets, are shown in solid lines. Highlighted in red is the connections we are trying to investigate in this study namely, that microRNA regulation of circadian rhythm-related genes influences chemosensitivity and patient survival
METHODS
Samples
TCGA is a comprehensive and coordinated effort to accelerate understanding of the molecular basis of many different cancer types through the application of genome analysis technologies.25 Genomic and clinical data from TCGA have been curated by the UCSC cancer genome browser project (https://genomecancer.ucsc.edu) and is available for download through a set of web-based tools.26 In this study, we extracted the clinical, miRNA expression, and mRNA expression data for TCGA colon adenocarcinoma from the UCSC cancer genome browser. The miRNA and mRNA expression were measured using HiSeq platform, and genome-wide characterizations of their expression patterns have been reported previously.27 For the clinical data, the survival information for 431 subjects is available, and the details can be found on TCGA database. There are 331 subjects that have both survival and miRNA expression data and 286 subjects that have both survival and mRNA expression data. All samples represent tissues taken from primary tumors of patients who have received no neoadjuvant therapy.
miRNA selection
The miRNA selected for this analysis was previously validated to target key genes associated with circadian rhythms. In addition, we considered miRNA for which we and others have validated as playing a role in chemoresistance. The validated mRNA target binding sites with miRNAs are illustrated in Figure 1.
Statistical analysis
We performed overall survival analyses for Stage II and Stage III or IV colon adenocarcinoma patients separately. We dichotomized the expression profile of miR-192 and miR-194 into two groups (high, low). For miR-192, the cut-off is 60%, i.e., expression greater than 60% of expression of the patients was denoted as “high;” otherwise, it was denoted as “low.” For miR-192, the cut-off is 55%. The cut-off values were chosen to yield the maximum separation of their survival distribution. Similarly, we dichotomized the expression profile of TS among the 286 patients into two groups (high, low), with the cut-off set as 45% of the expression. Subsequently, we performed log-rank tests between two groups and generated corresponding Kaplan–Meier curves.
We also examined the Pearson’s correlations between a small group of candidate miRNAs and mRNAs. If a candidate miRNA is inversely correlated with a candidate mRNA, it would indicate a possible regulatory mechanism of the miRNA on the mRNA as miRNA inhibit expression of their targets. The candidate miRNAs studied include miR-192, miR-194, miR-155, and the candidate mRNAs include Per1, Per2, Bmal1, TYMS, TS, DHFR. All statistical analyses were performed with R (Version 3.1.2, The R Foundation for Statistical Computing, Vienna, Austria). The statistical significance is described in the figures.
RESULTS
Inverse correlation between miRNA and target mRNA transcripts
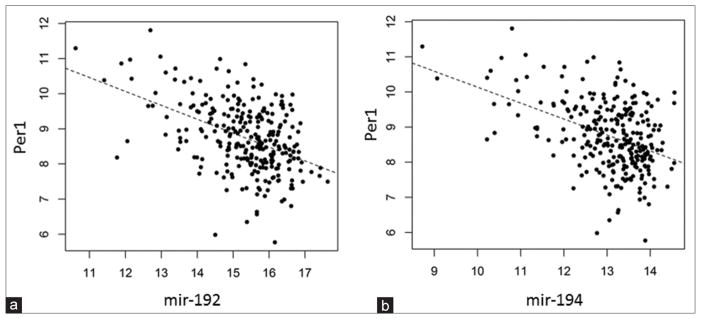
In order to assess the interactions between miRNAs and their target mRNAs in TCGA database, we calculated the pairwise correlations between the miRNAs and mRNAs. Our results show that the expression of miR-192 is significantly correlated with the Per1 expression level (P = 2.3 × 10−15) [Figure 3a]. The expression levels of miR-194 are also significantly (P = 1.5 × 10−15) correlated with the Per1 expression [Figure 3b]. The expression levels of TS and miR-192 are not correlated (data not shown). The lack of correlation between TS and miR-192 reflects the regulatory mechanism by which miR-192 suppresses the protein translation of TS without degrading the mRNA transcript.28 We did not observe any correlation between miR-155 and Bmal1 expression in colorectal cancer.
Figure 3.
(a) Inverse correlation of miR-192 expression with Per1 expression (P = 2.3 ×10−15) and (b) miR-194 expression with Per1 expression (P = 1.5 × 10−15)
Patient’s overall survival associated with miRNA and target transcript mRNA levels
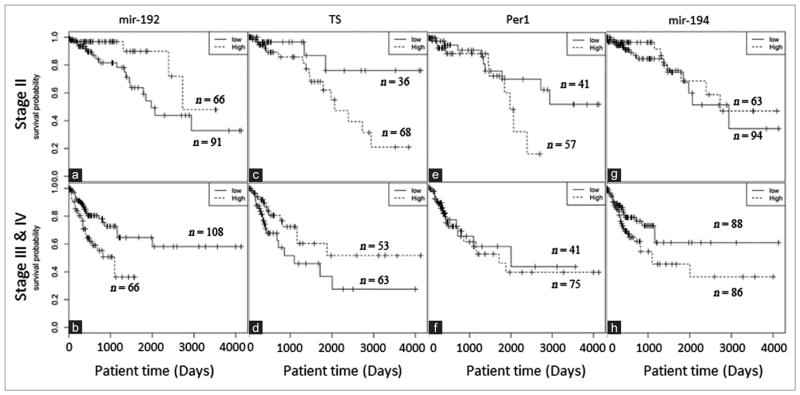
To determine the clinical relevance of miRNAs and target transcript levels, we performed survival analysis for different stages of colorectal cancer patients. Our results show that in Stage II colorectal cancer cohort, patients with a high level of miR-192 have better survival than the low expression group (P = 0.027) [Figure 4a]. Interestingly, in the more advanced cohort (Stages III and IV), high expression of miR-192 is also significantly (P = 0.003) associated with patient survival [Figure 4b], but with worse survival outcome.
Figure 4.
(a and b) Kaplan–Meier survival analysis of miR-192 (P = 0.027) (P = 0.003) (c and d) thymidylate synthase (P = 0.038) (P = 0.051) (e and f) Per1 (P = 0.10) (P = 0.79) (g and h) and miR-194 (P = 0.36) (P = 0.045) in Stage II, Stage III and IV colorectal cancer
In addition, the expression of miR-192 mediated target TS is also significantly associated with Stage II and Stage III/IV colorectal cancer survivals, with P values of 0.038 and 0.051, respectively [Figure 4c and d]. Interestingly, TS also shows opposite effects similar to those of miR-192, i.e., low TS expression corresponds to better overall survival in the Stage II cohort, but worse survival in Stage III/IV cohort. For the other target of miR-192, Per1, its high expression is marginally associated with worse survival outcomes in the Stage II cohort (P = 0.1) [Figure 4e], but it is not significant in Stage III/IV cohort [Figure 4f]. The levels of miR-155 are not significantly associated with colorectal cancer patient survival (data not shown). DHFR was also not significantly associated with survival of colorectal cancer patients (data not show). The expression of miR-194 levels significantly correlated (P = 0.045) with Stage III and IV colorectal tumor survival [Figure 4g and h].
DISCUSSION
In this study, we investigated the relationships between regulation of circadian rhythm by miRNAs and their target transcripts and clinical outcomes of colorectal cancer. This study is based on the miRNA and mRNA expression data of a large colorectal cancer patient cohort from TCGA.27 Our results clearly show that, among all miRNA circadian rhythm regulators, there is a significant correlation of miR-192 and miR-194 expression with colorectal cancer patient’s survival [Figure 4].
One important finding in this study is that the higher expression levels of miR-192 are associated with better survival in Stage II colorectal cancer while lower expression levels of miR-192 are significantly associated with better survival in more advanced Stage III and IV patients. This difference may be explained by the fact that miR-192 inhibits colon cancer cell proliferation.28 In Stage II colon cancer, slow proliferation of the cancer cells may be advantageous for patient outcomes as complete surgical resection of cancer may be possible. For Stage III and IV patients, the slower cell proliferation associated with higher miR-192 expression may reduce the effectiveness of chemotherapeutics such as 5-FU, which primarily target rapidly dividing cell populations. This finding is also highly consistent with miR-192 suppressed target proteins TS and Per1, as in Stage II colorectal cancer, patients with lower expression levels of TS and Per1 survived better.29,30 In contrast, in Stage III and IV patients, patients with high expression levels of TS had much better survival.30 It has been reported that in Stage II colon cancer, patients with low TS expression, survived better with surgery alone.29,30 Our results in Stage III and IV are also highly consistent with previous studies that show a high level of TS is associated with better survival than those with low TS in 5-FU treated patients.31 Although in that study, Stage II and III patients are combined for the survival correlation with TS.
The association of TS levels with 5-FU based chemotherapy has long been investigated. The rhythmic connection of TS levels with 5-FU toxicity and efficacy has been extensively studied and impacts the clinical management of colorectal cancer. In this study, we took a novel angle by investigating the impact of miRNA-mediated circadian rhythm gene expression on colorectal cancer survival using a bioinformatics approach. This is, in fact, the first report to show the potential clinical significance of miRNA, circadian rhythm, and colorectal cancer outcome.
Our results show that except for marginal association of Per1, other circadian proteins such as Per2, Per3, and Bmal1 that directly control circadian rhythm are not associated with colorectal cancer patient survival. This difference between the miRNA and target mRNA associations with survival may be due to the fact that miRNA can regulate multiple targets. The regulation of several other targets may have an effect on patient outcomes as well. It is not yet clear if the regulation of a few key targets by a miRNA or the regulation of a network of targets is more important to the biological functions of a miRNA. This is an important question that remains to be addressed in the miRNA field.32 In addition, it is also possible that using data from TCGA database, which represents a snapshot of expression of these genes at one moment in time, will miss the importance of changes in the dynamics of their expression that may influence cancer patient outcomes. This type of bioinformatics study cannot specifically address this concern, but does encourage continued exploration of this topic to address these questions. This could be explored through in vivo experiments aimed at studying changes in the expression of these genes over time.
In summary, we clearly demonstrated the significant correlation of circadian rhythm control miRNAs with overall survival of colorectal cancer patients. There is great potential for enhancing chemosensitivity and survival of colorectal cancer patients by modulating circadian rhythm miRNA regulator expression. Using the vast bioinformatics information available in TCGA, our findings suggest there may be some importance to miRNA regulation of circadian rhythm with regards to colorectal cancer patient outcomes. This study encourages further investigations to directly demonstrate the functional impact of miRNAs on circadian rhythm and chemosensitivity in colorectal cancer.
Acknowledgments
Financial support and sponsorship
This study was supported by the National Institute of Health/National Cancer Institute R01CA155019 (J. Ju), R33CA147966 (J. Ju).
Footnotes
For reprints contact: reprints@medknow.com
Conflicts of interest
There are no conflicts of interest.
References
- 1.Innominato PF, Lévi FA, Bjarnason GA. Chronotherapy and the molecular clock: clinical implications in oncology. Adv Drug Deliv Rev. 2010;62(9–10):979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Filipski E, Innominato PF, Wu M, Li XM, Iacobelli S, Xian LJ, Lévi F. Effects of light and food schedules on liver and tumor molecular clocks in mice. J Natl Cancer Inst. 2005;97(7):507–17. doi: 10.1093/jnci/dji083. [DOI] [PubMed] [Google Scholar]
- 3.Polidarová L, Olejníková L, Paušlyová L, Sládek M, Soták M, Pácha J, Sumová A. Development and entrainment of the colonic circadian clock during ontogenesis. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G346–56. doi: 10.1152/ajpgi.00340.2013. [DOI] [PubMed] [Google Scholar]
- 4.Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen B, Gratton E. In vivo single-cell detection of metabolic oscillations in stem cells. Cell Rep. 2015;10(1):1–7. doi: 10.1016/j.celrep.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innominato PF, Giacchetti S, Bjarnason GA, Focan C, Garufi C, Coudert B, Iacobelli S, Tampellini M, Durando X, Mormont MC, Waterhouse J, Lévi FA. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int J Cancer. 2012;131(11):2684–92. doi: 10.1002/ijc.27574. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal S, Chu E. Current therapies for advanced colorectal cancer. Oncology (Williston Park) 2005;19(5):589–95. [PubMed] [Google Scholar]
- 7.Saif MW, Choma A, Salamone SJ, Chu E. Pharmacokinetically guided dose adjustment of 5-fluorouracil: a rational approach to improving therapeutic outcomes. J Natl Cancer Inst. 2009;101(122):1543–52. doi: 10.1093/jnci/djp328. [DOI] [PubMed] [Google Scholar]
- 8.Danenberg PV. Thymidylate synthetase – A target enzyme in cancer chemotherapy. Biochim Biophys Acta. 1977;473(2):73–92. doi: 10.1016/0304-419x(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 9.Ju J, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci U S A. 1999;96(7):3769–74. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104(3):263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Focan C. Sequential chemotherapy and circadian rhythm in human solid tumours. A randomised trial. Cancer Chemother Pharmacol. 1979;3(3):197–202. doi: 10.1007/BF00262422. [DOI] [PubMed] [Google Scholar]
- 12.Lévi FA, Zidani R, Vannetzel JM, Perpoint B, Focan C, Faggiuolo R, Chollet P, Garufi C, Itzhaki M, Dogliotti L, lacobelli S, Adam R, Kunstlinger F, Gastiaburu J, Bismuth H, Jasmin C, Misset JL. Chronomodulated versus fixed-infusion-rate delivery of ambulatory chemotherapy with oxaliplatin, fluorouracil, and folinic acid (leucovorin) in patients with colorectal cancer metastases: a randomized multi-institutional trial. J Natl Cancer Inst. 1994;86(21):1608–17. doi: 10.1093/jnci/86.21.1608. [DOI] [PubMed] [Google Scholar]
- 13.Milano G, Chamorey AL. Clinical pharmacokinetics of 5-fluorouracil with consideration of chronopharmacokinetics. Chronobiol Int. 2002;19(1):177–89. doi: 10.1081/cbi-120002597. [DOI] [PubMed] [Google Scholar]
- 14.Marra G, Anti M, Percesepe A, Armelao F, Ficarelli R, Coco C, Rinelli A, Vecchio FM, D’Arcangelo E. Circadian variations of epithelial cell proliferation in human rectal crypts. Gastroenterology. 1994;106(4):982–7. doi: 10.1016/0016-5085(94)90757-9. [DOI] [PubMed] [Google Scholar]
- 15.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am J Pathol. 1999;154(2):613–22. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 2009;276(19):5447–55. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 17.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16(11):1544–50. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 18.Song B, Ju J. Impact of miRNAs in gastrointestinal cancer diagnosis and prognosis. Expert Rev Mol Med. 2010;12:e33. doi: 10.1017/S1462399410001663. [DOI] [PubMed] [Google Scholar]
- 19.Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J. miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res. 2008;14(24):8080–6. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju J. Beyond thymidylate synthase and dihydrofolate reductase: impact of non-coding microRNAs in anticancer chemoresistance. Curr Enzym Inhib. 2012;8(2):118–23. doi: 10.2174/157340812800793228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotoh T, Vila-Caballer M, Liu J, Schiffhauer S, Finkielstein CV. Association of the circadian factor period 2 to p53 influences p53’s function in DNA-damage signaling. Mol Biol Cell. 2015;26(2):359–72. doi: 10.1091/mbc.E14-05-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotoh T, Vila-Caballer M, Santos CS, Liu J, Yang J, Finkielstein CV. The circadian factor period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol Biol Cell. 2014;25(19):3081–93. doi: 10.1091/mbc.E14-05-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, Foley NH, Early JO, Chen L, Zhang H, Xue C, Geiger SS, Hokamp K, Reilly MP, Coogan AN, Vigorito E, FitzGerald GA, O’Neill LA. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A. 2015;112(23):7231–6. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman M, Craft B, Swatloski T, Ellrott K, Cline M, Diekhans M, Ma S, Wilks C, Stuart J, Haussler D, Zhu J. The UCSC cancer genomics browser: update 2013. Nucleic Acids Res. 2013;41(Database issue):D949–54. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9(8):2265–75. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 29.Donada M, Bonin S, Nardon E, De Pellegrin A, Decorti G, Stanta G. Thymidilate synthase expression predicts longer survival in patients with stage II colon cancer treated with 5-flurouracil independently of microsatellite instability. J Cancer Res Clin Oncol. 2011;137(2):201–10. doi: 10.1007/s00432-010-0872-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomiak A, Vincent M, Earle CC, Johnston PG, Kocha W, Taylor M, Maroun J, Eidus L, Whiston F, Stitt L. Thymidylate synthase expression in stage II and III colon cancer: a retrospective review. Am J Clin Oncol. 2001;24(6):597–602. doi: 10.1097/00000421-200112000-00014. [DOI] [PubMed] [Google Scholar]
- 31.Kornmann M, Schwabe W, Sander S, Kron M, Sträter J, Polat S, Kettner E, Weiser HF, Baumann W, Schramm H, Häusler P, Ott K, Behnke D, Staib L, Beger HG, Link KH. Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA expression levels: predictors for survival in colorectal cancer patients receiving adjuvant 5-fluorouracil. Clin Cancer Res. 2003;9(1111):4116–24. [PubMed] [Google Scholar]
- 32.Lai EC. Two decades of miRNA biology: lessons and challenges. RNA. 2015;21(4):675–7. doi: 10.1261/rna.051193.115. [DOI] [PMC free article] [PubMed] [Google Scholar]