Abstract
Puroindoline (Pina and Pinb) genes control grain texture or hardness in wheat. Wild-type/soft alleles lead to softer grain while a mutation in one or both of these genes results in a hard grain. Variation in hardness in genotypes with identical Pin alleles (wild-type or mutant) is known but the molecular basis of this is not known. We now report the identification of wheat genotypes with hard grain texture and wild-type/soft Pin alleles indicating that hardness in wheat may be controlled by factors other than mutations in the coding region of the Pin genes. RNA-Seq analysis was used to determine the variation in the transcriptome of developing grains of thirty three diverse wheat genotypes including hard (mutant Pin) and soft (wild type) and those that were hard without having Pin mutations. This defined the role of pin gene expression and identified other candidate genes associated with hardness. Pina was not expressed in hard wheat with a mutation in the Pina gene. The ratio of Pina to Pinb expression was generally lower in the hard non mutant genotypes. Hardness may be associated with differences in Pin expression and other factors and is not simply associated with mutations in the PIN protein coding sequences.
Introduction
Grain hardness or endosperm texture, defined as having a hard endosperm (hard wheat/s) or soft endosperm (soft wheat/s), is one of the prime determinants of wheat quality as it has a major impact on the milling properties and end-use quality of the wheat. For commercial trading purposes wheat is mainly classified into soft, hard and very hard wheats based on grain hardness. As grain hardness is a fundamental attribute of wheat quality it has been studied for more than a century, and is reported to be mainly under genetic control with the environment having a negligible role [1–3]. Grain hardness is predominantly controlled by the Puroindoline (Pin) genes, Pina and Pinb, which are part of only the D sub-genome and located on chromosome 5 at the Hardness (Ha) locus. Grain softness protein -1 (GSP-1), another gene tightly linked to the Pin genes on the Ha locus, was initially thought to be associated with grain hardness but later reports indicated otherwise [4–7]. During hexaploid wheat evolution, large genomic deletions in the short arm of chromosome 5 from the A and B sub-genomes caused the loss of the Pin genes but not the GSP-1 gene in these sub-genomes of hexaploid wheat [8].
Wheat genotypes with wild type Pina (Pina-D1a) and Pinb (Pinb-D1a) alleles display soft kernel texture whereas mutation in any of the Pin genes results in a hard phenotype. Several mutant alleles of Pina and Pinb have been reported in the last two decades [9–11]. Among the mutant Pin alleles, Pina-D1b and Pinb-D1b are the most frequently observed mutant alleles in common wheat, worldwide [11]. Pina-D1b is a null allele caused by gene deletion, and Pinb-D1b is a result of a Gly46Ser change relative to the wild type Pinb-D1a [12]. Genotypes with the Pina-D1b allele display harder grain texture than those with Pinb-D1b [11]. Transgenic experiments have demonstrated the role of puroindoines in grain softening. In hard red spring wheat with a mutant Pinb allele (Pina-D1a/Pinb-D1b), transformation with wild type Pin alleles has been shown to reduce grain hardness [13]. Similarly, transgenic expression of puroindoline in rice [14] and maize [15] has been shown to induce grain softness in these species which otherwise lack Pin genes.
Puroindoline protein isoforms, PINA and PINB, act together to form a friabilin protein [16] that binds to lipid molecules present on the starch surface through a hydrophobic tryptophan (trp) rich domain [17]. Friabilin prevents compact binding between starch and the protein matrix [18] which helps to soften the kernel texture. However, friabilin is less efficient in preventing this adhesion when composed of PIN proteins expressed by mutant Pin alleles resulting in grain hardness [19]. Mutation in Pinb has also been reported to reduce the amount of total PIN protein [20]. The Trp-rich domain in PINA contains 5 tryptophan residues whereas in PINB it contains 3 residues. Although PIN proteins act together their mode of action seems to be quite different most likely due to differences in their trp-rich domain. An in vitro study by Clifton, Sanders [21]shows that PINA forms self-assemblies in solution whereas PINB is dispersed in solution and PINB displays greater penetration into a lipid monolayer than PINA [17]. But, as the high resolution structures of these proteins is not yet known, the exact biochemical interactions between PIN proteins and starch remains unclear. However, it is believed that the trp-rich domain plays a key role in the biochemical action of the PIN proteins. According to in vitro studies this domain also provides antifungal and antibacterial properties to the PIN proteins [22].
The contribution of variations in Pin transcript abundance or patterns of Pina and Pinb expression to grain hardness remains unknown. Overexpression of wild type Pin alleles in hard wheat (Pina-D1a/Pinb-D1b) has been observed to reduce grain hardness [13]. In contrast, reduced expression of Pin genes through RNAi mediated silencing increases grain hardness [23]. Pin transcripts can be detected almost throughout the seed development period; from as early as 3 days post anthesis (DPA) until almost the end of maturity at 40 DPA. They are most abundant during the middle stages of seed development [13, 24–26]. Pina and Pinb have been reported to express at different levels (except Pina null) in some studies [12, 13, 27, 28], whereas, in other studies comparable levels (except Pina null) of gene expression has been reported [25, 28, 29]. This indicates the presence of different patterns of Pina and Pinb gene expression patterns in different genotypes. In this study we have studied these gene expression patterns in different genotypes.
In addition to Pinb on 5DS, several alleles of Pinb-2 variants have also been discovered on chromosome 7 ABD [30–32]. Pinb-2 variants share 57–60% homology with the Pinb. It has been suggested that they are likely to be involved in control of grain hardness [31]. However Giroux, Kim [33] showed that Pinb-2 transcripts are expressed at very low levels compared to Pina and Pinb and proposed that they are less likely to influence grain hardness.
The molecular basis for the variation in grain hardness within a particular grain texture class and containing identical Pin alleles still remains unexplained. Identifying the role, if any, of other genes in controlling grain hardness will increase our understanding of variation in grain hardness. These genes may express differentially between soft and hard wheats. In this study, RNA-seq data generated by Next Generation Sequencing (NGS) was analysed to identify differentially expressed genes (DEGs) between hard and soft wheats. We also analysed gene expression of the Pina, Pinb and GSP-1 ABD and Pinb-2 ABD genes in thirty four wheat genotypes at two stages of seed development, 14 DPA and 30 DPA. The objective was to determine the patterns of Pina, Pinb, GSP-1 ABD and Pinb-2 ABD expression and their association with levels of grain hardness and to identify other candidate genes that might influence hardness.
Materials and Methods
Plant material
A selection of 33 different wheat genotypes from the global wheat gene pool were used for this experiment; Amurskaja 75, Arnhem, Banks, Batavia, Beyrouth 3, Bobwhite, Bowebird, D.E.S. 367, Dollarbird, EGA Gregory, Ellison, Gabo, Giza 139, Greece 25, Huandoy, India 211, India 259, India 37, Iraq 46, Jing Hoang No.1, Kite, Lerma Rojo, Martonvasari 13T, NW108A, NW25A, NW51A, NW93A, Pelada, Punjab 7, Qalbis, Saturno, Sunco, Tunis 24. Seeds were obtained from the Australian Winter Cereal Collection, Tamworth, Australia. Plants were grown in a growth room at day and night temperatures of 20 0C and 18 0C with 12 hours of light, at Southern Cross University. The whole caryopsis was collected at two developmental time points; 14 DPA and 30 DPA. Separate sets of plants were grown in a glasshouse as duplicates, for genotypes Banks, Ellison, Gabo, Gregory, Kite and Sunco.
RNA isolation, cDNA preparation and NGS sequencing
Total RNA was isolated from the whole caryopsis using the Trizol reagent (Invitrogen, Carlsbad, USA) as published elsewhere [34]. To determine the total RNA concentration and quality, 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA) was used. cDNA was prepared and used to produce indexed Illumina NGS libraries which were then multiplexed to allow the sequencing of eight indexed libraries in one lane on a GA IIx Illumina sequencing platform. Seeds from the six duplicate genotypes grown in the glasshouse were also subjected to RNA isolation and NGS sequencing to provide replicate transcript profiles.
Grain hardness measurement
Grain hardness index (HI) was measured using a single kernel characterisation system (SKCS 4100 crushing machine) according to AACC Method 55–31 (AACC, 1999). Tests were performed at BRI, Sydney. SKCS HI was obtained from measurement of 300 matured wheat grains.
Quality analysis of sequence data
RNA sequencing of 33 genotypes generated a total of ~2.5 million to ~7.2 million reads varying with the genotype. All analysis of NGS data was undertaken using CLC Genomics Workbench V 8.0 (CLC Bio, Aarhus, Denmark). Illumina reads obtained from RNA-sequencing were trimmed for quality using default parameters to exclude calls with quality scores less than 20. Following trimming, all sequencing data was subjected to RNA Seq analysis using RNA-Seq tool.
Identification of Pin alleles and RNA seq analysis
Pina and Pinb gene sequences were identified from the consensus sequence obtained from RNA-seq. Using the RNA-Seq analysis tool available in CLC workbench, gene expression was measured using RPKM normalization. Triticum aestivum gene index (TaGI) dataset generated by Dana-Farber Cancer Institute (DFCI) (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Triticum_aestivum/) was used as a reference dataset for RNA-seq analysis. The parameters used for RNA–Seq analysis were, similarity fraction 0.9 and length fraction 0.8.
Identification of Pin alleles in wheat genotypes
The RNA-seq analysis files of all of the wheat genotypes were used to extract full length coding sequences of the Pina and Pinb genes. However, in some genotypes part of the 5’-region of the coding sequence (up to 5 bp including the ATG start codon) of the Pina and/or the Pinb gene was missing. The sequence identity of the missing region of the Pina and the Pinb genes was determined, as outlined below, by PCR amplification of the pin genes followed by Sanger sequencing in both direction. Genomic DNA was extracted from the wheat genotypes NW93A, NW25A, NW108A, Amurskaja 75, Lermarojo and Banks using method developed by Furtado [34].Full length Pina was amplified using forward primer 5’ CATCTATTCATCTCCACCTGC 3’ and reverse primer 5’ GTGACAGTTTATTAGCTAGTC 3’ [11], and full length Pinb was amplified using forward primer 5’ GAGCCTCAACCCATCTATTCATC 3’ and reverse primer 5’ CAAGGGTGATTTTATTCATAG 3’ [11]. Using a thermal cycler (Biorad T100), the Pin genes were amplified by PCR, first for 10 cycles by denaturing at 94°C for 30 s, followed by annealing at 41°C for 30s, and extension at 72°C for 2 min, followed by 25 cycles by denaturing at 94°C for 30 s, followed by annealing at 45°C for 30s, and extension at 72°C for 2 min. Amplified bands for the Pina (524 bp) and the Pinb (595 bp) genes were first confirmed by agarose gel electrophoresis and then sequenced in both directions by Sanger Sequencing.
The full length coding sequences Pina and Pinb genes of all wheat genotypes were aligned to the corresponding soft Pina or Pinb gene allele to determine the presence or absence of mutant Pin alleles in the wheat genotypes.
Identification of differentially expressed genes
The genotypes were divided into four groups based on grain hardness and presence of Pin mutant allele; Soft Non-Mutants, SNM (PinaD1a/Pinb-D1a; SKCS HI soft/ genetically soft), Hard Non-Mutants, HNM (PinaD1a/Pinb-D1a; SKCS HI hard/ genetically soft), Hard Pina-mutant, HPAM (PinaD1b/Pinb-D1a; SKCS HI hard/ genetically hard) and Hard Pinb-mutant, HPBM (PinaD1a/Pinb-D1b; SKCS HI hard/ genetically hard). These four groups were compared in nine different combinations to identify DEGs; SNM-HNM+HPAM+HPBM, SNM-HNM, SNM-HPAM, SNM-HPBM, HNM-HPAM, HNM-HPBM and non-mutants (Soft+HNM) vs HPAM+HPBM. ‘Empirical analysis of Differential Gene Expression’ test was performed to identify differentially expressed genes. Sequence ID’s with FDR p-value <1e-05 were selected as highly biologically significant differentially expressed sequences, candidate genes. These sequences were passed through NCBI BLASTx to find out similar protein sequences. BLAST hits were then functionally annotated using Blast2GO [35].
Functional annotation through Blast2GO
BLAST hits were passed through array of different tools in Blast2GO; mapping, annotation, InterProScan, Merge InterProScan, annex and GO slim (plant). This gave the information about their biological process, molecular function and cellular component.
Statistical analysis
Correlation between gene expression of Pina, Pinb, GSP-1 ABD and Pinb-2 ABD, and wheat grain hardness (SKCS HI) was evaluated based on linear and polynomial multiple regression analysis in RStudio software package. Data was grouped into 4 sets; Pina-mutant genotypes (Pina-D1b, Pinb-D1a), Pinb-mutant genotypes (Pina-D1a, Pinb-D1b), non-mutant genotypes (Pina-D1a, Pinb-D1a) and a last set included all the genotypes. A best fit model was selected by performing a stepwise Akaike information criterion (AIC) test in both directions mode followed by an Anova test.
Results
SKCS Hardness Index and Pin allele identification
Grain hardness as measured using a Single Kernel Characterisation System (SKCS) and the nature of the Pin alleles of the wheat genotypes included in this study are shown in Table 1. Genotypes were classified as soft wheat or hard wheat if their SKCS hardness index (SKCS HI) was below or above 50, respectively [36–38]. Using the full-length coding sequence of the Pina and Pinb genes obtained from RNA-Seq analysis or by PCR amplification, wheat genotypes were identified as consisting of no mutations in the Pina and Pinb genes (Pina-D1a/Pinb-D1a) or consisting of mutations in either one of the Pin genes but not both (Pina-D1a/Pinb-D1b or Pina-D1b/Pinb-D1a) (Table 1). Based on the absence or presence of mutant Pin alleles, the wheat genotypes were divided into three main groups; a non-mutant group (Pina-D1a/Pinb-D1a) of seventeen genotypes comprising both soft and hard (hard non-mutant, HNM) (genetically soft) wheats, a hard Pina-mutant (HPAM) group (Pina-D1b/Pinb-D1a) of nine hard genotypes, and a hard Pinb-mutant group (HPBM) (Pina-D1a/Pinb-D1b) of seven hard genotypes (Table 1). Within the non-mutant group, the genotypes; India 259, Greece 25, Beyrouth 3, Saturno, Amurskaja 75 and four genotypes from Nepal (NW108A, NW93A, NW25A and NW51A) were found to be hard (HNM) wheats even though these nine genotypes had no mutations in the Pin genes.
Table 1. Grain hardness Index (HI) based on presence of Pin alleles genotypes were grouped into three different groups.
SD—Standard deviation.
| Genotype | AUS Code | Country of origin | SKCS HI | SD + | Classified as S, soft; H, hard wheat | Pin alleles | Group | ||
|---|---|---|---|---|---|---|---|---|---|
| Huandoy | 14423 | PER | 39 | 16 | S | Pina-D1a (wild type) & Pinb-D1a (wild type) | Non- mutant group | ||
| Lerma Rojo | 473 | MEX | 42 | 19 | S | ||||
| D.E.S. 367 | 4385 | GRC | 42 | 15 | S | ||||
| Tunis 24 | 13160 | TUN | 43 | 16 | S | ||||
| Giza 139 | 12957 | EGY | 44 | 14 | S | ||||
| Pelada | 7449 | VEN | 44 | 23 | S | ||||
| Qalbis | 33372 | AUS | 49 | 14 | S | ||||
| India 211 | 15330 | IND | 50 | 12 | S | ||||
| NW108A | 15036 | NPL | 53 | 19 | H | ||||
| India 259 | 4838 | IND | 57 | 16 | H | ||||
| Greece 25 | 4606 | GRC | 57 | 14 | H | ||||
| Saturno | 24431 | MEX | 60 | 25 | H | ||||
| Beyrouth 3 | 4205 | LBN | 61 | 12 | H | ||||
| Amurskaja 75 | 20438 | SUN | 66 | 23 | H | ||||
| NW93A | 15022 | NPL | 74 | 17 | H | ||||
| NW51A | 14996 | NPL | 74 | 16 | H | ||||
| NW25A | 14981 | NPL | 83 | 17 | H | ||||
| Jing Hoang No. 1 | 17863 | CHN | 53 | 20 | H | Pina-D1a (wild type) & Pinb-D1b (mutant) | Pinb-mutant group | ||
| Batavia | 25271 | AUS | 71 | 15 | H | ||||
| EGA Gregory | 34283 | AUS | 71 | 15 | H | ||||
| Sunco | 23455 | AUS | 78 | 16 | H | ||||
| Martonvasari 13T | 24341 | HUN | 82 | 17 | H | ||||
| Banks | 20599 | AUS | 87 | 16 | H | ||||
| Kite | 16035 | AUS | 92 | 16 | H | ||||
| Ellison | 33371 | AUS | 82 | 15 | H | Pina-D1b (mutant) & Pinb-D1a (wild-type) | Pina-mutant group | ||
| Arnhem | 25607 | AUS | 83 | 16 | H | ||||
| India 37 | 4671 | IND | 87 | 13 | H | ||||
| Dollarbird | 23824 | AUS | 87 | 13 | H | ||||
| Punjab 7 | 879 | IND | 88 | 13 | H | ||||
| Gabo | 246 | AUS | 93 | 15 | H | ||||
| Iraq 46 | 28823 | IRAQ | 94 | 15 | H | ||||
| Bowerbird | 30434 | AUS | 96 | 16 | H | ||||
| Bobwhite S-26 | 30252 | MEX | 107 | 17 | H | ||||
Wheat Seeds were sourced from the Australian Winter Cereal Collection (AWCC), Tamworth, Australia.
Differentially expressed genes in hard wheats compared to soft wheats
Gene expression measurements were found to be comparable between replicate experiments for the same genotype (S3 Table). RNA-Seq analysis of expression data of soft wheats and of all hard wheats together i.e. Soft vs HNM+HPAM+HPBM, identified three genes differentially expressed at a FDR corrected value of p<1e-05 (Table 2A). However, the distribution of gene expression values as RPKM, for these three genes, across the two groups of wheat genotypes was similar except for one outlier, implying that these genes may not be significantly associated with grain hardness.
Table 2. Differentially expressed genes identified in the hard wheats when compared to the soft wheats.
2a. Soft vs HNM+HPAM+HPBM, 2b. Soft vs HPAM, 2c. Soft vs HPBM, 2d Soft vs HNM.
| TAGI seq. ID | Fold change (RPKM) | FDR p-value correction | Sequence Description |
|---|---|---|---|
| 2a. Soft vs HNM+HPAM+HPBM Differentially expressed genes in HNM+HPAM+HPBM group when compared to Soft group. | |||
| TC393450 | -15.99 | 2.52E-07 | high molecular weight glutenin subunit |
| TC388873 | -46.47 | 1.66E-06 | 60s ribosomal protein l23 |
| CV066856 | -35.82 | 3.23E-05 | ---NA--- |
| 2b. Soft VS HPAM Differentially expressed genes in HPAM group when compared to Soft group. | |||
| TC423373 | -1,768.05 | 3.60E-102 | puroindoline-a |
| TC434025 | -82.76 | 3.04E-15 | alpha- partial |
| TC393944 | -469.84 | 3.59E-15 | alpha- partial |
| TC401139 | -100.66 | 1.16E-08 | calreticulin interacted protein |
| NP9350187 | 248.04 | 1.72E-07 | low molecular weight glutenin |
| BE413821 | 124.96 | 2.33E-06 | alpha- partial |
| TC448640 | -135.24 | 3.42E-06 | ---NA--- |
| TC448029 | -49.14 | 7.01E-06 | proteasome subunit beta type-2-like |
| BJ221811 | 11.73 | 8.92E-06 | disease resistance protein rpm1 |
| BE398961 | -76.85 | 1.97E-06 | ---NA--- |
| 2c. Soft VS HPBM Differentially expressed genes in HPBM group when compared to Soft group. | |||
| CA602991 | 124.46 | 7.16E-10 | inactive poly |
| TC400917 | -100.24 | 8.05E-07 | histone h4 |
| TC372980 | -27.52 | 2.13E-06 | 40s ribosomal protein s12 |
| TC420043 | 3608.9 | 2.56E-05 | hypothetical protein TRIUR3_06809 |
| 2d. Soft VS HNM Differentially expressed genes in HNM group when compared to Soft group. | |||
| TC420856 | 98.71 | 9.31E-14 | ---NA--- |
| TC372034 | 71.66 | 5.22E-13 | eh domain-containing protein 1-like |
| CA636509 | 58.21 | 1.50E-06 | glycosyltransferase |
| TC427661 | 20.08 | 1.80E-06 | ---NA--- |
TAGI sequence ID = Triticum Aestivum Gene Indices sequence ID; RPKM = reads per kilobase per million; NA = not available; FDR = false discovery rate; HNM = hard non-mutant; HPAM = hard Pina-mutant; HPBM = hard Pinb-mutant.
Differentially expressed genes in hard Pina mutant (HPAM) wheats compared to soft wheats
When the HPAM group was compared with the soft group, 10 differentially expressed genes (DEGs) were identified (Table 2B). As expected, Pina was the most highly differentially expressed gene. The significance value and fold change value of no other gene was even close to that of Pina. In addition, none of the 10 DEGs identified in this analysis were common with DEGs identified in comparison of the soft group with the HNM+HPAM+HPBM group.
Differentially expressed genes in hard Pinb mutant wheats (HPBM) compared to soft wheats
When the HPBM group was compared with the soft group, DEGs were identified (Table 2C). Pinb (Pinb-D1b) was not identified as a DEG even up to a significance value at FDR corrected p-value of 0.01. The DEGs identified in this analysis were different to those identified in the soft vs HNM+HPAM+HPBM and the soft vs HPAM groups.
Differentially expressed in hard non-mutants (HNM) compared to soft, HPAM and HPBM groups
In the HNM group, grain hardness was observed but without mutation in either the Pina or the Pinb gene. To identify genes with expression contributing to grain hardness in the absence of the Pina/b mutation we compared the HNM group with other non-mutant wheats which are soft; soft vs HNM. In this comparison 4 DEGs were identified (Table 2D) which were different to the DEGs observed in the three earlier comparisons. Pin genes were not among the differentially expressed genes even at a FDR p-value up to 0.01.
The HNM group was also compared separately with the HPAM and the HPBM group, where 180 (173 up-regulated in HNM), 26 (23 up-regulated in HNM) DEGs were identified, respectively (S1 Table and Table 3). Among these two sets of DEGs, 9 DEGs were common and were up-regulated in HNM group (S1 Table). HNM vs HPAM and HNM vs HPBM comparisons had no DEGs in common with the SNM vs HNM comparison.
Table 3. Top down-regulated and up-regulated differentially expressed genes identified in HPAM (3a) and HPBM (3b) when compared to HNM group.
| TAGI seq. ID | Fold change (RPKM) | FDR p-value correction | Sequence Description |
|---|---|---|---|
| 3a. HNM vs HPAM Differentially expressed genes in HPAM group when compared to HNM group. | |||
| TC423373 | -1310.78 | 1.25E-43 | puroindoline-a |
| CD915505 | 902.62 | 9.63E-19 | low-molecular-weight glutenin subunit |
| NP9350187 | 166.51 | 5.41E-07 | low molecular weight glutenin |
| CJ520220 | -843.82 | 6.98E-07 | ---NA--- |
| BE413821 | 117.62 | 1.07E-06 | alpha- partial |
| TC393944 | -526.26 | 1.14E-06 | alpha- partial |
| TC458604 | -193.09 | 2.45E-06 | ---NA--- |
| TC393980 | 40.28 | 2.46E-06 | low-molecular-weight glutenin subunit |
| TC391918 | 28.3 | 6.57E-06 | formate dehydrogenase mitochondrial-like |
| TC412255 | -152.77 | 9.59E-06 | ---NA--- |
| 3b. HNM vs HPBM Differentially expressed genes in HPBM group when compared to HNM group. | |||
| CK194194 | -78.27 | 3.13E-17 | fructose-bisphosphate aldolase |
| CA602991 | 85.03 | 6.94E-08 | inactive poly |
| TC450096 | -66.92 | 8.68E-07 | ---NA--- |
| TC375635 | 11.6 | 1.91E-06 | dihydrolipoamide s-acetyltransferase |
| CK162779 | -233.06 | 2.66E-06 | lrr receptor-like serine threonine-protein kinase fls2 |
| CD897082 | -62.56 | 4.13E-06 | ---NA--- |
| TC457676 | -180.99 | 4.15E-06 | ---NA--- |
| TC420456 | 6.94 | 6.2E-06 | ac078948_18 serine protease |
TAGI sequence ID = Triticum Aestivum Gene Indices sequence ID; RPKM = reads per kilobase per million; NA = not available; FDR = false discovery rate; HNM = hard non-mutant; HPAM = hard Pina-mutant; HPBM = hard Pinb-mutant.
Differentially expressed genes in Pin mutant wheats (HPAM & HPBM) compared to non-mutants (soft & HNM)
When all the Non-mutant (soft and HNM) wheats were compared to HPAM and HPBM groups together, 345 differentially expressed genes were identified (S2 Table) and of these 337 DEGs were up-regulated in non-mutants. Comparison of these 345 DEGs with DEGs from soft VS HPAM, soft vs HPBM, HNM vs HPAM and HNM vs HPBM comparisons identified 2, 3, 128 and 15 common genes, respectively.
Differential expression of Pina, Pinb and GSP-1 ABD and Pinb-2 ABD at two stages of seed development
Differential expression of Pina, Pinb, GSP-1A, -1B, -1D and Pinb-2A, -2B, -2D for all genotypes at 14 DPA and 30 DPA is shown in Figs 1–3. The pattern but not the proportion of differential gene expression of Pina, Pinb, GSP-1 ABD and Pinb-2 ABD was roughly the same at both time points for most genotypes.
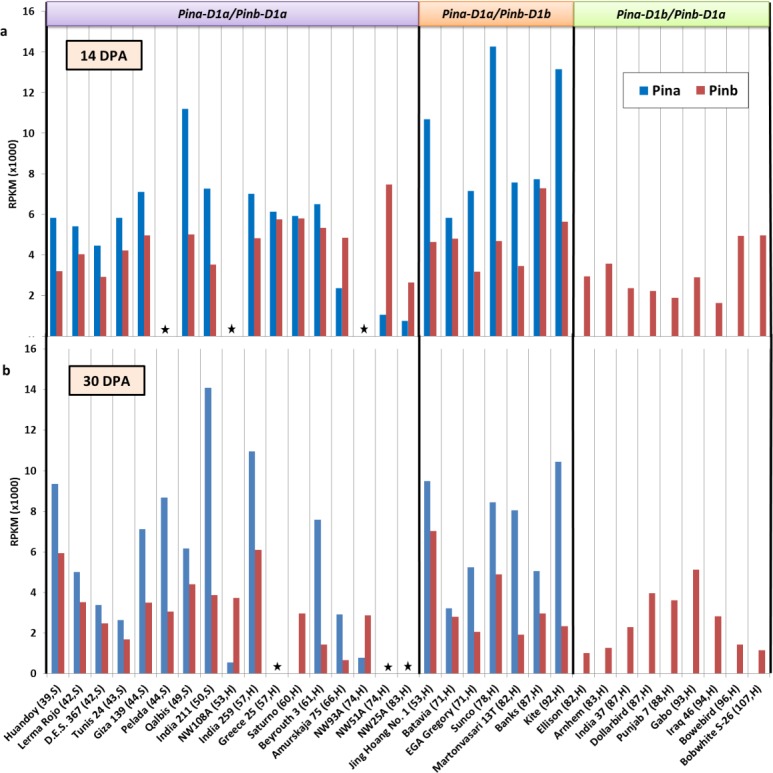
Fig 1. Expression of the Pina and Pinb genes in developing seeds of several wheat genotypes.
a,b gene expression data at 14 and 30 days post anthesis (dpa), respectively; RPKM, reads per kilo base per million mapped reads. Details in brackets after genotype names on the X-axis indicates grain hardness index and endosperm texture of genotypes classified as hard (H) or soft (S) wheats. Genotypes with HI above 50 were classified as Hard. Wheat genotypes in each group are arranged left to right with respect to increasing hardness index. Boxes on the top indicate Pin alleles present in the genotypes. cDNA prepared from RNA extracted from developing wheat seeds at 14 DPA, was subjected to next generation sequencing. RNA-seq analysis was undertaken using CLC Genomic Workbench V8 to determine gene expression of Pina and Pinb. The star symbol indicates data not available.
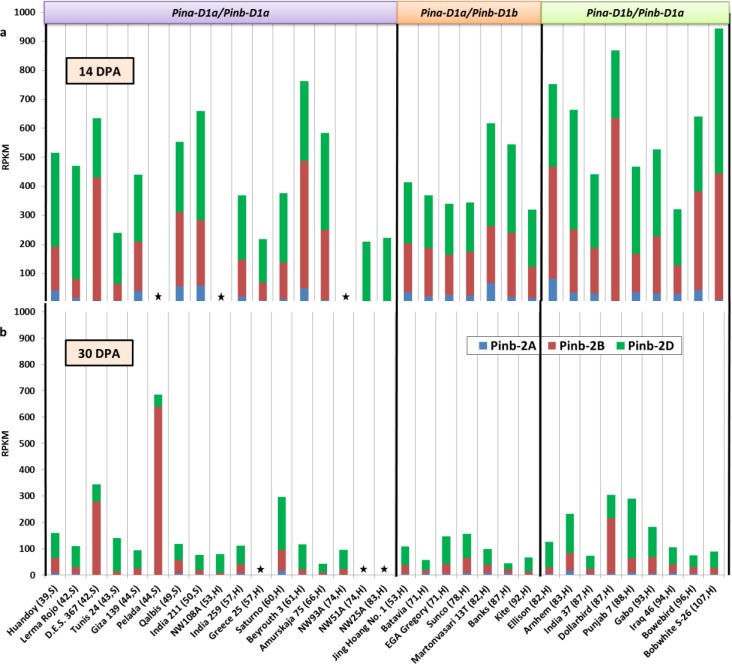
Fig 3. Expression of the Pinb-2A, -2B, -2D genes in developing seeds of several wheat genotypes.
a, b gene expression data at 14 and 30 days post anthesis (dpa) respectively; RPKM, reads per kilo base per million mapped reads. Boxes on the top indicate Pin alleles present in the genotypes. Details in brackets after genotype names on the X-axis indicates grain hardness index and endosperm texture of genotypes classified as hard (H) or soft (S) wheats. The asterisk indicates data not available.
In the non-mutant group, Pina (Pina-D1a) expression was found to be generally higher than Pinb expression (Fig 1). However, within the non-mutant group a specific pattern of Pin gene expression was observed at 14 DPA or 30 DPA in the hard wheat genotypes Amurskaja 75, NW25A, NW51A, NW93A and NW108A. The pattern of Pin gene expression when compared to the other genotypes in the non-mutant group reflected the reduced levels of Pina expression and in addition the increased level of Pinb expression when compared to Pina expression. Genotypes with increasing hardness in this group did not show an observable reduction in Pinb expression. However, reduced Pina expression was observed in those genotypes, as outlined above, with the highest hardness index in this group.
In the Pinb-mutant group, Pina (Pina-D1a) expression was found to be higher than Pinb expression (Fig 1). Genotypes with increasing hardness in this group did not show an observable reduction in the expression levels of Pina or Pinb-D1b.
In the Pina-mutant group, expression of Pina was not detected as expected, but the expression of Pinb was lower compared to the non-mutant and the Pinb-mutant group. Genotypes with increased hardness did not show an observable correlation with Pinb expression levels (Fig 1).
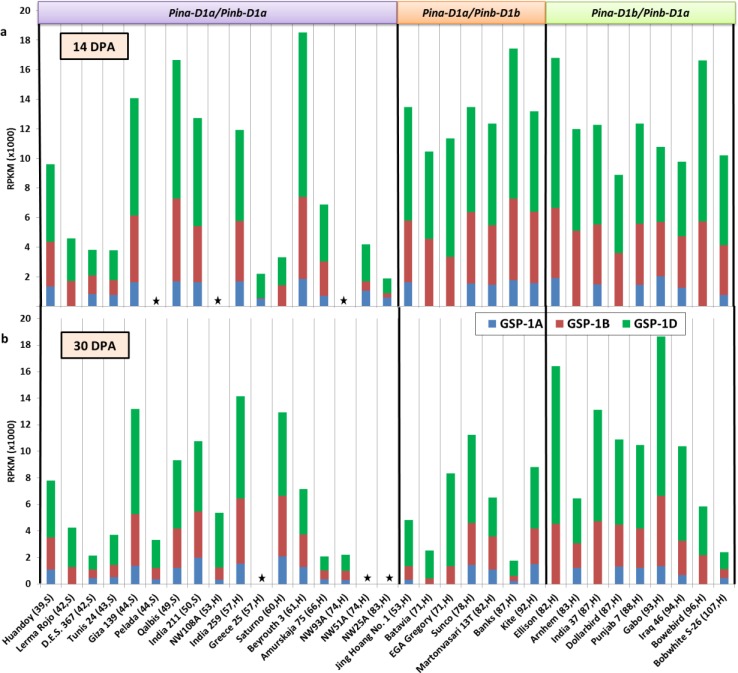
In case of GSP-1A, -1B, -1D and Pinb-2A, -2B, -2D, the highest expression was contributed by the D sub-genome allele followed by the B sub-genome allele and then the A sub-genome allele (Figs 2 and 3).
Fig 2. Expression of the GSP-1A, -1B, -1D genes in developing seeds of several wheat genotypes.
a, b gene expression data at 14 and 30 days post anthesis (DPA) respectively; RPKM, reads per kilo base per million mapped reads. Boxes on the top indicate Pin alleles present in the genotypes. Details in brackets after genotype names on the X-axis indicates grain hardness index and endosperm texture of genotypes classified as hard (H) or soft (S) wheats. The asterisk indicates data not available.
Total gene expression of Pina, Pinb and GSP-1 ABD and Pinb-2 ABD at two stages of seed development
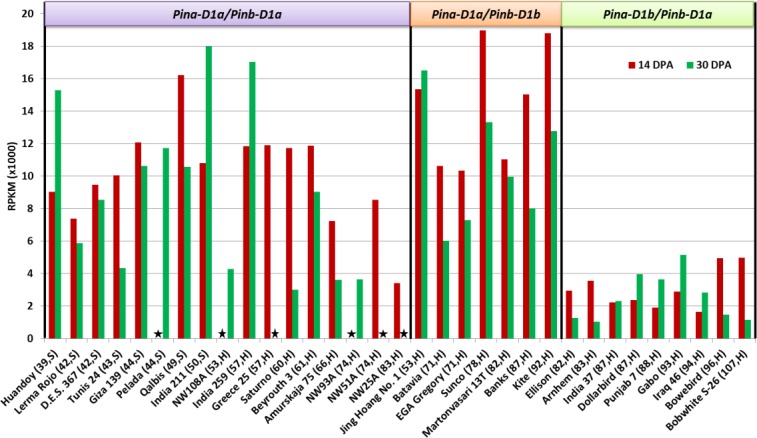
Total Pin gene expression in developing seeds at 14 and 30 DPA (Fig 4) was generally higher in most genotypes at 14 DPA than at 30 DPA. The total Pin gene expression, within each of the Pin-groups, showed no observable correlation with increasing SKCS-HI or with genotypes found to be soft or hard. In the Pina-mutant group, total Pin gene expression was much lower when compared to the non-mutant group or the Pinb-mutant group.
Fig 4. Total expression of Pin genes (Pina + Pinb) in developing wheat seeds of several wheat genotypes at 14- and 30-days post anthesis.
Details in brackets after genotype names on the X-axis indicates grain hardness index and endosperm texture of genotypes classified as hard (H) or soft (S) wheats. Grain hardness was measured by SKCS analysis and genotypes with HI above 50 were labelled as hard. Details in brackets after genotype names on the X-axis indicates grain hardness index and endosperm texture of genotypes classified as hard (H) or soft (S) wheats. Boxes on the top indicate Pin alleles present in the genotypes. Transcriptome analysis by next generation sequencing data was undertaken to determine total Pin gene expression which was then normalised and expressed as reads per kilo base per million mapped reads (RPKM). The asterisk indicates data not available.
Total GSP-1 expression was also observed to be slightly reduced at 30 DPA (Fig 2). Total Pinb-2 expression at 30 DPA was substantially reduced as compared to 14 DPA (Fig 3). Analysis showed a higher expression of GSP-1 in Pina- and Pinb- mutant genotypes than in non-mutant genotypes but no such pattern was observed for Pinb-2.
Statistical analysis of associations with hardness
The three groups of wheat genotypes, non-mutants (Soft and HNM), HPAM and HPBM, were analysed separately and together to find out which of the Pin, Pinb-2 or the GSP-1 genes had the greatest impact on SKCS HI. In a polynomial regression model which included all the genotypes and the individual genes, Pina, Pinb, GSP-1 ABD and Pinb-2 ABD, our data suggests that the expression of Pina + Pinb + GSP-1B + Pinb-2D explained 62% of the variation at 14 DPA with a p-value of 0.0001 (Table 4). The correlation between Pina gene expression and SKCS HI was the highest among all genes in a group containing all genotypes (Table 4). The combined or individual effects of GSP-1 ABD and Pinb-2 ABD alleles did not show significant correlation with SKCS HI in any group. Within the non-mutant group, an interactive effect of Pina x Pinb explained 80% (p-value 0.01) of the variation among SKCS HI.
Table 4. Association between Pina, Pinb and GSP-1 ABD and Pinb-2 ABD gene expression measurements and SKCS hardness index, at 14 DPA.
No significant association was observed at 30 DPA. Genotypes were grouped into four groups for statistical analysis; all genotypes, non-mutants, Pina-mutants and Pinb-mutants. Additive and interactive effects of gene expression were analysed using linear and polynomial regression model. Only significant associations are listed.
| Group | Genes | R2 value | P value |
|---|---|---|---|
| Allgenotypes | Pina+Pinb+GSP-1A+Pinb-2A # | 0.39 | 0.0007 |
| Pina+Pinb+GSP-1B+Pinb-2D * | 0.62 | 0.0001 | |
| Pina +Pinb * | 0.56 | 2.562e-05 | |
| Pina * | 0.53 | 7.808e-06 | |
| Pina X Pinb * | 0.57 | 0.00038 | |
| Non-mutant | Pina X Pinb # | 0.60 | 0.005 |
| Pina + Pinb * | 0.57 | 0.01 | |
| Pina X Pinb * | 0.81 | 0.01 | |
| Pina * | 0.53 | 0.005 |
# Linear regression
* Polynomial regression, + Additive effect of gene expression, X Interactive effect of gene expression
Discussion
In this study we have addressed two questions; firstly, which genes are differentially expressed in hard wheats compared to soft wheats and secondly, does differential expression of Pina and Pinb genes explain variation in wheat grain hardness.
The TAGI reference dataset was used for RNA-seq analysis in this study as it contains more genes compared to the IWGSC reference dataset. Recently, a gene responsible for good bread making, wheat bread making gene, was discovered using the TAGI database[39]. This gene is not present in the IWGSC database. The IWGSC survey sequence database is in the process of completion and is updated frequently by addition of new genes. The majority of grain hardness is explained by puroindolines, however, some variation still remains unexplained possibly due to the involvement of other genes playing a minor role in the control of grain hardness. A number of QTLs have been shown to be associated with grain hardness, on chromosome 4A, 4B [40], 1A, 2B, 2D, 3B, 7A, 7B [41], 6A [42], 1B [43]. However, no genes other than the Pin genes have been associated with grain hardness. We hypothesised that genes other than Pin genes that control grain hardness may be expressed differentially between the soft and the hard wheats. We identified these DEGs by analysing the transcriptome of several soft and hard wheat genotypes. Four groups of wheats; soft, HNM, HPAM and HPBM were compared with each other in different combinations to identify differentially expressed genes. This provided the group specific and the common DEGs within different groups.
The most significant differential expression of Pina in comparisons between soft vs HPAM and HNM vs HPAM supports the vital role of Pina in controlling grain hardness. However, Pina or Pinb was not identified as a differentially expressed gene in comparison of the soft vs HNM group. This indicates that although grain hardness is greatly influenced by mutation in the Pin genes, other factors may also be involved in control of this trait. In the genes which were differentially expressed between soft+HNM group and HPAM+HPBM group it was observed that most of the genes were down-regulated in hard wheats (HPAM+HPBM). This down-regulation of gene expression could be the influence of grain hardness or it may contribute to the grain hardness. No annotation was available for some of the significant DEGs. None of the other annotated DEGs identified in different comparisons has been previously reported to be associated with wheat grain hardness. Further study is required to determine the role of candidate genes identified in this study. Up-regulation of glutenins and gliadins (Table 2 and S1 Table) in the hard wheats is most likely a mere coincidence as the hard wheats have been selected for higher gluten content for bread-making. The differential expression of these genes may be because of human selection for different wheat quality traits in hard and soft wheats rather than their association with the determination of hardness.
Pinb-2 variants and GSP-1 have been suggested as possible candidates for control of grain hardness [7, 31]. Results of this study indicate that the GSP-1 and the Pinb-2 do not have a significant influence on grain hardness on their own as has been reported in other studies [7, 19, 33]. However, the possibility of a combined influence of the Pinb-2, GSP-1 and Pin genes in control of grain hardness is likely as the expression of Pina+Pinb+GSP-1B+Pinb-2D explained 62% of the variation in grain hardness whereas Pina+Pinb explained 56% across all genotypes in this study.
Pina and the Pinb still remain the only major genes to influence grain hardness. In the non-mutant group, the additive effect of Pina+Pinb expression explained 60% of the variation in grain hardness whereas the interactive effect explained 80%. However, the individual effect of Pina expression was the highest (53%) (Table 4). Higher expression of Pina rather than Pinb was one of the most consistent patterns of gene expression observed in these genotypes, except for the case of the Pina-mutants (Fig 1). This observation has been reported in earlier studies [12, 13, 27, 28]. At the protein level, a higher amount of PINA has been suggested to be associated with increased starch bound PIN, which is supposedly associated with increased grain softness [44]. We found some wheat genotypes with a hard grain texture without the presence of any mutant-Pina or -Pinb alleles. This is a very significant observation which provides an exception to the widely accepted view that wheats with wild-type Pin alleles display soft grain texture but a mutation in any of the Pin alleles leads to hard grain texture. Among the nine genotypes with hard grain texture within the non-mutant group, five genotypes showed a distinct pattern of the Pina and the Pinb gene expression at either 14 or 30 DPA (Fig 4). Generally, the Pina transcripts are expressed at a higher level than the Pinb but in these genotypes we found the opposite, with the Pinb expression levels exceeding the Pina expression levels. This result provides strong evidence that reduced expression of Pina and Pinb alleles, and not just protein structure, plays a critical role in determining grain hardness. In agreement with these results Swan, Meyer [45] reported that grain softness is explained better by the proportional amounts of the PINA and the PINB proteins rather than the total PIN content [45]. Hard grain texture in genotypes containing wild type Pina and Pinb has been reported earlier in Australian cultivars Cook and Diaz, with a PINA:PINB ratio of about 2:1 (ELISA test) [27]. However, gene expression wasn’t examined in that study.
A slightly lower Pinb expression was observed in Pina-mutants (HPAM) compared to other groups (Fig 1). Likewise, a lower amount of the PINB protein has been reported in wheat genotypes with a Pina null mutation [27]. Amoroso, Longobardo [28] suggested that PINB is expressed in Pina null genotypes but is later degraded due to the absence of the PINA which was suggested to stabilise PINB. In contrast, Wanjugi, Hogg [46] reported that PINA and PINB can act independently and produce intermediate grain texture but are more effective in producing soft grains when acting together. A transgenic study by Hogg, Sripo [13] showed that hard red spring wheat (Hi-line) with a mutant Pinb allele (Pina-D1a/Pinb-D1b), when transformed with wild type Pin-a or–b alleles produces soft grains. However, lines transformed with Pinb-D1a were softer than the lines transformed with Pina-D1a and in addition there was no correlation with levels of PIN expression and hardness. This demonstrates that excessive expression of one wild type Pin allele does not compensate for another mutant Pin allele and also indicated that PINA and PINB act together. Comparable levels of Pina and Pinb expression in some non-mutants and Pinb-mutants suggests that mutation in the Pin-b gene does not necessarily alter the expression levels of Pin genes. A study by Gasparis, Orczyk [23] using RNAi mediated silencing of Pin genes also reported that puroindoline transcript abundance or protein content is not altered by mutation. However, mutation may interfere with PIN protein functionality and stability. Greater association of Pin transcripts at 14 DPA with grain hardness is most likely due to the abundance of transcripts whereas less association at 30 DPA is most likely due to reduced gene expression. The total gene expression process starts to slow down as the seed proceeds towards maturity [47].
Study of 5’ and the 3’ sequences of the Pin genes may explain differences in expression levels of Pin genes. Mutations in these regions are likely to affect gene expression levels. Further understanding of grain hardness may allow selection of specific levels of gene expression to breed for specific level of grain hardness and not just for broader classes of hardness. A more complete understanding of the regulatory sequences at these loci may allow a more complete explanation of genetic variation in hardness in wheat and allow reliable selection for hardness based upon sequence specific markers.
Conclusions
Results of this study show that grain hardness in some wheat genotypes containing wild type Pin alleles is explained by higher expression of Pinb than Pina. Puroindoline genes still remain the major determinants of grain hardness. Expression of Pin genes at earlier stages of grain development determines grain hardness. Variation in grain hardness (SKCS HI) among the different genotypes within a particular class of hardness couldn’t be explained by patterns of Pina and Pinb expression. Several statistically significant differentially expressed genes were identified between soft wheats and hard wheats. Further investigation of these genes may provide more understanding of wheat grain hardness.
Supporting Information
Differentiallyexpressed genes identified between the HNM group and the HPAM group (1a), the HPBM group (1b) and common DEGs (1c).
(DOC)
(DOC)
Duplicates were grown in a glasshouse and analysed to exmine reprodctibility of expression patterns.
(DOC)
Acknowledgments
We thank Australian Research Council and Green Gold Seeds Ltd., Maharashtra, India for financial support.
Data Availability
We have uploaded minimal data set (list of all differentially expressed genes identified) underlying the findings in our study in the paper and supplementary files.
Funding Statement
Australian Research Council and Green Gold Seeds Ltd., Maharashtra, India provided funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Symes K. The inheritance of grain hardness in wheat as measured by the particle size index. Aust J Agr Res.1965;16(2):113–23. 10.1071/AR9650113. [DOI] [Google Scholar]
- 2.Igrejas G, Gaborit T, Oury F-X, Chiron H, Marion D, Branlard G. Genetic and Environmental Effects on Puroindoline-a and Puroindoline-b Content and their Relationship to Technological Properties in French Bread Wheats. J Cereal Sci. 2001;34(1):37–47. 10.1006/jcrs.2000.0381. [DOI] [Google Scholar]
- 3.Gazza L, Taddei F, Corbellini M, Cacciatori P, Pogna NE. Genetic and environmental factors affecting grain texture in common wheat. J Cereal Sci. 2008;47(1):52–58. 10.1016/j.jcs.2007.01.004 [DOI] [Google Scholar]
- 4.Turnbull KM, Turner M, Mukai Y, Yamamoto M, Morell MK, Appels R, et al. The organization of genes tightly linked to the Ha locus in Aegilops tauschii, the D-genome donor to wheat. Genome. 2003;46(2):330–338. 10.1139/g02-124 [DOI] [PubMed] [Google Scholar]
- 5.Doekes GJ, Belderok B. Kernel hardness and baking quality of wheat—A genetic analysis using chromosome substitution lines. Euphytica. 1976;25(1):565–576. 10.1007/BF00041594 [DOI] [Google Scholar]
- 6.Morris CF, DeMacon VL, Giroux MJ. Wheat Grain Hardness Among Chromosome 5D Homozygous Recombinant Substitution Lines Using Different Methods of Measurement. Cereal Chem. 1999;76(2):249–254. 10.1094/CCHEM.1999.76.2.249 [DOI] [Google Scholar]
- 7.Tranquilli G, Heaton J, Chicaiza O, Dubcovsky J. Substitutions and Deletions of Genes Related to Grain Hardness in Wheat and Their Effect on Grain Texture. Crop Sci. 2002;42(6):1812–1817. 10.2135/cropsci2002.1812 [DOI] [Google Scholar]
- 8.Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell. 2005;17(4):1033–1045. 10.1105/tpc.104.029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhave M, Morris C. Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol. 2008;66(3):205–219. 10.1007/s11103-007-9263-7 [DOI] [PubMed] [Google Scholar]
- 10.Li G, He Z, Lillemo M, Sun Q, Xia X. Molecular characterization of allelic variations at Pina and Pinb loci in Shandong wheat landraces, historical and current cultivars. J Cereal Sci. 2008;47(3):510–517. 10.1016/j.jcs.2007.06.003. [DOI] [Google Scholar]
- 11.Lillemo M, Chen F, Xia X, William M, Peña RJ, Trethowan R, et al. Puroindoline grain hardness alleles in CIMMYT bread wheat germplasm. J Cereal Sci. 2006;44(1):86–92. 10.1016/j.jcs.2006.03.004. [DOI] [Google Scholar]
- 12.Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P. Triticum aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol. 1994;25(1):43–57. [DOI] [PubMed] [Google Scholar]
- 13.Hogg AC, Sripo T, Beecher B, Martin JM, Giroux MJ. Wheat puroindolines interact to form friabilin and control wheat grain hardness. Theor Appl Genet. 2004;108(6):1089–1097. 10.1007/s00122-003-1518-3 [DOI] [PubMed] [Google Scholar]
- 14.Krishnamurthy K, Giroux MJ. Expression of wheat puroindoline genes in transgenic rice enhances grain softness. Nat Biotechnol. 2001;19(2):162–166. 10.1038/84435 [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Martin JM, Beecher B, Morris CF, Curtis Hannah L, Giroux MJ. Seed-specific expression of the wheat puroindoline genes improves maize wet milling yields. Plant Biotechnol J. 2009;7(8):733–743. 10.1111/j.1467-7652.2009.00438.x [DOI] [PubMed] [Google Scholar]
- 16.Oda S, Schofield JD. Characterisation of Friabilin Polypeptides. J Cereal Sci. 1997;26(1):29–36. [Google Scholar]
- 17.Clifton LA, Green RJ, Frazier RA. Puroindoline-b Mutations Control the Lipid Binding Interactions in Mixed Puroindoline-a:Puroindoline-b Systems†. Biochemistry. 2007;46(48):13929–13937. 10.1021/bi701680w [DOI] [PubMed] [Google Scholar]
- 18.Greenwell P, Schofield JD. A starch granule protein associated with endosperm softness in wheat. Cereal Chem. 1986;63:379–380. [Google Scholar]
- 19.Giroux MJ, Morris CF. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci USA. 1998;95(11):6262–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day L, Bhandari DG, Greenwell P, Leonard SA, Schofield JD. Characterization of wheat puroindoline proteins. FEBS Journal. 2006;273(23):5358–5373. 10.1111/j.1742-4658.2006.05528.x [DOI] [PubMed] [Google Scholar]
- 21.Clifton LA, Sanders MR, Castelletto V, Rogers SE, Heenan RK, Neylon C, et al. Puroindoline-a, a lipid binding protein from common wheat, spontaneously forms prolate protein micelles in solution. Phys Chem Chem Phys. 2011;13(19):8881–8888. 10.1039/c0cp02247k [DOI] [PubMed] [Google Scholar]
- 22.Alfred RL, Palombo EA, Panozzo JF, Bhave M. The Antimicrobial Domains of Wheat Puroindolines Are Cell-Penetrating Peptides with Possible Intracellular Mechanisms of Action. PLoS ONE. 2013;8(10):e75488 10.1371/journal.pone.0075488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasparis S, Orczyk W, Zalewski W, Nadolska-Orczyk A. The RNA-mediated silencing of one of the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other, and increases grain hardness. J Exp Bot. 2011;62(11):4025–4036. 10.1093/jxb/err103 [DOI] [PubMed] [Google Scholar]
- 24.Turnbull KM, Marion D, Gaborit T, Appels R, Rahman S. Early expression of grain hardness in the developing wheat endosperm. Planta. 2003;216(4):699–706. 10.1007/s00425-002-0911-5 [DOI] [PubMed] [Google Scholar]
- 25.Clarke B, Rahman S. A microarray analysis of wheat grain hardness. Theor. Appl. Genet. 2005;110(7):1259–1267. 10.1007/s00122-005-1962-3 [DOI] [PubMed] [Google Scholar]
- 26.Drea S, Leader DJ, Arnold BC, Shaw P, Dolan L, Doonan JH. Systematic spatial analysis of gene expression during wheat caryopsis development. Plant Cell. 2005;17(8):2172–2185. 10.1105/tpc.105.034058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbull K-M, Gaborit T, Marion D, Rahman S. Variation in puroindoline polypeptides in Australian wheat cultivars in relation to grain hardness. Funct Plant Biol. 2000;27(2):153–158. 10.1071/PP99061. [DOI] [Google Scholar]
- 28.Amoroso MG, Longobardo L, Capparelli R. Real Time RT-PCR and flow cytometry to investigate wheat kernel hardness: role of puroindoline genes and proteins. Biotechnol Lett. 2004;26(22):1731–1737. 10.1007/s10529-004-3745-3 [DOI] [PubMed] [Google Scholar]
- 29.Giroux MJ, Morris CF. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor Appl Genet. 1997;95(5–6):857–864. 10.1007/s001220050636 [DOI] [Google Scholar]
- 30.Chen F, Beecher B, Morris C. Physical mapping and a new variant of Puroindoline b-2 genes in wheat. Theor Appl Genet. 2010;120(4):745–751. 10.1007/s00122-009-1195-y [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson M, Wan Y, Tosi P, Leverington M, Snape J, Mitchell RAC, et al. Identification and genetic mapping of variant forms of puroindoline b expressed in developing wheat grain. J Cereal Sci. 2008;48(3):722–728. 10.1016/j.jcs.2008.03.007. [DOI] [Google Scholar]
- 32.Ramalingam A, Palombo EA, Bhave M. The Pinb-2 genes in wheat comprise a multigene family with great sequence diversity and important variants. J Cereal Sci. 2012;56(2):171–180. 10.1016/j.jcs.2012.02.006. [DOI] [Google Scholar]
- 33.Giroux MJ, Kim K-H, Hogg AC, Martin JM, Beecher B. The Puroindoline b-2 Variants are Expressed at Low Levels Relative to the Puroindoline D1 Genes in Wheat Seeds. Crop Sci. 2013;53(3):833–841. 10.2135/cropsci2012.06.0359 [DOI] [Google Scholar]
- 34.Furtado A. DNA Extraction from Vegetative Tissue for Next-Generation Sequencing In: Henry JR, Furtado A, editors. Cereal Genomics: Methods and Protocols. Totowa, NJ: Humana Press; 2014. p. 1–5. [DOI] [PubMed] [Google Scholar]
- 35.Conesa A, Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008. 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maghirang EB, Dowell FE. Hardness Measurement of Bulk Wheat by Single-Kernel Visible and Near-Infrared Reflectance Spectroscopy. Cereal Chem. 2003;80(3):316–322. 10.1094/CCHEM.2003.80.3.316 [DOI] [Google Scholar]
- 37.Beecher B, Bettge A, Smidansky E, Giroux M. Expression of wild-type pinB sequence in transgenic wheat complements a hard phenotype. Theor Appl Genet. 2002;105(6–7):870–877. 10.1007/s00122-002-1034-x [DOI] [PubMed] [Google Scholar]
- 38.Gaines CS, Finney PF, Fleegee LM, Andrews LC. Predicting a Hardness Measurement Using the Single-Kernel Characterization System. Cereal Chem. 1996;73(2):278–283. [Google Scholar]
- 39.Furtado A, Bundock PC, Banks PM, Fox G, Yin X, Henry RJ. A novel highly differentially expressed gene in wheat endosperm associated with bread quality. Scientific reports. 2015;5:10446 10.1038/srep10446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne BG, Turnbull KM, Anderssen RS, Rahman S, Sharp PJ, Appels R. The hardness locus in Australian wheat lines. Aus J Agr Res. 2001;52(12):1275–1286. 10.1071/AR01056. [DOI] [Google Scholar]
- 41.Campbell KG, Bergman CJ, Gualberto DG, Anderson JA, Giroux MJ, Hareland G, et al. Quantitative Trait Loci Associated with Kernel Traits in a Soft × Hard Wheat Cross. Crop Sci. 1999;39(4):1184–1195. 10.2135/cropsci1999.0011183X003900040039x [DOI] [Google Scholar]
- 42.Carter AH, Garland-Campbell K, Morris CF, Kidwell KK. Chromosomes 3B and 4D are associated with several milling and baking quality traits in a soft white spring wheat (Triticum aestivum L.) population. Theor Appl Genet. 2012;124(6):1079–1096. 10.1007/s00122-011-1770-x [DOI] [PubMed] [Google Scholar]
- 43.Breseghello F, Finney PL, Gaines C, Andrews L, et al. Genetic Loci Related to Kernel Quality Differences between a Soft and a Hard Wheat Cultivar. Crop Sci. 2005;45(5):1685–1695. [Google Scholar]
- 44.Capparelli R, Borriello G, Giroux MJ, Amoroso MG. Puroindoline A-gene expression is involved in association of puroindolines to starch. Theor Appl Genet. 2003;107(8):1463–1468. 10.1007/s00122-003-1392-z [DOI] [PubMed] [Google Scholar]
- 45.Swan CG, Meyer FD, Hogg AC, Martin JM, Giroux MJ. Puroindoline B Limits Binding of Puroindoline A to Starch and Grain Softness. Crop Sci. 2006;46(4):1656–1665. 10.2135/cropsci2005.06-0135 [DOI] [Google Scholar]
- 46.Wanjugi HW, Hogg AC, Martin JM, Giroux MJ. The Role of Puroindoline A and B Individually and in Combination on Grain Hardness and Starch Association. Crop Sci. 2007;47(1):67–76. 10.2135/cropsci2006.05.0310 [DOI] [Google Scholar]
- 47.McIntosh S, Watson L, Bundock P, Crawford A, White J, Cordeiro G, et al. SAGE of the developing wheat caryopsis. Plant Biotechnol J. 2007;5(1):69–83. 10.1111/j.1467-7652.2006.00218.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentiallyexpressed genes identified between the HNM group and the HPAM group (1a), the HPBM group (1b) and common DEGs (1c).
(DOC)
(DOC)
Duplicates were grown in a glasshouse and analysed to exmine reprodctibility of expression patterns.
(DOC)
Data Availability Statement
We have uploaded minimal data set (list of all differentially expressed genes identified) underlying the findings in our study in the paper and supplementary files.