Abstract
As fundamental processes in mitochondrial dynamics, mitochondrial fusion, fission and transport are regulated by several core components, including Miro. As an atypical Rho-like small GTPase with high molecular mass, the exchange of GDP/GTP in Miro may require assistance from a guanine nucleotide exchange factor (GEF). However, the GEF for Miro has not been identified. While studying mitochondrial morphology in Drosophila, we incidentally observed that the loss of vimar, a gene encoding an atypical GEF, enhanced mitochondrial fission under normal physiological conditions. Because Vimar could co-immunoprecipitate with Miro in vitro, we speculated that Vimar might be the GEF of Miro. In support of this hypothesis, a loss-of-function (LOF) vimar mutant rescued mitochondrial enlargement induced by a gain-of-function (GOF) Miro transgene; whereas a GOF vimar transgene enhanced Miro function. In addition, vimar lost its effect under the expression of a constitutively GTP-bound or GDP-bound Miro mutant background. These results indicate a genetic dependence of vimar on Miro. Moreover, we found that mitochondrial fission played a functional role in high-calcium induced necrosis, and a LOF vimar mutant rescued the mitochondrial fission defect and cell death. This result can also be explained by vimar's function through Miro, because Miro’s effect on mitochondrial morphology is altered upon binding with calcium. In addition, a PINK1 mutant, which induced mitochondrial enlargement and had been considered as a Drosophila model of Parkinson’s disease (PD), caused fly muscle defects, and the loss of vimar could rescue these defects. Furthermore, we found that the mammalian homolog of Vimar, RAP1GDS1, played a similar role in regulating mitochondrial morphology, suggesting a functional conservation of this GEF member. The Miro/Vimar complex may be a promising drug target for diseases in which mitochondrial fission and fusion are dysfunctional.
Author Summary
Mitochondrial dynamics including fusion, fission and transport are essential for energy supply in eukaryotic cells; and defects in mitochondrial dynamics often result in premature aging and diseases such as Parkinson's disease (PD). In mitochondrial transport machinery, the Miro/Milton complex loads mitochondria onto microtubule through kinesin motor proteins; and regulates mitochondrial fusion and fission through unknown mechanisms. As a small GTPase, the exchange of GTP/GDP in Miro requires a specific guanine nucleotide exchange factor (GEF). However, the GEF for Miro has not been identified. In this study, we identified Vimar as a new regulator of mitochondrial dynamics in Drosophila. We found that loss of vimar promoted mitochondrial shortening; and this function was mediated through Miro. As a GEF, Vimar partially localized on mitochondria and could physically interact with Miro. In the pathophysiological conditions, including a Pink1 mutant to model PD and a calcium-overload induced stress to model neuronal necrosis in Drosophila, loss of vimar suppressed both aberrant mitochondrial fusion and fragmentation in PD and necrosis, respectively. As the mammalian homolog of Vimar, RAP1GDS1 function was similar to Vimar. Therefore, Vimar/ RAP1GDS1 may be a great drug target to deal with diseases caused by defective mitochondrial dynamics.
Introduction
Mitochondrial fission, fusion and transport play important roles for the function of this organelle [1, 2]. The balance between fusion and fission controls mitochondrial morphology, which is mediated by series of large dynamin-related GTPases [3]. Among these GTPases, mitofusin1/mitofusin2 (MFN1/MFN2) and optic atrophy protein1 (OPA1) are the core components that are responsible for mitochondrial fusion [4–7], whereas dynamin-related protein 1 (Drp1) is the core component that is responsible for mitochondrial fission [8, 9]. In addition to these GTPases in dynamin-related family, mitochondrial Rho (Miro), an atypical member of the Rho small GTPase family, has a well-known function of transporting the mitochondria along microtubules [10, 11]. Miro also regulates mitochondrial morphology via inhibition of fission under physiological Ca2+ conditions, although the mechanism is not that clear [12–16]. Large GTPases such as dynamin-like GTPase family members hydrolyze GTP and exchange GTP and GDP without the assistance from other regulators [17, 18]. However, members of the small GTPase family often require other proteins to help release their tightly bound GDP or enhance their low GTPase activities. These proteins are referred to as guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), respectively [19]. To date, most small GTPases require unique GEFs or GAPs [19].
An understanding of the regulation of mitochondrial dynamics may help us to address many human diseases. For instance, mutations in OPA1 or MFN2 result in dominant optic atrophy or Charcot-Marie-Tooth neuropathy type 2A [20, 21]. Abnormal mitochondrial fission also promotes aging and cell death [22, 23]. In necroptosis, the formation of the necrosome promotes mitochondrial fission through dephosphorylation of Drp1 [24]. In neuronal excitotoxicity, calcium ions are overloaded, resulting in reduced levels of the MFN2 protein, which enhances mitochondrial fission and leads to neuronal necrosis [25, 26]. In addition, other components such as Miro may participate in this process [26]. Miro has two EF hand motifs that bind calcium; thus, Miro can couple calcium increase with reduced mitochondrial motility to meet the locally increased energy demands [16, 27]. Interestingly, Miro also promotes fission in the presence of excess calcium, which is distinct from its inhibitory role in fission under normal calcium concentrations [16]. It is unclear whether Miro plays a functional role in neuronal necrosis [26].
The mitochondrial morphology represents a transient balance between mitochondrial fusion and fission [28]. Using a systematic genetic screen in yeast covering approximately 88% of genes, 117 genes that regulate mitochondrial morphology were identified [29]. Similarly, a screen of 719 genes that are predicted to encode mitochondrial proteins in worms demonstrated that more than 80% of these genes regulate mitochondrial morphology [30]. Although many genes may regulate mitochondrial morphology, their relationships to the core mitochondrial fusion and fission components are unclear.
In studying mitochondrial morphology, we accidently discovered that the loss of vimar (visceral mesodermal armadillo-repeats), which encodes an atypical GEF [31–33], promoted mitochondrial fission in Drosophila flight muscle cells. Furthermore, we found that vimar was capable of interacting with Miro in vitro. Genetically, vimar required normal GDP- or GTP-bound activity of Miro to affect mitochondrial morphology, suggesting vimar is likely the Miro GEF. In addition, we found that the Miro/vimar complex suppressed mitochondrial fission during necrosis and mitochondrial fusion in PINK1 mutant model of Parkinson’s disease (PD), making vimar a potential drug target.
Results
Vimar is a novel regulator of mitochondrial morphology in Drosophila under normal conditions
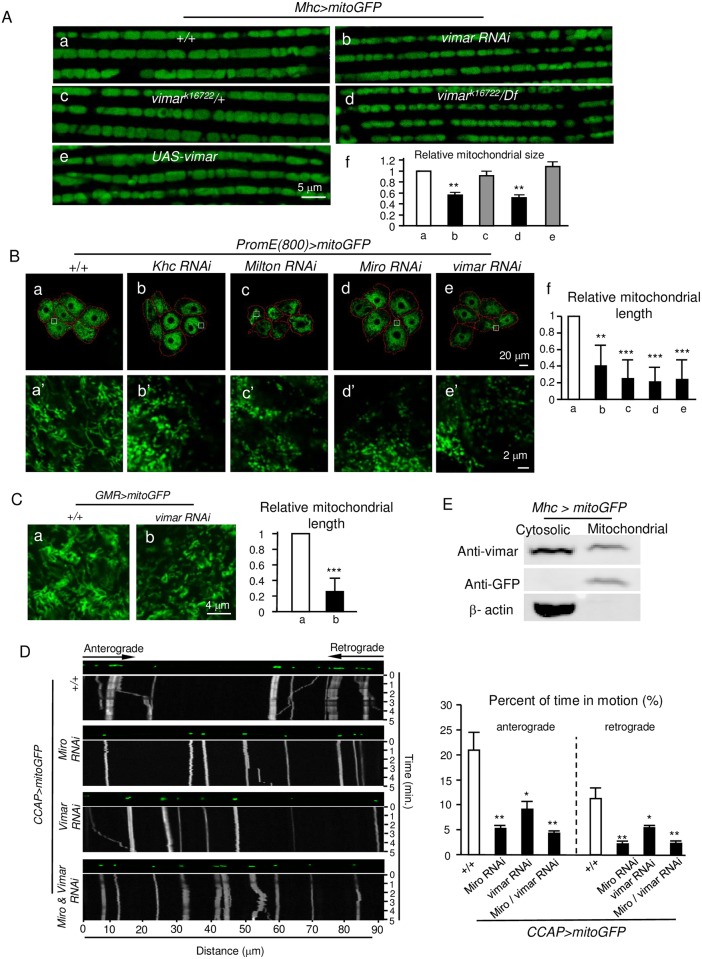
To identify novel regulators of mitochondrial morphology, we studied the flight muscle in Drosophila adults, because they have a stereotypic distribution of mitochondria in the longitudinal myofibers [34]. To visualize the mitochondria in the muscle cells, a muscle-specific promoter, Mhc-Gal4, was used to drive a mitochondria-targeted GFP (UAS-mitoGFP); the progenies were referred to as Mhc>mitoGFP. The mitochondrial morphology was clearly observed (Fig 1Aa and 1Af). Using these flies, we accidently observed that mitochondrial fission was enhanced when a vimar (visceral mesodermal armadillo-repeats) RNAi was expressed (Fig 1Ab and 1Af). To further confirm the loss-of-function (LOF) effect of vimar, we tested vimark16722, a P-element mutant with the mobile element inserted into the 5’-UTR region of the vimar gene. Again, we observed the trend of enhanced mitochondrial fission in the heterozygous vimark16722 mutant (Fig 1Ac and 1Af). Because the homozygous vimark16722 mutant was embryonic lethal, we selected a deficient mutant (Df(2R) ED1612) covering the vimar locus and generated a trans-heterozygous vimar (vimark16722/Df) mutant to further test the effect of vimar. In these flies, the mitochondria exhibited a stronger fission morphology compared to the heterozygous mutant (Fig 1Ad and 1Af). These results indicate that vimar plays a dominant role in regulating mitochondrial morphology in a dosage-dependent manner. To confirm the mitochondrial defect was generated from loss of vimar, we tried to rescue vimark16722/Df by tubulin-Gal4/UAS-vimar (tubulin-Gal4 is a ubiquitously expressed promoter). The result showed that the shortened mitochondria in the vimark16722/Df mutant were rescued by vimar overexpression (S1A Fig); while overexpression of vimar alone did not affect the mitochondrial morphology (Fig 1Ae and 1Af), suggesting that the levels of the vimar protein may be saturated under normal physiological condition. Using a polyclonal antibody of vimar, we confirmed that the protein levels of vimar were reduced in vimark16722 and vimar RNAi, and increased in the vimar overexpression line (S1B and S1C Fig).
Fig 1. Drosophila vimar affects mitochondrial morphology under normal conditions.
(A) a-e, Live imaging of the mitochondrial morphology in the flight muscle of adult flies. The mitochondria are labeled with UAS-mitoGFP driven by Mhc-Gal4 (Mhc>mitoGFP). The genotype is indicated on each micrograph. f, To quantify the mitochondrial size, the averaged mitochondrial size of the control (+/+) is set as 1, and the relative ratios of the other genotypes to the control are shown. Five thoraces from each genotype were quantified. Bar graphs throughout all figures are means ± SD. The white bar represents the control, the gray bar represents no statistical different from the control, and the black bar represents significantly different from the control. * for p<0.05; ** for p<0.01; ***for p<0.001. (B) Mitochondrial distribution and morphology in larval oenocytes. a, The mitochondria are labeled with PromE(800)> mitoGFP. b-e, The effects of Khc, Milton, Miro and vimar RNAi are shown. The dotted red lines denote the cell boundaries, which were determined by the mitoGFP background. a'-e', Enlarged view of the white box labeled area in the upper panel. f, To quantify the mitochondrial length, the averaged mitochondrial length of the control (+/+) is set as 1, and the relative ratios of the other genotypes to the control are shown. Mitochondrial length of five oenocytes was quantified per genotype and shown as means ± SD. (C) Live imaging of mitochondria in eye disc after knocking down vimar by GMR>mitoGFP. Three eye discs were analyzed for each genotype. (D) Effect of vimar on mitochondrial transport. The mitochondria are labeled with mitoGFP (CCAP>mitoGFP), and their movements in the axons were recorded and transformed into kymographs. Mitochondria motion in ten axons from five larvae was analyzed for each genotype. The quantification is shown on the bar graph. (E) Subcellular vimar protein distribution by protein fractionation. The proteins from adult thoraces (Mhc>MitoGFP) were separated into cytosolic and crude mitochondrial fractions. The vimar protein enrichment was analyzed by immunobloting with the anti-vimar antibody. The mitoGFP protein was detected by the anti-GFP antibody; and β-actin is a cytosolic protein.
To examine mitochondrial distribution, we studied Drosophila larval oenocytes because of their stereotypical location and morphology. The wild type mitochondria, labeled with UAS-mitoGFP driven by an oenocytes-specific promoter, PromE(800)-Gal4, were evenly distributed in the cytosol (Fig 1Ba). As positive controls, we knocked down Khc (kinesin heavy chain), Milton (an adaptor protein to link Khc to mitochondria) and Miro, which are the core components of mitochondrial transport machinery [35]. The results showed that mitochondrial spreading was greatly reduced in the cytosol, and resulted in accumulation in the perinuclear region (Fig 1Bb–1Bd). Interestingly, knocking down vimar by RNAi showed a similar distribution pattern (Fig 1Be). For mitochondrial morphology, loss of Khc, Milton, Miro and vimar resulted in mitochondrial shortening (Fig 1Ba'–1Be' and 1Bf). Similarly, mitochondria in the eye disc of GMR>mitoGFP/vimar RNAi was also shortened (Fig 1C). These results suggest that vimar regulates mitochondrial morphology in different cell types, such as muscle, oenocyte and eye disc.
Transport of mitochondria along the axon can be quantified in Drosophila neurons in vivo [36]. As a positive control, the CCAP-Gal4>Miro RNAi line (CCAP-Gal4 is a promoter labeling a single axon within a neuron bundle) displayed reduced flux of mobile mitochondria in both anterograde (soma to synapse) and retrograde (synapse to soma) transport (Fig 1D). The CCAP-Gal4>vimar RNAi line showed a similar result (Fig 1D). RNAi of both vimar and Miro resulted in a similar reduction of mitochondrial transport as Miro RNAi alone (Fig 1D), suggesting vimar and Miro may function in the same pathway.
To test vimar subcellular localization, proteins from the thoraces of adult flies (Mhc>MitoGFP) were extracted and separated into cytosolic and mitochondrial fractions. The Western blot data showed that endogenous vimar was present in both cytosol and mitochondria (Fig 1E). Apart from mitochondria, to test whether Vimar can distribute in other subcellular compartments, we fractionized organelles of ER, lysosome and Golgi apparatus, and found that Vimar was also enriched in the ER fraction, as well as in the cytosol (S2A Fig). This result is consistent with reports suggesting that Miro protein is localized and function in the site of mitochondria-ER junction [37, 38].
Vimar functions through Miro to regulate mitochondrial morphology
We asked whether vimar regulates mitochondrial morphology through controlling the GTP/GDP exchange of Miro, because Miro is a well-known small GTPase that regulates mitochondrial transport and morphology [10, 14].
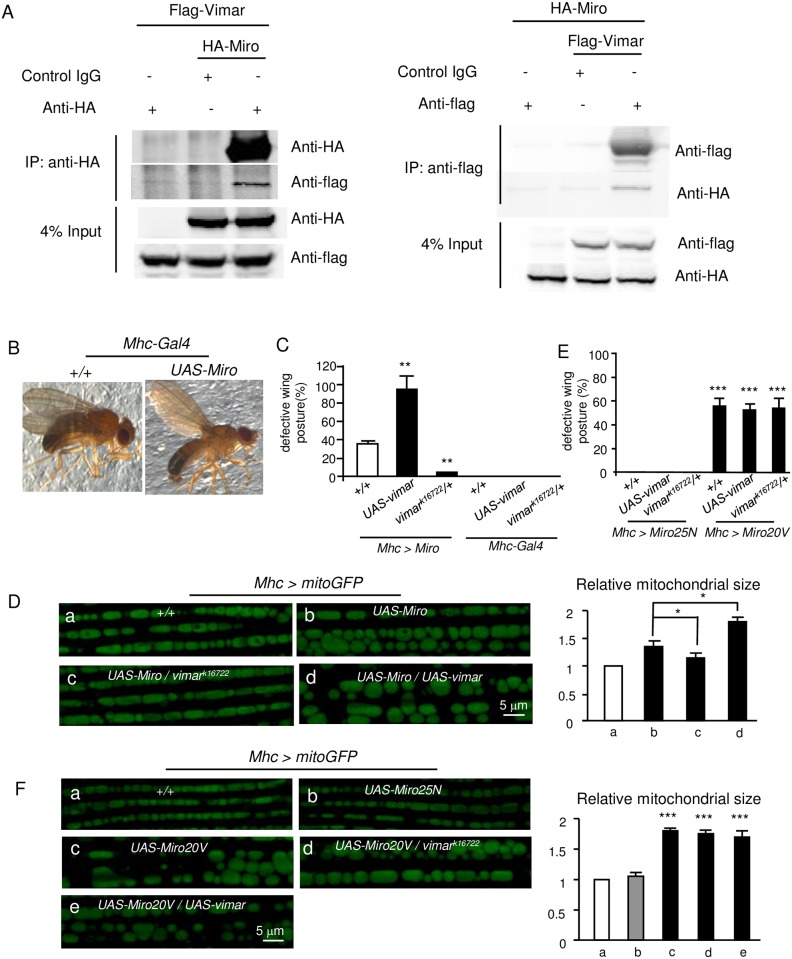
First, we evaluated their physical interactions. The Flag-tagged Vimar (Vimar-Flag) and HA-tagged Miro (Miro-HA) were ectopically expressed in the HEK293T cells. By co-immunoprecipitation (co-IP) assays with anti-HA and anti-Flag antibodies, Miro and Vimar could pull down with each other (Fig 2A). This result suggests that Miro and Vimar can bind with each other, at least under the overexpression conditions.
Fig 2. Interaction of vimar and Miro.
(A) Co-Immunoprecipitation of vimar and Miro. The proteins were collected from the HEK293T cells that expressed both Flag-tagged Vimar (Flag-Vimar) and HA-tagged Miro (HA-Miro). Then, the proteins were precipitated with a HA (left panel) or Flag antibody (right panel). The control IgG is shown as a negative control. The total protein input is shown as the protein loading control. (B) An example of the defective wing posture. Compared to the control, overexpression of Miro in the adult flight muscle (Mhc>Miro) resulted in an upright fly wing posture. (C) Quantification of defective wing posture in the Miro overexpression background or in the Mhc-Gal4 background. Trial N = 3, with 100–150 flies examined in each experiment. (D) Live imaging of the mitochondrial morphology in the fly flight muscle. The genotype of each fly muscle is labeled on the micrograph. Five thoraces were quantified for each genotype. (E) Quantification of defective wing posture in the Miro20V and Miro25N overexpression background. Trial N = 3, with 100–150 flies examined in each experiment. (F) Live imaging of the mitochondrial morphology in the fly flight muscle in the indicated genotypes. Five thoraces were quantified for each genotype.
Next, we tested their genetic interactions. The wing posture defects underline dysfunctional flight muscles that control wing position and movement [39]. It has been reported that overexpression of Miro induces mitochondrial enlargement [13, 15, 16]. Consistently, we observed this mitochondrial change in the Miro overexpression condition. Meanwhile, the wing posture defects of Mho>Miro flies increased progressively after eclosion and reached the maximum to approximately 30% at the seventh day after eclosion (Fig 2B and 2C). Interestingly, vimark16722 almost completely abolished the wing defect induced by the Miro overexpression; while vimar overexpression greatly enhanced the wing posture defect. As controls, vimar overexpression alone or vimar mutant (vimark16722) had no wing posture defect (Fig 2C). For the mitochondrial morphology in the Mhc>mitoGFP flies, Miro overexpression resulted in aberrant mitochondrial size enlargements (Fig 2Da and 2Db), and these defects could be rescued by the heterozygous vimark16722 mutant (Fig 2Dc). Moreover, vimar overexpression further enhanced mitochondrial size increase under the Miro overexpression background (Fig 2Dd). These results suggest that vimar may genetically interact with Miro.
We cannot test effect of GOF vimar under the Miro RNAi background, because Miro RNAi did not induce the wing posture defects in the flight muscles. To further test Miro/vimar interaction, we generated transgenes of constitutively GDP-bound or GTP-bound mutant of Miro. The rational is that GOF or LOF vimar should not affect these mutant phenotypes if vimar functions as a Miro GEF. Based on a previous report [40], the amino acid substitutions of A20V (Miro20V) and T25N (Miro25N) should render Miro constitutively GTP-bound and GDP-bound, respectively. As expected, a Miro25N overexpression in the flight muscle (Mhc>Miro25N) did not affect the wing posture (Fig 2E) or mitochondria morphology (Fig 2Fa and 2Fb). In contrast, a strong wing posture defect (Fig 2E) and enlarged mitochondria size (Fig 2Fc) were observed in the Miro20V overexpression line (Mhc>Miro20V). Importantly, GOF or LOF vimar failed to affect the defects in the Miro20V overexpression line (Fig 2E, 2Fd and 2Fe). We also examined vimar effect on mitochondrial transport in the GOF MiroWT, Miro20V and Miro25N background. However, we found that almost no mitochondria were distributed in the axons in the GOF MiroWT or Miro20V background. This data is consistent with previous reports indicating GOF Miro strongly increased mitochondrial length and reduced transportation [41, 42]. We could not examine their mitochondrial transports. In contrast, mitochondrial transport was unaltered under GOF vimar background or combined with Miro25N expression (S2B Fig). Together, these results suggest that vimar requires the normal GTP/GDP binding activity of Miro for its function.
To test whether the vimar/Miro interaction depends on the GTPase activity of Miro, we co-transfected vimar-Flag with inactive (Miro25N) and active (Miro20V) form of Drosophila HA-Miro in the HEK293T cells. The co-IP results showed that the vimar/Miro interaction was unaffected by these Miro mutants (S2C Fig). This result suggests that Miro/vimar interaction is not regulated by the GTPase activity of Miro. For mitochondrial distribution of vimar under the LOF Miro background (Mhc>mitoGFP/Miro RNAi), we observed that the mitochondrial fraction of vimar was unaltered (S3A Fig). This result indicates that vimar may attach with mitochondria by itself or with other partners.
It has been reported that mitochondrial shortening caused by Miro loss required the function of Drp1 [16]. Therefore, we could expect that loss of Drp1 might rescue the mitochondrial shortening in the muscle of Miro RNAi background. Indeed, it is the case (S3B Fig). Regarding the interaction between Drp1 and Vimar, our data showed that loss of Drp1 also rescued the mitochondrial shortening of the Vimar mutant (S3B Fig). This result indicates that Miro/Vimar complex is likely to regulate mitochondrial fission through Drp1.
To study whether Miro/vimar affected the Drp1 recruitment to mitochondria under Miro RNAi or vimar RNAi backgrounds, we used a transgene with a 9.35 kb genomic DNA insertion, which contains an endogenous Drp1 gene labeled by a HA tag (Flag-FlAsH-HA-Drp1) [43, 44]. The result showed that the mitochondria fraction of Drp1 monomer was unaltered in these RNAi conditions (S3C and S3D Fig). This result indicates that loss of Miro/vimar may not affect the recruitment of Drp1 to mitochondria, and how Miro/vimar affects Drp1 function is unclear.
Vimar promotes mitochondrial fission in response to high calcium concentrations
Miro plays distinct roles in regulating mitochondrial morphology under normal and high calcium conditions [16]. In normal conditions, Miro increases mitochondrial size through inhibition of Drp1 function [13, 15, 16]; however, it promotes mitochondrial fission in high calcium conditions by increasing Drp1 activity, such as in depolarized neurons [16, 35]. If vimar functions through Miro, we expect that vimar may promote mitochondrial fission in high calcium conditions.
To test Miro/vimar response at high calcium state, We had previously established a fly model to study the high calcium-induced cellular response, and accomplished calcium overload by expressing a leaky cation channel, the glutamate receptor 1 Lurcher mutant (GluR1Lc) [45, 46]. This fly model (simplified as the AG model) contained Appl-Gal4 (a neuron-specific promoter), UAS-GluR1Lc and tub-Gal80ts (an inhibitor of Gal4 at 18°C, which lost its function at 30°C). Thus, the AG flies were normal at 18°C, and calcium overload was induced upon a shift to 30°C [45]. Following the time progression after the GluR1Lc induction, calcium accumulates and neuronal necrosis increases gradually in the AG flies [45].
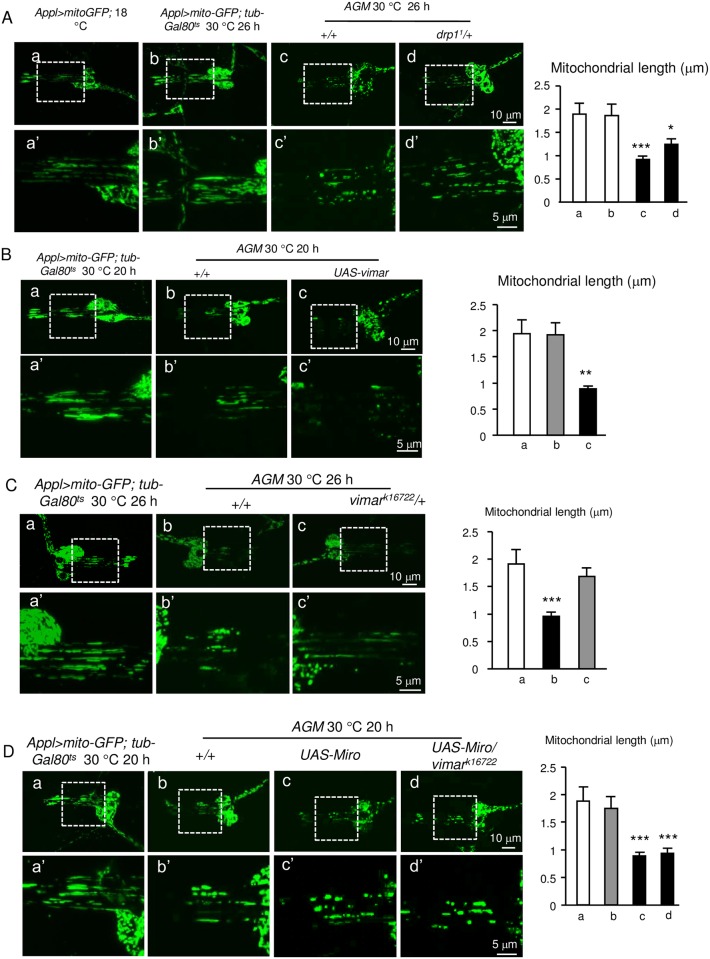
It is well known that mitochondrial fragmentation occurs upon calcium overloaded [47]. To recapitulate this phenomenon and observe mitochondrial morphology by live cell imaging, we added UAS-mitoGFP to the AG flies (simplified as the AGM model). After the AGM larval flies were raised at 30°C to induce calcium influx for 20 hours, mitochondrial fragmentation in the chordotonal neurons showed subtle fission compared to control; while at the 26 hour, the mitochondria in the AGM dendrites underwent dramatic fragmentation (Fig 3A–3D). For the rescue effect of a given genetic manipulation, we showed the 26 hour time point (to rescue the more severe defects); and for the enhancer effect of a given genetic manipulation, we showed the 20 hour time point (to enhance a less defective phenotype). As a positive control, a LOF mutant of Drp1, drp11, which possessed an A186V amino acid substitution at the Dynamin-GTPase domain [44], strongly suppressed the mitochondrial fission defect (Fig 3Ac and 3Ad). These results suggest that the AGM model can be adopted to study the mitochondrial morphology in high calcium conditions.
Fig 3. Vimar modulates the mitochondrial length in the high calcium condition.
(A) Effect of the Drp1 mutation on mitochondrial fission in high calcium conditions. a and b, Mitochondrial dendrites in the larval chordotonal neurons in the control flies (Appl>mitoGFP at 18°C and Appl>mitoGFP, tub-Gal80ts at 30°C). c and d, Mitochondrial morphology in the AGM and AGM/drp11 flies. a'-d' are the enlarged view from the boxed area in a-d, and the mitochondrial lengths in a'-d' were quantified. 10 to 16 chordotonal organs from each genotype were examined. (B) Effect of the vimar overexpression on the mitochondrial length after induction of AGM expression for 26 hours. 10 to 16 chordotonal organs were examined for each genotype. (C) Effect of vimar mutant (vimark16722) on the mitochondrial fragmentation of the AGM flies. 10 to 16 chordotonal organs were examined for each genotype. (D) Effect of Miro overexpression and the vimar mutant on the mitochondrial fragmentation of the AGM flies. 10 to 16 chordotonal neurons were examined for each genotype.
Because Miro promotes mitochondrial fission in the high calcium conditions [16], we expected that vimar might enhance Miro function under high calcium concentrations; and the LOF vimar might rescue the mitochondrial fission defect in the AGM flies. Indeed, GOF vimar enhanced mitochondrial fission (Fig 3Bb and 3Bc); and vimark16722 rescued mitochondrial fission in the AGM flies (Fig 3Cb and 3Cc). In the high calcium state, the mitochondrial localization of vimar was unaltered (S4 Fig), indicating recruitment of vimar on mitochondria is likely independent on calcium level. To further test the role of Miro/vimar complex, we examined effect of the GOF Miro transgene in the vimark16722 background. The result demonstrated that the GOF Miro enhanced mitochondrial fission (Fig 3Db and 3Dc), whereas vimark16722 could not rescue the defect (Fig 3Dd), indicating that basal function of Miro may be partially independent from vimar. Together, these results suggest that vimar functions through Miro to regulate mitochondrial morphology in high calcium conditions.
The loss of vimar suppressed both necrotic cell death and muscle defects in a Drosophila PD model
Mitochondrial fission may enhance calcium overload-induced necrotic cell death in neuron cultures [47]. However, there is still insufficient genetic evidence to demonstrate that mitochondrial fission plays a causal role in neuronal necrosis [48]. To study this question, we previously showed that we could quantify necrosis in the AGG flies (the AG flies containing UAS-GFP) at single cell resolution [45]. The result showed that Drp11 could rescue necrosis in the chordotonal neurons (Fig 4A). In addition, the function of these neurons could be assessed at the behavioral level by quantifying adult fly death [45]; Drp11 rescued the lethality of the AG flies (Fig 4B). Strikingly, vimark16722 exhibited a rescue effect in the AGG flies at both the cellular and behavioral levels (Fig 4C and 4D). In contrast, the GOF vimar transgene had the opposite effect (Fig 4E and 4F). This result is consistent with the suppression of mitochondrial fission in this mutant. Furthermore, the GOF Miro transgene enhanced necrosis; however, vimark16722 did not rescue the GOF Miro phenotype (Fig 4G and 4H), similar to its effect on mitochondria. These results indicate that Miro has the dominant role in the Miro/vimar complex and that the Miro/vimar complex plays a functional role in neuronal necrosis.
Fig 4. Vimar suppresses neuronal necrosis and muscle degeneration induced by the Pink1 mutant.
(A) Effect of the Drp1 mutant on neuronal necrosis. The micrographs showed the live images from larval chordotonal neurons. The control (Appl>GFP;tub-Gal80ts) displays the cell bodies of the wild type chordotonal neurons, which form a cluster containing 6 neurons. In the AGG background, the wild type (+/+) flies showed swollen cell bodies, weakened GFP intensity and neuronal cell loss; and these defects were rescued under the Drp1 mutant (Drp11) background. The right panel shows the quantification of the cell loss. For all quantification of neuronal necrosis, trial N = 5, with 10–15 flies were examined in each trial in this figure. (B) Effect of the Drp1 mutant on the survival of the AG adult flies. For all quantification of AG lethality, trial N = 3, with 100–150 flies were examined for each trial. (C) Effect of vimar mutant on neuronal necrosis. (D) Effect of the vimar mutant on the survival of the AG flies. (E) Effect of vimar overexpression on neuronal necrosis. (F) Effect of vimar overexpression on the survival of the AG flies. (G) Effect of Miro overexpression on neuronal necrosis. The result showed that Miro overexpression enhanced neuronal necrosis; and the vimar mutant had no rescue effect on this defect. (H) Effect of Miro overexpression on the survival of the AG flies. (I) Effect of the vimar mutant (vimark16722) on PINK1 mutant induced mitochondrial defect. The live image showed the mitochondrial morphology in the PINK1 mutant (PINK15) and under the vimar mutant background. Ten thoraces were analyzed for each genotype. (J) Effect of the vimar mutant (vimark16722) on the wing posture defect of the PINK1 mutant (PINK15). Trial N = 3, with 100–150 flies were examined in each trial.
In the PINK1 mutant of the Parkinson's disease (PD) Drosophila model, mitochondrial fusion is enhanced, and the LOF Miro mutant could suppress this mitochondrial defect [13]. Therefore, we speculate that the LOF vimar mutant might rescue the defective mitochondrial fusion in the PINK1 mutant. To test this hypothesis, we studied a PINK1 mutant, PINK15 [49]. In the PINK15 flies, the mitochondria are abnormally elongated and fused (Fig 4I), and the fly wing posture is defective (Fig 4J). Strikingly, vimark16722 could rescue both the mitochondrial morphology and wing posture defects (Fig 4I and 4J). Together, these results indicate that LOF of the Miro/vimar complex suppressed both mitochondrial fragmentation during necrosis and PINK1 mutant of Drosophila PD model.
Furthermore, we found that vimark16722 and UAS-vimar had no effect on classical apoptosis induced by Hid expression [50] (S5 Fig), suggesting that vimar may specifically affect PD and necrosis, but does not regulate apoptosis.
The mammalian homolog of vimar (RAP1GDS1) plays a similar role in mitochondrial morphology and cell death
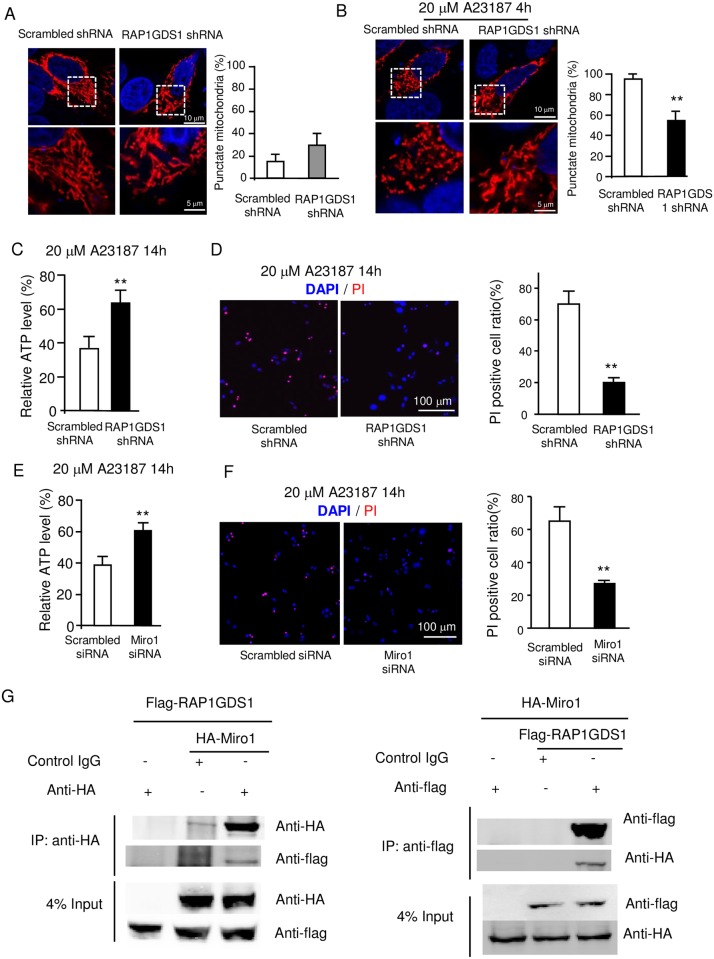
A protein sequence comparison showed that Drosophila vimar shares great similarity with the mammalian protein RAP1GDS1 (S6 Fig); however, it is not clear whether vimar is a functional homolog of RAP1GDS1 [51]. Here, we further investigated the role of RAP1GDS1 in mitochondrial morphology. First, we used a lentivirus to transfect a RAP1GDS1 shRNA into HEK293T cells and established a stable cell line. As expected, the protein level of the RAP1GDS1 was significantly reduced in the shRNA line (S7A Fig). Then, this shRNA line was transiently transfected with a mitochondrial reporter, mitoDsRed. We found that the mitochondrial length showed a trend of reduction in the RAP1GDS1 shRNA cells (Fig 5A). Next, we studied the effect of RAP1GDS1 on necrosis. Necrotic cell death was induced by a calcium ionophore (A23187), which causes calcium overloading and necrosis [52]. As expected, the calcium ionophore induced mitochondrial fragmentation, and the RAP1GDS1 shRNA rescued the mitochondrial defect (Fig 5B). To quantify the cell death, we measured cellular ATP level and performed propidium iodide (PI) staining [53]. The result showed that RAP1GDS1 shRNA rescued necrosis in both assays (Fig 5C and 5D). Moreover, we tested the RAP1GDS1 shRNA in another human cell line, the SH-SY5Y neuroblastoma cells. Similar to the HEK293T cells, the RAP1GDS1 shRNA protected the SH-SY5Y cells from calcium overload (S7B and S7C Fig). In addition, we examined the effect of a Miro-1 siRNA on calcium ionophore induced necrosis. The result showed that it also rescued the cell death (Fig 5E and 5F, and the Miro-1 siRNA effect is shown in S7D Fig). Furthermore, the HA-tagged Miro1 and the Flag-tagged RAP1GDS1 could co-immunoprecipitate in vitro (Fig 5G). Together, these results indicate that the function of the Miro1/RAP1GDS1 complex in regulating mitochondrial morphology and necrosis is conserved with the Drosophila Miro/vimar complex.
Fig 5. The conserved role of RAP1GDS1 in mammalian cells.
(A) Effect of RAP1GDS1 knock down on the mitochondrial morphology in HEK293T cells. The mitochondria in the cells that stably express the RAP1GDS1 shRNA are labeled with a transiently transfected MitoDsred expression vector. The cells were classified as tubular-shape or punctate-shape based on differences in their mitochondrial lengths. The ratio of punctate-shape mitochondria is shown in the right panel. The result showed that RAP1GDS1 shRNA had a trend to increase the punctate-shape mitochondria (not statistically different from the control shRNA). Trial N = 3, with 100 cells were quantified in each trial. (B) Effect of RAP1GDS1 knocking down on the mitochondrial fragmentation under calcium overload stress. The HEK293T cells were treated with 20 μM calcium ionophore (A23187) for 4 hours. The result showed that RAP1GDS1 shRNA reduced fragmented mitochondria upon calcium ionophore treatment. Trial N = 3, with 100 cells were quantified in each trial. (C) Effect of the RAP1GDS1 shRNA on calcium ionophore-induced necrosis. The HEK293T control and RAP1GDS1 shRNA stable cell lines were treated with 20 μM A23187 for 14 hours. Then, the cell death was quantified by the ATP assay. The result indicated that less cell death occurred in the RAP1GDS1 shRNA expressing cells. Trial N = 3. (D) Effect of the RAP1GDS1 shRNA on calcium ionophore-induced necrosis. The PI and DAPI staining patterns are shown. The red signals indicate the PI-positive cells and the blue channel indicates the DAPI staining. Trial N = 3. (E) Effect of the Miro1 siRNA on calcium ionophore induced necrosis determined by the ATP assay. The Miro1 siRNA was transiently transfected in HEK293T cells for 48 hours. Trial N = 3. (F) Effect of the Miro1 siRNA on calcium ionophore induced necrosis determined by the PI staining assay. The PI and DAPI staining patterns are shown. The same result was observed as in E. Trial N = 3. (G) Co-Immunoprecipitation of RAP1GDS1 and Miro1. The proteins were collected from the HEK293T cells that expressed Flag-tagged RAP1GDS1 (Flag-RAP1GDS1) and HA-tagged Miro1 (HA-Miro1). The control IgG is shown as a negative control. The total protein input is shown as the protein loading control. Trial N = 3.
Discussion
Vimar is a novel regulator of mitochondrial morphology
Mitochondrial function can be assessed by the enzymatic activity of citrate synthase (CS), the first enzyme in the Krebs cycle that converts acetyl-CoA and oxaloacetate to citrate [54]. In cultured Drosophila S2 cells, vimar knock down by RNAi resulted in reduced CS activity [54], indicating that vimar may positively regulate mitochondrial function. Because mitochondrial fission has generally been associated with reduced mitochondrial respiration [55], the decreased CS activity may be a result of mitochondrial fission. Consistent with this notion, our results demonstrated that the LOF of vimar promoted mitochondrial fission. In addition, a GOF vimar transgene had a minimal effect on mitochondrial morphology, indicating that vimar activity might be saturated under normal physiological conditions.
Vimar functions through Miro to regulate mitochondrial morphology
Because Vimar has been predicted to be a GEF, we hypothesized that vimar may regulate mitochondrial morphology by affecting a small GTPase, which requires a GEF to help with the GTP/GDP exchange process [19]. Interestingly, Miro is one such small GTPase that is known to play important roles in mitochondrial fission and transport [10, 14, 16]. We propose that vimar and Miro may function as a complex. First, a fraction of the vimar protein was localized to the mitochondria, possibly indicating a functional role on mitochondria. Interestingly, the mitochondrial localization of vimar seems not dependent on Miro, because LOF Miro did not affect the mitochondrial fraction of vimar. This indicates that vimar may directly bind with mitochondria or through other scaffolding proteins. Second, vimar and Miro could physically interact with each other, at least in vitro. Their interaction seems not affected by the GTPase activity of Miro, because the constitutively GDP- or GTP-bound Miro mutants did not affect their interactions. Third, vimar genetically interacted with Miro. This included the result demonstrating that the LOF vimar mutant reduced the effect of Miro on mitochondrial fission inhibition and the GOF vimar transgene had the opposite effect. Moreover, in the constitutive GFP-bound or GDP-bound Miro mutants, the effect of the GOF or LOF vimar was abolished. Therefore, vimar requires the normal GDP/GTP binding activity of Miro to function. It is also known that Miro1 overexpression increase mitochondrial size partially by suppression of the Drp1 function [15, 16]. Consistently, increased mitochondrial fission in the LOF of Miro or vimar was abolished by loss of Drp1, suggesting the Miro/vimar complex depends on Drp1 to regulate mitochondrial morphology.
The Miro/vimar complex may regulate PD and neuronal necrosis through mitochondrial fusion and fission
Familial PD caused by mutations in PINK1 or Parkin results in a series of mitochondrial dysfunctions, particularly the failure to eliminate damaged mitochondria through mitophagy [56, 57]. In these PINK1 or Parkin mutants, the key proteins involved in mitochondrial fusion and fission, such as Marf/Mitofusin and Miro, accumulate [13, 58]. In the PINK1 mutant flies, the flight muscle is damaged, resulting in wing posture defects [59]. Similarly, we observed that Miro overexpression in the flight muscle resulted in a strong wing posture defect. This result may explain the wing posture defect in the PINK1 mutant, in which the levels of the Miro protein are increased [13]. Our result demonstrated that the LOF of vimar could rescue the wing defect in the PINK1 mutant, consistent with the hypothesis that vimar functions through Miro.
When the intracellular calcium level is high, Miro switches from promoting mitochondrial fission inhibition to enhancing mitochondrial fission [16]. The mechanism for this switch is unclear, although alterations of Drp1 function could be one possibility [16]. Interestingly, Gem1, the yeast homolog of Miro GTPase, has been reported to function as a negative regulator for ER-mitochondria contacts, where Drp1 aggregates and cleaves mitochondria into smaller units [37]. This may serve as the mechanism for Miro to regulate mitochondrial morphology via Drp1. In addition to affect mitochondrial fission, Miro also regulates mitochondrial transport in a calcium dependent manner. For mitochondrial transport, Miro forms protein complexes with Milton, a kinesin adaptor, and with motor proteins, such as kinesin and dynein [35]. In high calcium conditions, Miro alters its binding patterns and results in reduced transport activity [27, 60, 61]. Based on these reports, we proposed that the Miro/vimar complex acted together to affect mitochondrial morphology: at normal condition, Miro/vimar inhibits fission via Drp1; at high calcium state, Ca2+ bound Miro switches its function to promote fission. Indeed, vimar responds to the calcium change in the same way as Miro (Fig 6). In addition, our data demonstrated that knocking down RAP1GDS1 and Miro1 increased mitochondrial fission and could rescue calcium overload induced necrosis, similar to the loss of vimar or Miro in Drosophila. These data support the hypothesis that RAP1GDS1 is the mammalian homolog of vimar, supporting a previous prediction [51].
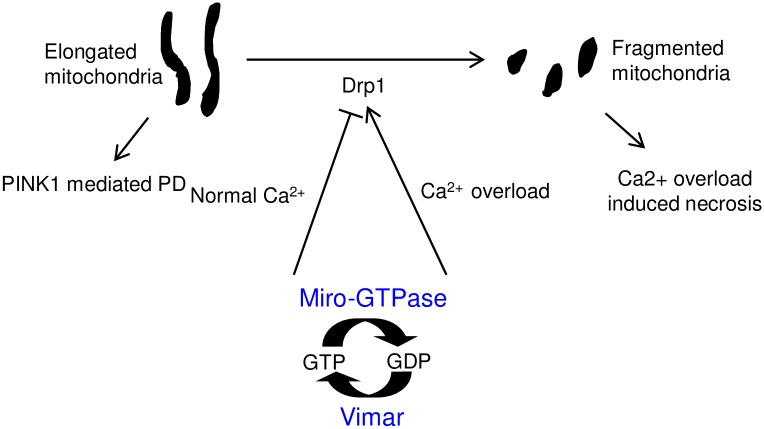
Fig 6. A schematic model of Miro/vimar function on mitochondrial morphology.
In normal calcium conditions, the Miro/vimar complex promotes mitochondrial fission inhibition, and their GOF results in elongated mitochondria. Increased mitochondrial fusion is known to occur in the PINK1 mutant flies, and this defect can be rescued by LOF Miro/vimar. In the high calcium state, the Miro/vimar complex promotes mitochondrial fragmentation, which accelerates neuronal necrosis. Regardless of the intracellular calcium level, vimar enhances the function of Miro, because vimar is likely the GEF to promote Miro's GTP/GDP exchange.
Mitochondrial fission plays important role in apoptosis by promoting mitochondrial outer-membrane permeabilization (MOMP) to release cytochrome c from the mitochondria [62]. The use of the Drp1 inhibitor mdivi to block fission has been shown to be an effective treatment for stroke [47], and the function of mitochondrial fission on necrotic cell death has been well documented [24, 26, 48]. The uncertainty lies in the lack of genetic evidence and downstream mechanism of mitochondrial fission in necrosis [48]. Our data demonstrated that mitochondrial fragmentation occurred in necrotic neurons, and the LOF Drp1 and vimar mutants both suppressed neuronal necrosis.
Much evidence suggests that the mitochondrial fusion and fission defects are directly linked to many human diseases [22], and strategies that target the Miro/vimar complex may affect a broad spectrum of diseases. For instance, mutations in the fragile X mental retardation 1 (FMR1) gene, which result from expansion of trinucleotide repeat in the 5′ untranslated region, often cause enhanced mitochondrial fission and mental retardation syndrome [63]. Likewise, aberrant mitochondrial fusion was observed in a Drosophila Alzheimer's disease model induced by the ectopic expression of a human tau mutant (tauR406W) [43]. In this case, the tau mutant may promote excessive actin stabilization to decrease Drp1 recruitment to the mitochondria, which results in excessive mitochondrial fusion and neurodegeneration [43, 64]. Due to the dual function of the Miro/vimar complex in high-Ca2+ induced necrosis and PINK1 mutant induced PD, a drug to target this complex may benefit both disease states. As a modulator, it may be safer to target vimar/ RAP1GDS1.
Supporting Information
(A) a-c, Live imaging of the mitochondrial morphology in the flight muscle of adult flies. The mitochondria are labeled with UAS-mitoGFP driven by Tubulin-Gal4 (Tubulin>mitoGFP). The genotype is indicated on each micrograph. d, The averaged mitochondrial size of the control (+/+) is set as 1, and the relative ratios of the other genotypes to the control are shown. Five thoraces from each genotype were quantified. Bar graphs throughout all figures are means ± SD. The white bar represents the control, the gray bar represents no statistical different from the control, and the black bar represents significantly different from the control. * for p<0.05; ** for p<0.01; ***for p<0.001. (B) and (C) Vimar protein level in the adult thoraces. The Western blot shows immunobloting with a vimar antibody, with the genotype listed on each lane. β-actin is shown as the protein loading control. The quantified data is shown as means ± SD. Trial N = 3.
(PDF)
(A) Vimar distribution in other subcellular compartments. The fly homogenate was separated into cytosol, lysosome, Golgi apparatus and ER. Vimar protein level was determined by immunobloting using a vimar antibody. Calnexin, Lamp1, GM130 and β-actin are markers for ER, lysosome, Golgi apparatus and cytoplasm, respectively. (B) Effect of vimar overexpression on mitochondrial transport. The mitochondria are labeled with mitoGFP (CCAP>mitoGFP), and their movements in the axons were recorded and transformed into kymographs. Overexpression of Miro25N, vimar or both of them had no effects towards mitochondria transport. Mitochondria motion in ten axons from five larvae was analyzed for each genotype. (C) Effect of Drosophila Miro mutants on its interaction with vimar in vitro. The HA-tagged Miro, Miro20V (a constitutive GTP-bound mutant) and Miro25N (a constitutive GDP-bound mutant) was individually co-transfected with Flag-tagged vimar. The co-IP experiment showed that GTP or GDP state of Miro did not affect its interaction with vimar. Trial N = 3. IgG is shown as a negative control. The total protein input is shown as the protein loading control.
(PDF)
(A) Effect of LOF Miro on the mitochondrial localization of vimar. In the Mhc>mitoGFP/Miro RNAi flies, the mitochondrial fraction of vimar was unaltered as the control Mho>mitoGFP flies. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2. (B) Live image of mitochondria in flight muscle. Miro RNAi and vimar RNAi resulted in shortened mitochondria, which could be blocked by Drp1 RNAi. Five thoraces from each genotype were quantified. (C) and (D) Drp1 recruitment to mitochondria in Miro RNAi or vimar RNAi background. Thoracic homogenate was separated into cytosol and mitochondria and Drp1 protein level in different fractions was immunoblotted. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2.
(PDF)
The AG fly heads were homogenized and separated in cytoplasmic fraction and crude mitochondria. Vimar level was labeled by a vimar antibody. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2.
(PDF)
The apoptotic flies (GMR-Gal4;GMR-Hid) showed smaller eye size. Addition of UAS-P35, a known apoptosis inhibitor, is shown as a positive control, which rescued the smaller eye size defect. However, vimark16722 or UAS-vimar showed no effect on the eye size defect.
(PDF)
The alignment is generated from CLUSTAL alignment algorithm.
(PDF)
(A) Effect of the RAP1GDS1 shRNA on the level of the RAP1GDS1 protein in the stable HEK293T cells. β-actin was used as the loading control. (B) Effect RAP1GDS1 shRNA on necrosis in the stable SH-SY5Y cells. The cells were treated with 20 μM A23187 for 1 hour. The bright field images of the cells showed less cell death in the RAP1GDS1 shRNA cells upon calcium ionophore treatment. Trial N = 4. (C) Quantification of necrosis by the ATP assay. The stable SH-SY5Y cell lines were treated with 20 μM A23187 for 6 hours. The result showed that less cell death occurred in the RAP1GDS1 shRNA cells than the control (scrambled shRNA) cells. Trial N = 3. (D) The efficiency of the Miro1 siRNA on Miro transcripts in 293T cells. The transcript level of Miro1 was determined by qRT-PCR. The result showed that Miro1 siRNA significantly knocked down Miro1 transcripts. Trial N = 3.
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by the Chinese Ministry of Science and Technology (Grant No. 2013CB530700; http://www.most.gov.cn/) to LL and the National Science Foundation (Grant No. 81325007; http://www.nsfc.gov.cn/) for Distinguished Young Scholars to XJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sheng ZH, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 13(2):77–93. Epub 2012/01/06. doi: nrn3156 [pii] 10.1038/nrn3156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8(11):870–9. 10.1038/nrm2275 . [DOI] [PubMed] [Google Scholar]
- 3.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. 10.1146/annurev.cellbio.22.010305.104638 . [DOI] [PubMed] [Google Scholar]
- 4.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11(12):872–84. 10.1038/nrm3013 . [DOI] [PubMed] [Google Scholar]
- 5.Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26(2):211–5. 10.1038/79944 . [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101(45):15927–32. Epub 2004/10/29. doi: 0407043101 [pii] 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1(5):298–304. 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–58. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47(3):379–93. 10.1016/j.neuron.2005.06.027 . [DOI] [PubMed] [Google Scholar]
- 11.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173(4):545–57. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J Cell Biol. 2004;167(1):87–98. 10.1083/jcb.200405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, et al. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 8(3):e1002537 Epub 2012/03/08. 10.1371/journal.pgen.1002537 PGENETICS-D-11-02331 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fransson A, Ruusala A, Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem. 2003;278(8):6495–502. 10.1074/jbc.M208609200 . [DOI] [PubMed] [Google Scholar]
- 15.Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344(2):500–10. 10.1016/j.bbrc.2006.03.163 . [DOI] [PubMed] [Google Scholar]
- 16.Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, et al. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A. 2008;105(52):20728–33. 10.1073/pnas.0808953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13(2):75–88. 10.1038/nrm3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10(6):423–9. 10.1038/nrm2689 . [DOI] [PubMed] [Google Scholar]
- 19.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiological reviews. 2013;93(1):269–309. 10.1152/physrev.00003.2012 . [DOI] [PubMed] [Google Scholar]
- 20.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26(2):207–10. 10.1038/79936 . [DOI] [PubMed] [Google Scholar]
- 21.Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116(1–2):23–7. 10.1007/s00439-004-1199-2 . [DOI] [PubMed] [Google Scholar]
- 22.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends in cell biology. 2013;23(2):64–71. 10.1016/j.tcb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Annals of the New York Academy of Sciences. 2008;1147:283–92. 10.1196/annals.1427.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1–2):228–43. 10.1016/j.cell.2011.11.030 . [DOI] [PubMed] [Google Scholar]
- 25.Martorell-Riera A, Segarra-Mondejar M, Munoz JP, Ginet V, Olloquequi J, Perez-Clausell J, et al. Mfn2 downregulation in excitotoxicity causes mitochondrial dysfunction and delayed neuronal death. EMBO J. 2014;33(20):2388–407. 10.15252/embj.201488327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Zhang F, Li L, Tang F, Siedlak SL, Fujioka H, et al. MFN2 couples glutamate excitotoxicity and mitochondrial dysfunction in motor neurons. J Biol Chem. 2015;290(1):168–82. 10.1074/jbc.M114.617167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Schwarz TL. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136(1):163–74. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazier AE, Kiu C, Stojanovski D, Hoogenraad NJ, Ryan MT. Mitochondrial morphology and distribution in mammalian cells. Biological chemistry. 2006;387(12):1551–8. 10.1515/BC.2006.193 . [DOI] [PubMed] [Google Scholar]
- 29.Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Molecular biology of the cell. 2005;16(11):5410–7. 10.1091/mbc.E05-07-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichishita R, Tanaka K, Sugiura Y, Sayano T, Mihara K, Oka T. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J Biochem. 2008;143(4):449–54. 10.1093/jb/mvm245 . [DOI] [PubMed] [Google Scholar]
- 31.Kikuchi A, Kaibuchi K, Hori Y, Nonaka H, Sakoda T, Kawamura M, et al. Molecular cloning of the human cDNA for a stimulatory GDP/GTP exchange protein for c-Ki-ras p21 and smg p21. Oncogene. 1992;7(2):289–93. . [PubMed] [Google Scholar]
- 32.Kaibuchi K, Mizuno T, Fujioka H, Yamamoto T, Kishi K, Fukumoto Y, et al. Molecular cloning of the cDNA for stimulatory GDP/GTP exchange protein for smg p21s (ras p21-like small GTP-binding proteins) and characterization of stimulatory GDP/GTP exchange protein. Molecular and cellular biology. 1991;11(5):2873–80. 10.1128/MCB.11.5.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto T, Kaibuchi K, Mizuno T, Hiroyoshi M, Shirataki H, Takai Y. Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. The Journal of biological chemistry. 1990;265(27):16626–34. . [PubMed] [Google Scholar]
- 34.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100(7):4078–83. 10.1073/pnas.0737556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birsa N, Norkett R, Higgs N, Lopez-Domenech G, Kittler JT. Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Biochemical Society transactions. 2013;41(6):1525–31. 10.1042/BST20130234 . [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Schwarz TL. Imaging axonal transport of mitochondria. Methods in enzymology. 2009;457:319–33. 10.1016/S0076-6879(09)05018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(34):14151–6. 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beall CJ, Fyrberg E. Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J Cell Biol. 1991;114(5):941–51. 10.1083/jcb.114.5.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babic M, Russo GJ, Wellington AJ, Sangston RM, Gonzalez M, Zinsmaier KE. Miro's N-terminal GTPase domain is required for transport of mitochondria into axons and dendrites. J Neurosci. 2015;35(14):5754–71. 10.1523/JNEUROSCI.1035-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, et al. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS genetics. 2012;8(3):e1002537 10.1371/journal.pgen.1002537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo GJ, Louie K, Wellington A, Macleod GT, Hu F, Panchumarthi S, et al. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29(17):5443–55. 10.1523/JNEUROSCI.5417-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DuBoff B, Gotz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75(4):618–32. 10.1016/j.neuron.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47(3):365–78. 10.1016/j.neuron.2005.06.018 . [DOI] [PubMed] [Google Scholar]
- 45.Liu K, Ding L, Li Y, Yang H, Zhao C, Lei Y, et al. Neuronal necrosis is regulated by a conserved chromatin-modifying cascade. Proc Natl Acad Sci U S A. 2014;111(38):13960–5. 10.1073/pnas.1413644111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci. 2000;3(4):315–22. 10.1038/73877 . [DOI] [PubMed] [Google Scholar]
- 47.Grohm J, Kim SW, Mamrak U, Tobaben S, Cassidy-Stone A, Nunnari J, et al. Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ. 2012;19(9):1446–58. 10.1038/cdd.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jahani-Asl A, Germain M, Slack RS. Mitochondria: joining forces to thwart cell death. Biochimica et biophysica acta. 2010;1802(1):162–6. 10.1016/j.bbadis.2009.09.006 . [DOI] [PubMed] [Google Scholar]
- 49.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–6. 10.1038/nature04779 . [DOI] [PubMed] [Google Scholar]
- 50.Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95(3):331–41. 10.1016/S0092-8674(00)81765-1 . [DOI] [PubMed] [Google Scholar]
- 51.Lo PC, Frasch M. bagpipe-Dependent expression of vimar, a novel Armadillo-repeats gene, in Drosophila visceral mesoderm. Mech Dev. 1998;72(1–2):65–75. 10.1016/S0925-4773(98)00016-1 . [DOI] [PubMed] [Google Scholar]
- 52.Karch J, Kwong JQ, Burr AR, Sargent MA, Elrod JW, Peixoto PM, et al. Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. eLife. 2:e00772 Epub 2013/08/31. 10.7554/eLife.00772 00772 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nature chemical biology. 2005;1(2):112–9. 10.1038/nchembio711 . [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Shi X, Padmanabhan R, Wang Q, Wu Z, Stevenson SC, et al. Identification of novel modulators of mitochondrial function by a genome-wide RNAi screen in Drosophila melanogaster. Genome Res. 2008;18(1):123–36. Epub 2007/11/29. doi: gr.6940108 [pii] 10.1101/gr.6940108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochimica et biophysica acta. 2012;1817(10):1833–8. 10.1016/j.bbabio.2012.02.033 . [DOI] [PubMed] [Google Scholar]
- 56.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8(1):e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189(2):211–21. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107(11):5018–23. 10.1073/pnas.0913485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–61. 10.1038/nature04788 . [DOI] [PubMed] [Google Scholar]
- 60.Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61(4):541–55. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, Sheng ZH. Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. The Journal of cell biology. 2013;202(2):351–64. 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14(2):193–204. 10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross-Inta C, Omanska-Klusek A, Wong S, Barrow C, Garcia-Arocena D, Iwahashi C, et al. Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. The Biochemical journal. 2010;429(3):545–52. 10.1042/BJ20091960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, et al. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol. 2007;9(2):139–48. 10.1038/ncb1528 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) a-c, Live imaging of the mitochondrial morphology in the flight muscle of adult flies. The mitochondria are labeled with UAS-mitoGFP driven by Tubulin-Gal4 (Tubulin>mitoGFP). The genotype is indicated on each micrograph. d, The averaged mitochondrial size of the control (+/+) is set as 1, and the relative ratios of the other genotypes to the control are shown. Five thoraces from each genotype were quantified. Bar graphs throughout all figures are means ± SD. The white bar represents the control, the gray bar represents no statistical different from the control, and the black bar represents significantly different from the control. * for p<0.05; ** for p<0.01; ***for p<0.001. (B) and (C) Vimar protein level in the adult thoraces. The Western blot shows immunobloting with a vimar antibody, with the genotype listed on each lane. β-actin is shown as the protein loading control. The quantified data is shown as means ± SD. Trial N = 3.
(PDF)
(A) Vimar distribution in other subcellular compartments. The fly homogenate was separated into cytosol, lysosome, Golgi apparatus and ER. Vimar protein level was determined by immunobloting using a vimar antibody. Calnexin, Lamp1, GM130 and β-actin are markers for ER, lysosome, Golgi apparatus and cytoplasm, respectively. (B) Effect of vimar overexpression on mitochondrial transport. The mitochondria are labeled with mitoGFP (CCAP>mitoGFP), and their movements in the axons were recorded and transformed into kymographs. Overexpression of Miro25N, vimar or both of them had no effects towards mitochondria transport. Mitochondria motion in ten axons from five larvae was analyzed for each genotype. (C) Effect of Drosophila Miro mutants on its interaction with vimar in vitro. The HA-tagged Miro, Miro20V (a constitutive GTP-bound mutant) and Miro25N (a constitutive GDP-bound mutant) was individually co-transfected with Flag-tagged vimar. The co-IP experiment showed that GTP or GDP state of Miro did not affect its interaction with vimar. Trial N = 3. IgG is shown as a negative control. The total protein input is shown as the protein loading control.
(PDF)
(A) Effect of LOF Miro on the mitochondrial localization of vimar. In the Mhc>mitoGFP/Miro RNAi flies, the mitochondrial fraction of vimar was unaltered as the control Mho>mitoGFP flies. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2. (B) Live image of mitochondria in flight muscle. Miro RNAi and vimar RNAi resulted in shortened mitochondria, which could be blocked by Drp1 RNAi. Five thoraces from each genotype were quantified. (C) and (D) Drp1 recruitment to mitochondria in Miro RNAi or vimar RNAi background. Thoracic homogenate was separated into cytosol and mitochondria and Drp1 protein level in different fractions was immunoblotted. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2.
(PDF)
The AG fly heads were homogenized and separated in cytoplasmic fraction and crude mitochondria. Vimar level was labeled by a vimar antibody. Anti-ATP5A is shown as the mitochondria marker and actin as cytosolic marker. Trial N = 2.
(PDF)
The apoptotic flies (GMR-Gal4;GMR-Hid) showed smaller eye size. Addition of UAS-P35, a known apoptosis inhibitor, is shown as a positive control, which rescued the smaller eye size defect. However, vimark16722 or UAS-vimar showed no effect on the eye size defect.
(PDF)
The alignment is generated from CLUSTAL alignment algorithm.
(PDF)
(A) Effect of the RAP1GDS1 shRNA on the level of the RAP1GDS1 protein in the stable HEK293T cells. β-actin was used as the loading control. (B) Effect RAP1GDS1 shRNA on necrosis in the stable SH-SY5Y cells. The cells were treated with 20 μM A23187 for 1 hour. The bright field images of the cells showed less cell death in the RAP1GDS1 shRNA cells upon calcium ionophore treatment. Trial N = 4. (C) Quantification of necrosis by the ATP assay. The stable SH-SY5Y cell lines were treated with 20 μM A23187 for 6 hours. The result showed that less cell death occurred in the RAP1GDS1 shRNA cells than the control (scrambled shRNA) cells. Trial N = 3. (D) The efficiency of the Miro1 siRNA on Miro transcripts in 293T cells. The transcript level of Miro1 was determined by qRT-PCR. The result showed that Miro1 siRNA significantly knocked down Miro1 transcripts. Trial N = 3.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.