Abstract
IMPORTANCE
Oxaliplatin added to fluorouracil plus leucovorin therapy for patients with colon cancer has been shown to provide significant but modest absolute benefit for disease-free survival. However, acute and chronic neurotoxic effects from this regimen underscore the need for markers that predict oxaliplatin benefit.
OBJECTIVE
To test our hypothesis that molecular subtypes of colon cancer would be associated with differential prognosis and benefit from oxaliplatin added to fluorouracil plus leucovorin therapy.
DESIGN, SETTING, AND PARTICIPANTS
Participants in the NSABP C-07 trial were divided into discovery (n = 848) and validation (n = 881) cohorts based on the order of tissue block submission. A reestimated centroid using 72 genes was used to determine Colorectal Cancer Assigner subtypes and their association with oxaliplatin benefit in the discovery cohort. The validation cohort was examined with a locked-down algorithm for subtype classification and statistical analysis plan. Post hoc analysis included examination of the entire cohort with Colorectal Cancer Assigner, Colorectal Cancer Subtype (CCS), and Consensus Molecular Subtype (CMS) methods.
INTERVENTIONS
Fluorouracil plus leucovorin with or without oxaliplatin.
MAIN OUTCOMES AND MEASURES
Percent recurrence-free survival.
RESULTS
Among 1729 patients, 744 (43%) were female and mean (SD) age was 58 (11) years. Although C-07 participants with stage III disease with an enterocyte subtype showed a statistically significant benefit from oxaliplatin in the discovery cohort (hazard ratio, 0.22 [95% CI, 0.09–0.56]; P = .001 [N = 65]), no statistically significant benefit was observed in the validation cohort (hazard ratio, 0.53 [95% CI, 0.22–1.24]; P = .14 [N = 70]). The stemlike subtype was associated with poor prognosis and lack of benefit from oxaliplatin treatment (HR, 0.99 [95% CI, 0.73–1.34]; P = .96 [N = 367]). Examination of the different subtyping methods shows that all 3 methods robustly identified patients with poor prognosis (stemlike, CCS-3, and CMS-4) in both stage II and III.
CONCLUSIONS AND RELEVANCE
Patients with stemlike tumors may be appropriate for clinical trials testing experimental therapies because stemlike tumors were robustly identified and associated with a poor prognosis regardless of stage or chemotherapy regimen. The clinical utility of using subtyping for the identification of patients for treatment with oxaliplatin requires validation in independent clinical trial cohorts.
TRIAL REGISTRATION
clinicaltrials.gov Identifier: NCT00004931
The MOSAIC1 and National Surgical Adjuvant Breast and Bowel Project (NSABP) C-07/NRG Oncology (referred to hereafter as NSABP C-07)2 clinical trials showed that oxaliplatin added to fluorouracil and leucovorin therapy significantly improved disease-free survival (DFS) and established oxaliplatin as part of the standard of care for the adjuvant treatment of patients with early-stage colon cancer. Patients treated with the addition of oxaliplatin in the NSABP C-07 trial showed superior DFS (hazard ratio [HR], 0.80 [95% CI, 0.69–0.93]; P = .002). However, the acute and chronic neurotoxic effects associated with exposure to oxaliplatin3 emphasize the importance of prospectively identifying patients who will benefit from oxaliplatin therapy so that patients can avoid adverse effects of a noneffective treatment. Likewise, it is clinically meaningful to identify patients who do not receive benefit from oxaliplatin despite a poor prognosis, because they represent the ideal candidates for clinical trials testing experimental therapeutics. However, previous efforts to develop a predictive test for these patients using gene expression or mutation profiling have been unsuccessful.4,5
Recently, several studies6–9 have used unsupervised clustering methods to develop genomic signatures to classify colorectal cancer (CRC) into different subtypes and have shown that each subtype has distinct molecular features and prognosis. Different studies have identified different numbers of clusters, presumably due to using different methods and training data sets. For example, the CRC Assigner (CRCA) classifier categorized CRC into 5 distinct subtypes: enterocyte, gobletlike, inflammatory, stemlike, and transit amplifying (TA)7; and the Colon Cancer Subtypes (CCS) classifier identified 3 groups: CCS1, CCS2, and CCS3.6 Several studies have shown that different classifiers are highly correlated; for example, for CCS and CRCA classifiers, most CCS1 tumors are classified as TA or enterocyte, most CCS2 tumors are classified as inflammatory and gobletlike tumors, and most CCS3 tumors are classified as stemlike tumors.10,11 Different subtypes were also shown to have different prognosis. Particularly, stemlike or CCS3 subtypes are associated with high risk of relapse despite fluorouracil-based chemotherapy.6
On the basis of biological differences among identified subtypes, we hypothesized that colorectal subtypes would differ in residual risk after fluorouracil-leucovorin adjuvant chemotherapy and the degree of benefit derived from the addition of oxaliplatin in NSABP C-07. The C-07 tumor samples were classified into CCS6 and CRCA subtypes7 and tested for their association with prognosis and interaction with oxaliplatin.
Methods
Study Participant Protection
The institutional review board (IRB) of record for the C-07 clinical protocol with embedded tissue banking and molecular profiling studies was the Allegheny-Singer Research Institute, and local IRBs approved the C-07 protocol at the enrolling sites. The molecular profiling study itself was considered to be exempt by the Chesapeake IRB. All patients included signed informed consent for biomedical research using archived tumor tissue. All assays were performed by investigators blinded to clinical outcome using deidentified specimens. All merged data sets containing clinical and molecular data were anonymized by an honest broker.
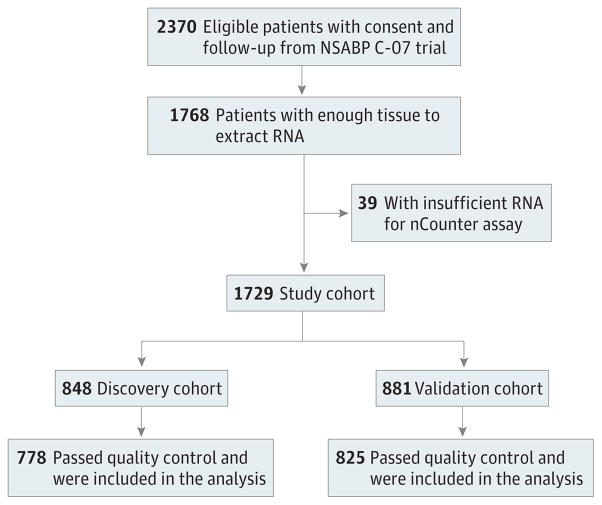
Initiated in 2000, NSABP C-07 was closed to follow-up in 2012 and included 2370 eligible patients with consent and follow-up. This study included 1729 cases with a median follow-up of 10.22 (interquartile range, 9.5–11.2) years. Clinical variables were well balanced between the 2 populations (eTable 1 in the Supplement).
Study Design
We followed the principle of “prospectively designed retrospectively tested” according to Simon et al12 with a locked-down algorithm for subtype classification and prespecified direction of interaction of subtypes with oxaliplatin as shown in Figure 1. To test whether molecular subtypes are predictive of oxaliplatin benefit, the entire data set was split into discovery and validation cohorts of similar sample size. For historical reasons (details in eMethods 1 in the Supplement), the original selection of our discovery and validation cohorts was based primarily on the temporal order in which they were entered into the trial. As presented in eTable 2 in the Supplement, without considering missing data, most clinical variables were balanced between the discovery and validation cohorts, except that fewer patients in the discovery cohort had positive lymph nodes and obstruction. The discovery cohort was used to identify the subtype that benefited from oxaliplatin therapy, and this interaction term was tested in the validation cohort.
Figure 1.
Consolidated Standards of Reporting Trials Diagram, National Surgical Adjuvant Breast and Bowel Project (NSABP) C-07
Clinical data from the validation cohort were not available to us until we had a locked-down algorithm and received approval from the Protocol Review Committee of the Cancer Therapeutics Evaluation Program of the National Cancer Institute.
We recognized that the statistical power for the validation of this study was not optimal. Therefore, if the signature failed validation within C-07, we planned to use the entire C-07 data set as a new discovery cohort for hypothesis generation. Validation would then require an independent data set.
Gene Expression Profiling
The C-07–customized nCounter code set (Colo-295) consisted of 295 genes selected from prognostic genes from our internal data and from the literature (details in eMethods 2 in the Supplement) plus 6 positive and 8 negative technical control genes (gene list in eTable 3 in the Supplement). The quality control metrics and the details of the analytical performance of nCounter mRNA expression profiling are shown in eMethods 3 and 4 and eFigure 1 in the Supplement.
Redevelopment of Subtype Identification Algorithm Using a Custom nCounter Probe Set
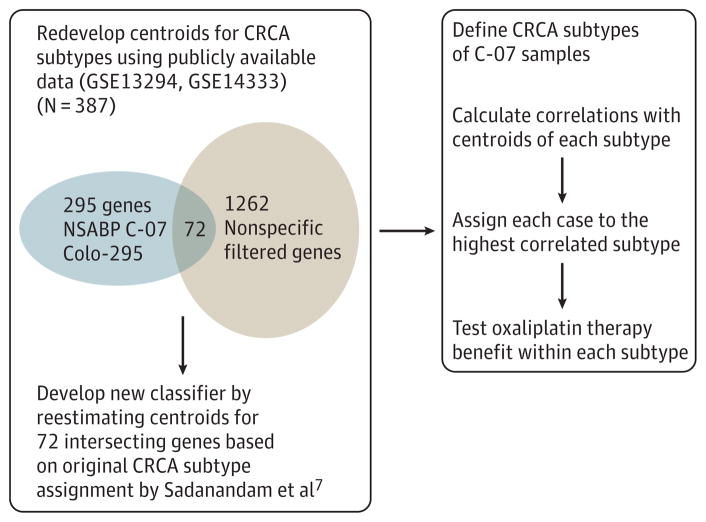
Because the custom nCounter assay code (Colo-295) set was designed on the basis of candidate prognostic genes before the publication of the articles describing intrinsic subtypes, only a small portion of genes in the subtype classifiers (56 of 786 genes in the CRCA classifier and 10 of 146 genes in the CCS classifier) were included in our nCounter code set. To overcome the obstacle of subtype identification caused by the limited number of genes, we redeveloped the CRCA classifier for the nCounter data by taking advantage of the publicly available gene expression microarray data from the core training data set (N = 387) in which the original CRCA classifier was developed by Sadanandam et al.7 Of 1262 genes with high dynamic range in the core training data set, 72 were included in our nCounter code set of 295 genes (Figure 2). Based on the expression profile of 72 genes and the original CRCA subtype assignment for the 387 samples in the CRCA core training data set, we generated the centroid using these 72 genes for each subtype using the prediction analysis of microarray method.13 The number of genes is selected by 10-fold cross-validation. The redeveloped centroids of the CRCA subtypes are presented in eTable 4 and eFigure 2 in the Supplement. The overall cross-validation error rate is 0.157, ranging from 0.052 for stem-like subtype to 0.302 for gobletlike subtype (eTable 5 and eMethods 5 in the Supplement). Analysis was carried out using the pamr packages implemented in R. To assign C-07 samples to a CRCA subtype, we calculated the Spearman rank correlation between each sample and the redeveloped centroids for each subtype and assigned the sample to the most correlated subtype. Similarly, centroids were redeveloped for the CCS classifier as described in eMethods 6 in the Supplement and are presented in eTable 6 in the Supplement.
Figure 2.
Procedure for Redeveloping Colorectal Cancer Assigner (CRCA) Classifier Using 72 Genes Included in the nCounter Data Profiled With the Customized nCounter Code Set (Colo-295)
Statistical Analysis
Associations of clinical variables and mutations with subtypes were analyzed by means of the χ2 test.
The primary end point in this study was time to recurrence (time from random assignment to recurrence, censored for death [competing risk] or last follow-up). For each variable, we assessed (1) prognostic significance using univariate Cox models and (2) predictive values for oxaliplatin therapy benefit using Cox models with an interaction term between treatment and tested variable. For subtypes, we further tested prognostic value using multivariate analysis with adjustment for clinical variables age, sex, tumor stage, stage, grade, perforation, and obstruction.
In the validation cohort, the prespecified primary hypothesis to be tested was the enterocyte-oxaliplatin interaction term in a Cox model with or without adjustment of clinical variables. All reported P values are 2 sided, and the statistical significance level was set to less than .05. All statistical analyses were performed in R.
We calculated the effect sizes that would be necessary to achieve adequate power in the validation cohort. For the enterocyte subtype, with the present sample size (n = 70), with an a level of .05, the HR for oxaliplatin treatment must be less than 0.20 to achieve 80% power. The enterocyte-oxaliplatin interaction needed to be less than 0.30 to achieve 80% power (details in eMethods 7 and 8 in the Supplement).
Results
In C-07, among 2370 eligible patients with consent and follow-up, 1768 cases with available tumor blocks were used in this study (Figure 1). An additional 39 cases were lost to insufficient amount of RNA.
Prognostic and Predictive Values of Clinical Variables in the Entire C-07 Cohort
Stage, tumor stage, perforation, and obstruction are significantly associated with recurrence-free survival, but among all analyzed clinical variables (eTable 7 in the Supplement), only obstruction was predictive of oxaliplatin benefit regardless of whether we examined only stage III or combined patients with stage II and III disease into one category (eFigure 3 in the Supplement). Because obstruction was not associated with oxaliplatin benefit in the MOSAIC trial,1 we did not pursue this observation further.
Current National Comprehensive Cancer Network (NCCN) guidelines recommend that all patients with stage III or high-risk stage II CRC be treated with oxaliplatin. However, analysis in C-07 indicated that the NCCN guideline was not predictive for oxaliplatin benefit (details in eResults 1 and eFigure 4 in the Supplement).
Prognostic and Predictive Values of Subtypes in the Discovery Cohort
Using redeveloped centroids, we identified the CRCA subtypes7 and CCS subtypes.6 Among 778 cases classifiable with CRCA, 91 (11.7%) were classified as enterocyte, 63 (8.1%) as goblet-like, 202 (26.0%) as inflammatory, 239 as stemlike (30.7%), and 183 (23.5%) as TA subtype. For CCS, 258 (33.2%), 200 (25.7%), and 320 (41.1%) were classified as CCS1, 2, and 3, respectively. Consistent with other studies,10 the CRCA subtype and CCS subtype were correlated with each other, suggesting that our newly developed classifier performed as expected; that is, most CCS1 tumors were subtyped as TA or enterocyte, most CCS2 tumors as inflammatory, and most CCS3 tumors as stemlike (eTable 8 in the Supplement). Association of subtypes and other clinical pathological variables was also similar to what has been found in other studies; for example, the inflammatory subtype was enriched with clinical variables associated with good prognosis (stage II, undifferentiated, deficient mismatch repair), whereas the stemlike subtype was enriched with clinical variables associated with poor prognosis (stage III, T3, or T4 tumor stage) (eTable 9 in the Supplement).
We assessed the prognostic value of subtypes. For both univariate and multivariate analysis with adjustment of clinical variables, time to first recurrence varied among the CCS subtypes and CRCA subtypes (eFigure 5 and eTables 10 and 11 in the Supplement). Consistent with other studies, stemlike or CCS3 was associated with the shortest time to first recurrence.
We further assessed the predictive value of subtypes. The CCS classifier did not identify any subtype with a significant benefit from oxaliplatin treatment and therefore was not pursued further for validation (eFigure 3 in the Supplement). For CRCA subtypes, only patients with the enterocyte subtype received significant benefit from oxaliplatin added to fluorouracil-leucovorin treatment (HR, 0.32 [95% CI, 0.14–0.70]; P = .005 [N = 91]) (eFigure 6 in the Supplement). Further analysis suggested that oxaliplatin benefit in the enterocyte subtype may be restricted to patients with stage III disease (eFigure 7 in the Supplement).
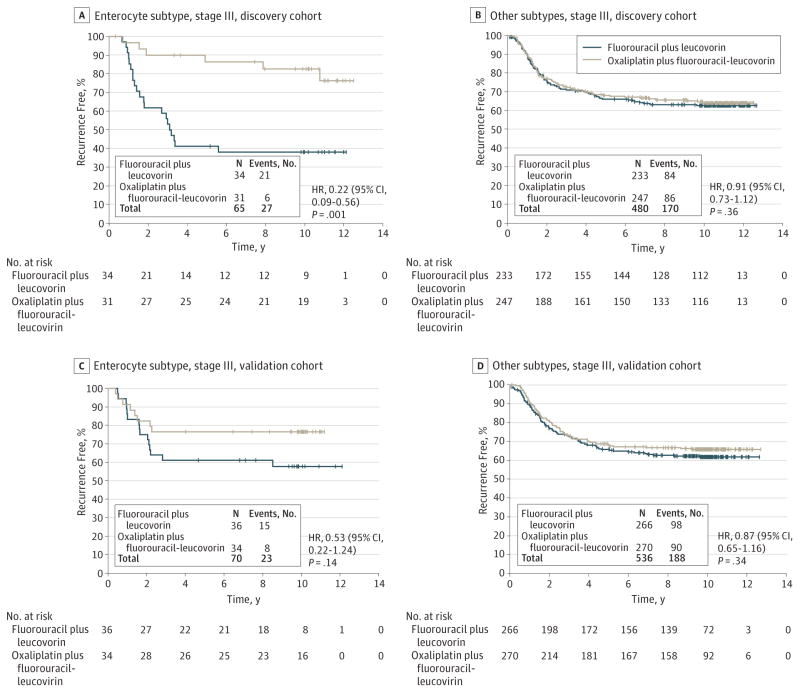
Because only patients with stage III, enterocyte type tumors received benefit from oxaliplatin (HR, 0.22 [95% CI, 0.09–0.56]; P = .001 [N = 65]), the 4 other subtypes, gobletlike, inflammatory, stemlike, and TA, were combined as a nonbenefit group (HR, 0.91 [95% CI, 0.73–1.12]; P = .36 [N = 480]) (Figure 3A and B). A significant enterocyte-oxaliplatin interaction (interaction HR, 0.24; P = .002) was observed.
Figure 3.
Kaplan-Meier Plots for Recurrence-Free Survival of Stage III Patients
Prospectively Designed Retrospective Testing of the Predictive Value of CRCA Subtypes in the Validation Cohort
Among 825 classifiable cases in the validation cohort, 93 (11.3%) were classified as enterocyte, 79 (9.6%) as gobletlike, 203 (24.6%) as inflammatory, 239 (29.0%) as stemlike, and 211 (25.6%) as TA subtype.
The prespecified primary analysis plan dictated primary comparison of oxaliplatin benefit in stage III patients in the enterocyte group vs all other subtypes combined (test of the enterocyte-oxaliplatin interaction). Tumors from 70 of 606 classifiable stage III patients (11.6%) were classified as enterocyte subtype. In the validation cohort, there was a nonsignificant finding of oxaliplatin benefit with an HR of 0.53 ([953% CI, 0.22–1.24]; P = .14 [N = 70]) for the enterocyte subtype. In the other subtypes combined, the HR was 0.87 ([95% CI, 0.65–1.16]; P = .34 [N = 536]). The interaction between enterocyte subtype and oxaliplatin treatment was not significant (interaction P = .32) (Figure 3C and D).
Examination of time to recurrence for other individual subtypes revealed that stemlike subtype had the worst prognosis regardless of treatment as observed in the discovery cohort (eTable 12 in the Supplement). Patients with gobletlike (HR, 0.97 [95% CI, 3.42–2.21]; P = .94 [N = 60]), stemlike (HR, 1.16 [95% CI, 0.76–1.80]; P = .48 [N = 185]), and TA (HR, 0.79 [95% CI, 0.46–1.35]; P = .39 [N = 166]) subtypes did not gain significant benefit from oxaliplatin treatment (eFigure 8 in the Supplement). There was a nonsignificant finding of oxaliplatin benefit for the inflammatory subtype in the validation cohort (HR, 0.50 [95% CI, 0.23–1.08]; P = .08 [N= 125]), but in the discovery cohort the HR was much higher (HR, 1.09 [95% CI, 0.51–2.32]; P = .83) (eFigure 7 in the Supplement).
Therefore, we failed to validate our primary hypothesis that enterocyte subtype is a marker that predicts benefit from oxaliplatin therapy. Post hoc power calculation suggested that the power is less than 40% with the findings in the validation cohort (eMethod 8 in the Supplement).
Post Hoc Analyses of the Entire C-07 Cohort
According to our plan to use the entire cohort for new exploratory analysis for hypothesis generation in the likely case of failure to validate enterocyte-oxaliplatin interaction due to lack of statistical power, we analyzed the entire cohort for not only CRCA and CCS but also the Consensus Molecular Subtype (CMS) classifier,11 which was published after the approval of the validation protocol (eResults 2 in the Supplement).
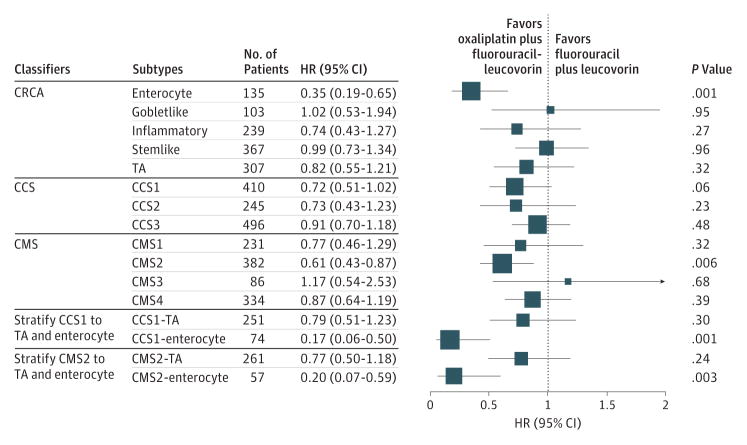
As shown in Figure 4, enterocyte subtype from the CRCA classifier, CCS1 subtype from the CCS subtype classifier, and CMS2 from the CMS classifier were associated with benefit from oxaliplatin treatment. Further analysis suggested that both CCS1 and CMS2 consist of enterocyte and TA subtypes based on the CRCA classifier (eTable 13 in the Supplement). Significant benefit was seen only in the CCS1-enterocyte (HR, 0.17 [95% CI, 0.06–0.50]; P = .001) and the CMS2-enterocyte (HR, 0.20 [95% CI, 0.07–0.59]; P = .003) and not in the CCS1-TA (HR, 0.79 [95% CI, 0.51–1.23]; P = .30) or in the CMS2-TA (HR, 0.77 [95% CI, 0.50–1.18]; P = .23). This suggests that the CRCA classifier may represent the best way to subclassify tumors to identify patients who will receive the greatest benefit from oxaliplatin. Examination of stage III patients in the entire cohort suggested significant enterocyte-oxaliplatin benefit (interaction HR, 0.38; P = .003) (eFigure 9 in the Supplement).
Figure 4.
Exploratory Analyses: Forest Plot of Treatment Benefit for Subtypes Identified by Different Classifiers for C-07 Participants With Stage III Disease
The box size is proportional to the precision. CCS indicates Colorectal Cancer Subtype; CRCA, Colorectal Cancer Assigner; CMS, Consensus Molecular Subtype; HR, hazard ratio; and TA, transit amplifying.
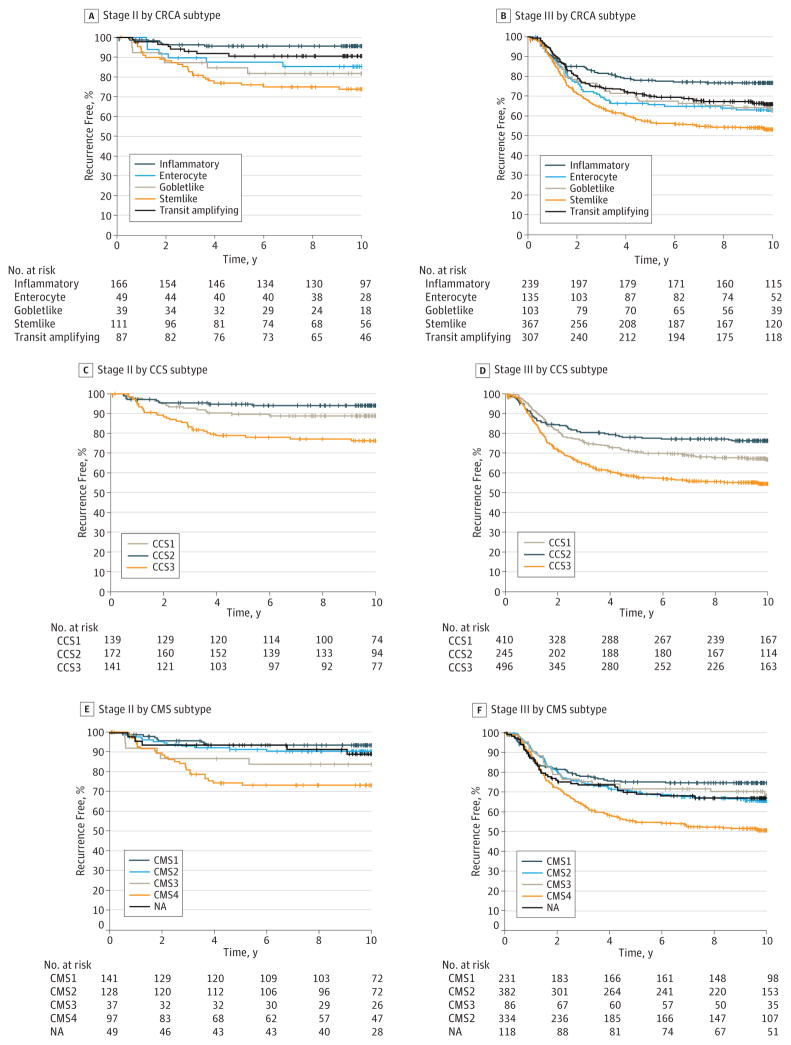
We also examined the prognostic implications of each subtyping method within patients with stage II or stage III disease. Regardless of clinical stage, all 3 subtyping methods identified similar tumors (stemlike, CCS3, and CMS4) as the worst prognostic group (P < .001 for each) (Figure 5).
Figure 5.
Kaplan-Meier Plots for Recurrence-Free Survival of Entire Cohort (Stage II or Stage III Patients) According to Colorectal Cancer Assigner (CRCA), Colorectal Cancer Subtype (CCS), and Consensus Molecular Subtype (CMS) Subtypes
NA indicates not assignable to a subtype.
Discussion
In this prospectively designed, retrospectively tested study of molecular subtype–by–oxaliplatin interaction, we observed a striking interaction between the enterocyte subtype with the CRCA classifier and oxaliplatin therapy benefit in the C-07 discovery cohort. However, the interaction was not statistically significant in the validation arm despite a nonsignificant finding in the same direction. Lack of statistical power could be one reason for our failure to validate the enterocyte-oxaliplatin interaction. In addition, our rederived classifier for CRCA did not have many robust classifier genes for enterocyte subtype, resulting in a high classification error rate. It is worth noting that the enterocyte subtype identification is inherently challenging, with a high classification error rate. Therefore, this subtype is bundled with the TA subtype in other subtype classification systems such as CCS classifier and CMS classifier.
Despite these limitations, the fact that subtype classifiers were originally developed outside C-07 makes it less likely that our data were overfitted. Thus, it is possible that the interaction of oxaliplatin therapy benefit with the enterocyte subtype could be validated with greater power and more genes that better defined the enterocyte subtype.
Our data also demonstrated heterogeneity of clinical outcome after fluorouracil-leucovorin therapy, as well as degree of benefit from the addition of oxaliplatin to fluorouracil-leucovorin in colon cancer according to molecularly defined subtypes. In particular, the stemlike subtype (or equivalently CCS3 subtype in the CCS classifier and CMS4 in the CMS classifier) was not only associated with poor prognosis in patients treated with fluorouracil-leucovorin but also in patients treated with oxaliplatin plus fluorouracil-leucovorin regardless of clinical stage. Therefore, we can conclude that C-07 data validated the poor prognosis of the stemlike subtype even after standard adjuvant chemotherapy in both stage II and III colon cancer.
The observation that patients with stemlike tumors have a poor prognosis is consistent with other studies, but our data demonstrate the possibility that, unlike the patients with a poor prognosis in breast cancer, the stemlike subtype does not receive a significant benefit from chemotherapy with a high residual risk even after oxaliplatin-based chemotherapy. Roepman et al9 demonstrated that CCS3 (ie, the stemlike subtype) is characterized by high expression of epithelial mesenchymal transition markers and low expression of proliferation markers and is resistant to adjuvant fluorouracil-leucovorin treatment, albeit in a nonrandomized retrospective cohort comparison. Surprisingly, the epithelial mesenchymal transition gene expression signature is mostly contributed by the stromal cells driven by transforming growth factor β signaling rather than by the cancer cells.14 Therefore, clinical and biological evidence suggests that stemlike subtype colon cancer is resistant to chemotherapy and attacking cancer cells alone may not be the optimal therapeutic approach for these tumors largely driven by microenvironmental interaction.
For future clinical trials in colon cancer, it may be reasonable to stratify patients according to stemlike subtype, not only because of this subtype’s poor prognosis, but also because the stemlike subtype has been most consistently identified across all gene expression profiling studies and the performance of classifier genes for this subtype is robust with a low misclassification rate (which varies among subtypes). Therefore, the development of alternative therapeutic strategies for patients with the stemlike subtype of colon cancer should be a priority.
Conclusions
This study suggested that patients with different subtypes of colon cancer have distinct prognoses and may obtain differential benefit from oxaliplatin added to fluorouracil-leucovorin therapy. Whether patients with enterocyte subtype are the only ones who derive a significant clinical benefit from oxaliplatin needs to be tested further in external cohorts due to the failure of validation in this study. To further investigate the predictive value of colon cancer subtypes for determining oxaliplatin therapy benefit, we plan to develop a better panel of genes for subtype identification, resulting in a more accurate identification of subtypes. Subtyping of patients with CRC may provide a rationale for the assignment of patients to different treatment regimens, such as oxaliplatin-based chemotherapy for patients with enterocyte tumors, immune checkpoint inhibitors for inflammatory cancer, anti-MUC1 antibodies for goblet cancers, and new targeted therapies for stemlike cancers.
Supplementary Material
Key Points.
Question
Do patients with different colorectal cancer subtypes have different prognosis and do their responses differ with regard to treatment with oxaliplatin when it is combined with fluorouracil plus leucovorin?
Findings
This secondary analysis of a randomized clinical trial validated the finding that patients with a stemlike or Colorectal Cancer Subtype 3, or Consensus Molecular Subtype 4 have a poor prognosis, regardless of stage or chemotherapy regimen. The observation that patients with the enterocyte subtype received significant benefit from oxaliplatin was not validated in an independent cohort.
Meaning
This study highlights the importance of subtyping colorectal cancer with gene expression profiling for prognostic information and of conducting future clinical trials to test the clinical utility of subtyping to determine oxaliplatin treatment benefit.
Acknowledgments
Funding/Support: This work was supported by the National Cancer Institute at the National Institutes of Health, US Department of Health and Human Services, Public Health Service grants U10-CA180868, U10-CA180822, and U24-CA196067; Korea Health Technology R&D Project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant HI13C2162 to Dr Paik); under a grant from the Pennsylvania Department of Health; and by Sanofi-Synthelabo Inc.
Footnotes
Conflict of Interest Disclosures: Dr Yothers has acted in a consulting/advisory role for Pharmacyclics. No other disclosures are reported.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Disclaimer: The Pennsylvania Department of Health specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Previous Presentation: This study was presented at the American Society of Clinical Oncology annual meeting; June 6, 2016; Chicago, Illinois.
Additional Information: All NSABP legacy trials are now part of the NRG Oncology portfolio.
Additional Contributions: We thank the following NSABP staff members for their valuable contributions: Melanie Finnigan, BS, for data and tissue block management; William Hiller, ASCP (deceased), and Teresa A. Oeler, BS, Paralegal Cert, for histologic analysis; Wendy L. Rea, BA, Christine I. Rudock, and Barbara C. Good, PhD, for manuscript editing and preparation; and Teresa A. Bradley, PhD, and Ethan Barry, BA, CCRC, for regulatory affairs related to this manuscript. These contributors were not compensated beyond their usual salaries for this work. We also thank NSABP members who contributed tissue blocks, as well as patients who enrolled in the study.
Author Contributions: Drs Pogue-Geile and Paik had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Song and Pogue-Geile shared equally in the work of this manuscript.
Study concept and design: Song, Pogue-Geile, Yothers, Johnson, Wolmark, Paik.
Acquisition, analysis, or interpretation of data: Song, Pogue-Geile, Gavin, Yothers, Kim, Lipchik, Allegra, Petrelli, O’Connell, Paik.
Drafting of the manuscript: Song, Pogue-Geile, Gavin, Kim, Paik.
Critical revision of the manuscript for important intellectual content: Pogue-Geile, Gavin, Yothers, Kim, Johnson, Lipchik, Allegra, Petrelli, O’Connell, Wolmark, Paik.
Statistical analysis: Song, Gavin, Yothers, Paik.
Obtained funding: Paik.
Administrative, technical, or material support: Pogue-Geile, Gavin, Yothers, Kim, Johnson, Lipchik, Allegra, Wolmark, Paik.
Study supervision: Pogue-Geile, Paik.
References
- 1.André T, Boni C, Mounedji-Boudiaf L, et al. Multicenter International Study of Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of Colon Cancer (MOSAIC) Investigators. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350(23):2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 2.Kuebler JP, Wieand HS, O’Connell MJ, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABP C-07. J Clin Oncol. 2007;25(16):2198–2204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 3.Cersosimo RJ. Oxaliplatin-associated neuropathy: a review. Ann Pharmacother. 2005;39(1):128–135. doi: 10.1345/aph.1E319. [DOI] [PubMed] [Google Scholar]
- 4.Yothers G, O’Connell MJ, Lee M, et al. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31(36):4512–4519. doi: 10.1200/JCO.2012.47.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavin PG, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18(23):6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Sousa E Melo F, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 7.Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marisa L, de Reyniès A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadanandam A, Wang X, de Sousa E Melo F, et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle. 2014;13(3):353–357. doi: 10.4161/cc.27769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.