Membrane-bound enzyme relocates to the cell nucleus to modify chromatin, inducing cancer resistance to radiotherapy.
Keywords: Cell biology, DNA repair, oncogenes, TIE2, ABL1, ANG1, NHEJ
Abstract
DNA repair pathways enable cancer cells to survive DNA damage induced after genotoxic therapies. Tyrosine kinase receptors (TKRs) have been reported as regulators of the DNA repair machinery. TIE2 is a TKR overexpressed in human gliomas at levels that correlate with the degree of increasing malignancy. Following ionizing radiation, TIE2 translocates to the nucleus, conferring cells with an enhanced nonhomologous end-joining mechanism of DNA repair that results in a radioresistant phenotype. Nuclear TIE2 binds to key components of DNA repair and phosphorylates H4 at tyrosine 51, which, in turn, is recognized by the proto-oncogene ABL1, indicating a role for nuclear TIE2 as a sensor for genotoxic stress by action as a histone modifier. H4Y51 constitutes the first tyrosine phosphorylation of core histones recognized by ABL1, defining this histone modification as a direct signal to couple genotoxic stress with the DNA repair machinery.
INTRODUCTION
DNA repair pathways enable malignant gliomas to survive DNA damage that is induced after genotoxic therapies (1, 2). For that reason, the development of new therapeutic strategies requires the identification of key molecular pathways that regulate the cancer-resistant phenotype. The abnormal function of tyrosine kinase receptors (TKRs) is a hallmark of malignant gliomas (3), and TKRs have been reported as regulators of the DNA repair machinery (4). The TKR TIE2 is overexpressed in human surgical glioma specimens, at levels that correlate with the degree of increasing malignancy (5), and in brain tumor stem cells (BTSCs) (6). Here, we described a new mechanistic link between TIE2 and DNA repair machinery involving epigenetic histone modification and the proto-oncogene ABL1.
RESULTS AND DISCUSSION
To determine the biological role of TIE2 in the resistance of malignant gliomas to radiotherapy, we subjected a subset of BTSCs, derived from human malignant glioma surgical specimens, and a TIE2 isogenic system to ionizing radiation (IR). We found that TIE2-expressing cultures were more resistant to IR-induced cell death and retained their ability to divide after IR treatment at a higher rate than were cells that did not express the TKR (Fig. 1, A to C, and fig. S1, A and B). In agreement with these data, down-regulation of TIE2 expression using small interfering RNA (siRNA) resulted in the acquisition of a radiosensitive phenotype (Fig. 1D and fig. S1, C to E). In an attempt to delineate the mechanism underlying the role of TIE2 in radioresistance, we first analyzed the expression of TIE2 by confocal microscopy and observed that, upon IR treatment, TIE2 protein localized to the nucleus rather than its normal cytoplasmic localization (Fig. 1E). Consistently, we observed nuclear TIE2 expression in experiments involving subcellular fractionation (Fig. 1F). These results were confirmed both in IR-exposed endothelial cultures, which express high levels of endogenous TIE2 (fig. S1, F and G), and in brains of mice bearing glioma stem cell (GSC)–derived intracranial xenografts after IR treatment (Fig. 1G and fig. S1, H and I). These observations led us to identify a region that endows TIE2 protein with the ability to translocate to the nucleus to further explore specific functions of TIE2 in this subcellular compartment, as pursued with several TKRs (7). We refer to this putative region as a nuclear localization signal (NLS) sequence (580-RRSVQKS-586; fig. S2A) and performed a series of mutational analyses (fig. S2B). We observed that cells expressing mutant Tie2 forms d585-86 and SS582/86AA were more sensitive to IR and exhibited a deficient nuclear TIE2 translocation (Fig. 1, H to J). These results suggest that nuclear translocation of TIE2 is involved in the radioresistance of TIE2-expressing glioma cells.
Fig. 1. Nuclear TIE2 localization is associated with the resistance of glioma to IR.
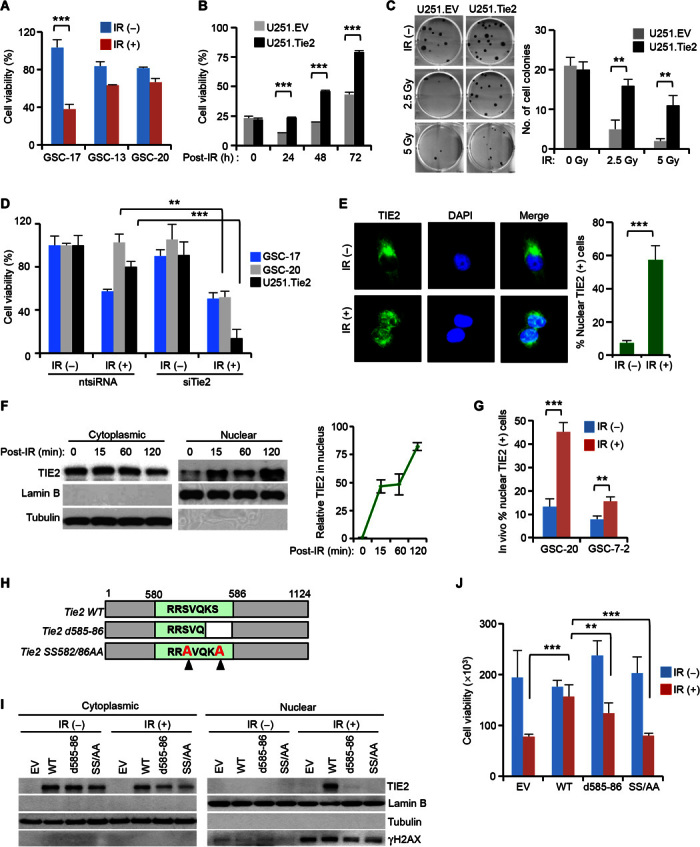
(A and B) Cell viability assay to determine the response to IR of (A) TIE2-expressing GSCs (GSC-13 and GSC-20), TIE2-nonexpressing GSC (GSC-17), and (B) TIE2 isogenic U251 cultures in a time-point experiment. (C) Colony-forming assay of isogenic U251 cells upon IR treatment. (D) TIE2 silencing results in the radiosensitization of GSCs and U251.Tie2 cells. ntsiRNA, nontargeting siRNA. (E and F) TIE2 localizes in the nucleus of U251 cells upon IR, as assessed by (E) immunofluorescence and confocal microscopic analysis and (F) Western blot analysis. (G) TIE2 localizes in the nucleus of GSCs upon in vivo IR of intracranial xenografts. (H) Schematic representation of Tie2 constructs with mutations within the NLS sequence. WT, wild type. (I and J) NLS mutations jeopardize TIE2 (I) nuclear translocation upon IR, and (J) U251 glioma radioresistance. Data represent means ± SD; **P ≤ 0.01, ***P ≤ 0.001. EV, empty vector.
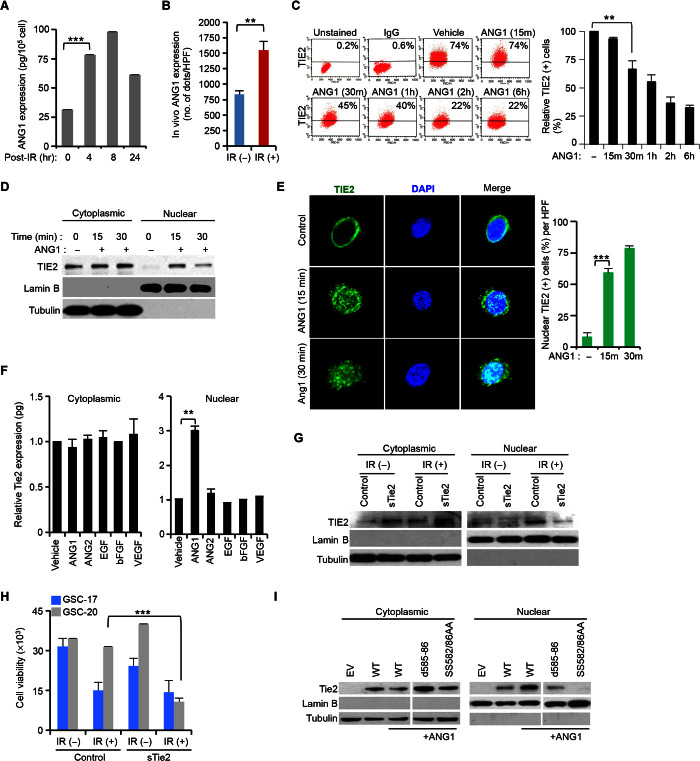
To ascertain whether TIE2 trafficking is ligand-dependent, we quantified the expression levels of its ligands ANG1 and ANG2 (8) using enzyme-linked immunosorbent assay (ELISA), and observed increased levels of ANG1 (Fig. 2A), but not ANG2, after IR exposure. We also observed that, after IR, Ang1 mRNA levels incremented (fig. S3A). Corroborating the in vitro data, the ANG1 protein expression in sections from GSC-derived intracranial xenografts increased after IR treatment (Fig. 2B). To determine the role of ANG1 in TIE2 trafficking, we performed a fluorescence-activated cell sorting (FACS) analysis and detected a decrease in the expression levels of membrane-bound TIE2 after ANG1 exposure (Fig. 2C); however, ANG2 or epidermal growth factor (EGF) treatment did not modify these levels (fig. S3B). Although it has been previously reported in endothelial cells (9) that ANG1, but not ANG2, was responsible for TIE2 internalization and degradation, we observed that, upon ANG1 exposure, TIE2 was present in the nucleus (Fig. 2, D and E, and fig. S3, C and D). This phenomenon was ligand/receptor-specific, as tested in experiments involving exposure to different ligands, including ANG2, and in an analysis of the subcellular localization of the other Tie member TIE1 upon incubation with ANG1 and ANG2 ligands (Fig. 2F and fig. S3E). Notably, although Tie1 and Tie2 are highly homologous, sharing 49% amino acid identity, Tie1 did not contain a similar putative NLS motif (fig. S2, C and D). Consistent with a role of ANG1 in TIE2 nuclear localization, the addition of sTIE2 as a decoy receptor jeopardized both the presence of TIE2 in the nucleus upon IR (Fig. 2G) and the radioresistance of TIE2-expressing BTSC cultures (Fig. 2H). Additional data using antibodies recognizing several domains suggested that full-size, phosphorylated TIE2 protein was localized in the nucleus upon ANG1 exposure (fig. S3, F and G). Similar to our previous observations using IR (Fig. 1I), TIE2 nuclear translocation was blocked when cells expressed mutant TIE2 forms d585-86 and SS582/86AA (Fig. 2I and fig. S4, A to D), suggesting a role for S582, S586, and K585 in this TIE2 cellular trafficking.
Fig. 2. ANG1 induces TIE2 nuclear localization.
(A) Increase of total ANG1 protein levels in U251.Tie2 cells in response to IR. (B) ANG1 protein levels of expression upon in vivo IR treatment of GSC-20–derived intracranial xenografts. HPF, high-power field. (C) Decrease of cell membrane–bound TIE2 upon ANG1 exposure in U251.Tie2 cultures. IgG, immunoglobulin G. (D) ANG1 exposure results on TIE2 nuclear translocation. (E) TIE2 localizes in the nucleus of human umbilical vein endothelial cells (HUVECs) upon ANG1 exposure, as assessed by immunofluorescence and confocal microscopic analysis. DAPI, 4′,6-diamidino-2-phenylindole. (F) TIE2 protein levels in cytoplasmic and nuclear U251.Tie2 cellular compartments after exposure to several ligands. bFGF, basic fibroblast growth factor; VEGF, vascular endothelial growth factor. (G and H) Soluble TIE2 (sTIE2) (G) jeopardizes IR-induced TIE2 nuclear translocation and (H) sensitizes GSCs to IR. (I) NLS mutations jeopardize TIE2 nuclear translocation upon ANG1 exposure. Data represent means ± SD; **P ≤ 0.01, ***P ≤ 0.001.
To further understand the biological significance of nuclear TIE2 translocation and its association with radioresistance, we analyzed γH2AX expression, a key component of the DNA damage response (DDR) (10), in cultures differing in their TIE2 status. Upon IR treatment, the persistence of γH2AX (11) was prolonged in TIE2-negative cells (U251.EV) or in cells harboring a mutant TIE2 (U251.Tie2SS/AA), and it was unable to translocate to the nucleus, when compared with isogenic wild-type (U251.Tie2) Tie2 cells (Fig. 3A). Furthermore, after DNA/protein cross-linking and DNA precipitation (fig. S5A), we found the presence of TIE2 in the γH2AX-containing DNA/protein complexes generated after genotoxic stress (Fig. 3, B and C, and fig. S5, B and C), together with components of the DNA repair machinery, BRG1 (12, 13), DNA-PK, Ku70, and Ku80 (Fig. 3D) (14). On the basis of our observation that TIE2 binds to DNA repair complexes (Fig. 3, B to D), we investigated the role of TIE2 in DNA repair by focusing on one of the main pathways involved in DNA double-strand break (IR-induced), namely, nonhomologous end joining (NHEJ) (15). Using a fluorescent reporter (16) (fig. S6, A and B), we observed that TIE2-expressing cells displayed a more efficient DNA repair mechanism than did their TIE2-negative counterparts (Fig. 3, E and F, and figs. S6C and S7, A and B). Notably, we did not detect significant differences in the proliferation rate or cell cycle distribution between these two cultures that may justify their dissimilar NHEJ efficiencies (fig. S8, A and B). NHEJ competence was inhibited by using siRNA against TIE2 or by genetically impairing the nuclear translocation (Fig. 3, G to I, and fig. S6, D and E) or the catalytic kinase domain of the protein (fig. S9, A and B). However, a similar experiment designed to test the role of TIE2 in homologous recombination (HR) did not yield positive results (fig. S10, A to C). Collectively, these data suggest that nuclear TIE2 plays a role in DNA repair, specifically NHEJ, upon genotoxic stress.
Fig. 3. Nuclear TIE2 regulates DNA repair through a NHEJ mechanism.
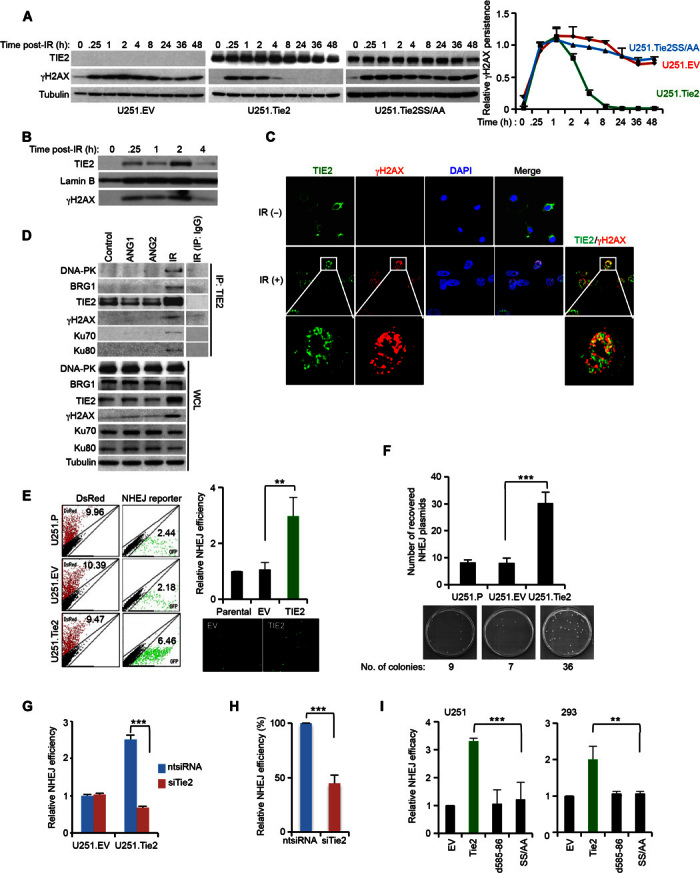
(A) γH2AX persistence in U251.EV, U251.Tie2, and U251.Tie2SS/AA cells in response to IR. (B) Formation of TIE2/DNA complexes in U251.Tie2 in response to IR, analyzed after DNA/protein cross-linking and protein elution from precipitated DNA. (C) γH2AX/TIE2 colocalization in HUVECs after IR treatment, as assessed by immunofluorescence and confocal microscopy. (D) TIE2 complexes in HUVECs in response to ANG1, ANG2, and IR stimuli. IP, immunoprecipitation; WCL, whole-cell lysate. (E) Increased NHEJ efficiency in TIE2-expressing cells. GFP, green fluorescent protein. (F) Increased recovery of NHEJ reporter plasmids in TIE2-expressing cells. (G) TIE2 silencing results in decreased NHEJ efficiency. (H) Silencing endogenous TIE2 in U87 MG cells results in decreased NHEJ efficiency. (I) NHEJ efficiency is jeopardized in cytoplasmic-sequestered TIE2. Data represent means ± SD; **P ≤ 0.01, ***P ≤ 0.001.
Because posttranslational modification of histones is important for signaling the position of DNA damage, recruiting the DNA repair proteins to the site of damage, and creating an open structure such that the repair proteins can access the site of damage (17, 18), we investigated whether TIE2 directly phosphorylates histones. To this end, purified TIE2 and core histones were used for in vitro kinase assays; following analysis by autoradiography or Western blot, we detected tyrosine phosphorylation of histones, specifically H4, in the presence of active TIE2 (Fig. 4, A to C). Furthermore, TIE2 phosphorylation of H4 was confirmed by the detection of a mobility shift in the presence of Mn2+ ion, which was abolished by alkaline phosphatase treatment (Fig. 4D). We observed TIE2/H4 complexes in the chromatin fraction, after ANG1 or IR stimulus (Fig. 4E). IR- or ANG1-treated nucleosomes were analyzed by mass spectrometry (MS), and the tandem mass spectrum of H4 matched the indicated amino acid sequence, suggesting that TIE2 directly phosphorylated the highly conserved Y51 residue (Fig. 4F and fig. S11, A to C). To validate these data, we generated a specific H4pTyr51 antibody. A dot blot assay and an in vitro kinase assay performed using purified histone proteins confirmed the specificity of the antibody (Fig. 4G and fig. S11, D and E) and that TIE2 phosphorylation of H4 at Tyr51 preferentially occurs in the presence of core histones.
Fig. 4. ABL1 is a reader of the TIE2-modified H4Y51 histone mark.
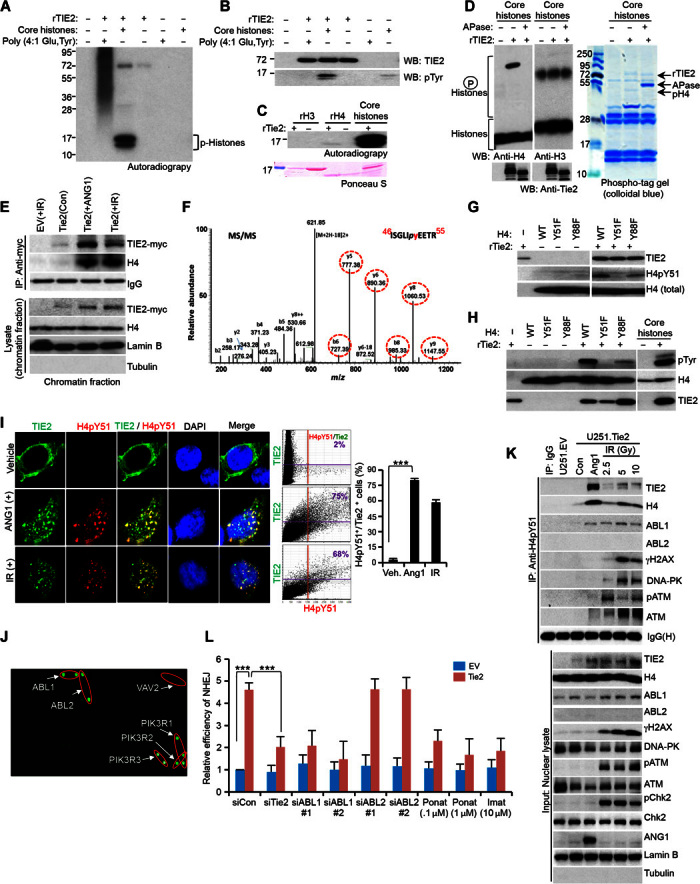
(A) rTIE2 potentially phosphorylates core histones. (B) rTIE2 potentially phosphorylates core histones at tyrosine residues. WB, Western blotting. (C) rTIE2 potentially phosphorylates rH4 but not rH3. (D) rTIE2 phosphorylates H4 as assayed using a phospho-tag gel. APase, alkaline phosphatase. (E) Cross-linked chromatin contains TIE2/H4 complexes upon ANG1 or IR stimulus. (F) Detection of phosphorylation of H4Tyr51 in the nucleosomes isolated from HEK293.Tie2 cells after ANG1 and IR exposure. Mass spectrometric analysis of a tryptic fragment at m/z mass/charge ratio of 630.7990 (mass error: 2.71 ppm) matched to the doubly charged phosphopeptide ISGLIpyEETR, suggesting that Y6 was phosphorylated. Mascot ion score was 52, with an expectation value of 6.5 × 10−5. Phosphotyrosine-containing peptide fragments are shown in dotted circles. MS/MS, tandem mass spectrometry. (G and H) Tyrosine phosphorylation of H4 analyzed using H4pY51 (G) and pTyr (H) antibodies with purified H4 mutant proteins. (I) Colocalization of TIE2 and H4pY51 in HEK293.Tie2 cells upon ANG1 and IR stimuli, as assessed by confocal microscopy. Colocalization was quantified with Olympus FluoView version 3.1a software. (J) rH4pY51 peptide binds to specific SH2 domain–containing proteins. (K) H4pY51 complexes with a panel of DNA repair proteins, including ABL1, in U251.Tie2-myc cells after IR exposure. (L) TIE2-induced NHEJ DNA repair is jeopardized by inhibiting ABL1 but not ABL2. Data represent means ± SD; **P ≤ 0.01, ***P ≤ 0.001.
An in vitro kinase assay with recombinant H4 mutant proteins demonstrated that TIE2 specifically phosphorylates H4Y51 (Fig. 4, G and H, and fig. S11F). Confocal analyses revealed that TIE2 colocalized with H4pY51 in the nucleus of IR- or ANG1-stimulated cells (Fig. 4I). Interestingly, H4Y51 was not detected in irradiated TIE2-negative cells (fig. S12) or in cells harboring a deleted or point-mutated catalytic kinase domain (fig. S9, C and D) (19, 20). We screened a high-density microarray searching for SH2 (Src homology 2) domain proteins (21, 22) that may act as readers of the H4Y51 phosphorylation (fig. S13, A and B). The array revealed that, among the 76 proteins tested, 6 of them interacted preferentially with H4pY51 compared to the nonphosphorylated H4 (Fig. 4J and fig. S13C). Among these proteins, we focused on ABL1 because of previous reports describing its role in DDR (23–25). The interaction was further verified by immunoprecipitation analysis, which revealed that H4pY51 binds ABL1, but not ABL2 [which does not contain an NLS (24)], in ANG1- or IR-treated cells (Fig. 4K). Corroborating these results, immunofluorescence analyses revealed TIE2/ABL1 complexes after these treatments (fig. S14). Notably, whereas ANG1 was sufficient to induce TIE2 nuclear translocation and H4Y51 modification with subsequent ABL1 recruitment, DDR-related proteins such as pATM, γH2AX, and DNA-PK (15, 26) were found in the H4Y51 immunoprecipitates only after irradiation treatment (Fig. 4K). Immunoprecipates of H3pS28, a DNA damage–unrelated mark (27), did not contain TIE2/DNA repair complexes in irradiated cells, showing specificity for the H4Y51 mark (fig. S15). Supporting an important role of the TIE2-ABL1 pathway, inhibition of ABL1 expression by siRNA, or using the ABL chemical inhibitor ponatinib or imatinib (24) (fig. S16, A, C, and D), resulted in decreased NHEJ efficiency upon IR only in TIE2-expressing cells (Fig. 4L and fig. S16, B and E). Decreasing ABL2 expression levels by using siRNA did not result in modification of the NHEJ cellular activity upon IR (Fig. 4L and fig. S16E), suggesting a specific role for ABL1 as reader of the H4pY51 mark.
This study identifies a new H4 modification, the phosphorylation of Tyr51 by the TKR TIE2. The H4pY51 mark is read by the SH2-containing protein ABL1. Furthermore, our data highlight the functional significance of the tyrosine kinase activity of TIE2, encouraging the search for other substrates for this TKR in the nucleus. The H4pY51 mark is being added to the tyrosine code of histones, which have been reported to be related to the cellular response to DNA damage (H2A.X Y142 and H4Y72), histone turnover (H3Y99) and transcription (H2BY37 and H2AY57), and chromatin architecture (H3Y41) (28–33). Different from the H2A.X Y142 and H4Y72 modifications, H4Y51 is a positive mark, directly read by the ABL1 DNA damage signaling protein, functioning as a functional component of the DNA repair machinery. The identification of specific sites that favor chromatin/ABL1 complexes may guide the development of adjuvant agents to overcome radiation resistance for patients with malignant gliomas and other cancers.
MATERIALS AND METHODS
Cell culture, cytokine stimulation, and IR
U251 human glioma cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin/streptomycin. HUVECs were maintained in EGM-2 BulletKit medium (Lonza). GSC-7-2, GSC-13, GSC-17, and GSC-20, characterized as previously described (34), were maintained in DMEM/F-12 supplemented with B27 (Life Technologies), EGF, and bFGF (20 ng/ml each) (Sigma-Aldrich). Human embryonic kidney 293 (HEK293) and HeLa cells were cultured in DMEM supplemented with 10% FBS (v/v) and 1% penicillin/streptomycin. U251.EV, U251.Tie2, U251.Tie2-myc, U251.Tie2-mycSS/AA, HEK293.EV, HEK293.Tie2-myc, and HEK293.Tie2-mycSS/AA cell lines were maintained in the presence of G418 (300 μg/ml) (Corning). Flag-H2AX– and Flag-H2AX S139A–expressing 293T cells (13) were maintained in DMEM supplemented with 5% FBS (v/v), 1% penicillin/streptomycin, and puromycin (0.5 μg/ml). U2OS.DR-GFP cells (35) were maintained in McCoy’s 5A medium supplemented with 10% FBS (v/v), 1% penicillin/streptomycin, and hygromycin (200 μg/ml). For cytokine-mediated stimulation, cells were starved for 6 to 24 hours in serum-free medium and then incubated in fresh serum-free medium containing ANG1 (400 ng/ml), ANG2 (400 ng/ml), VEGF (400 ng/ml) (R&D Systems), EGF (20 ng/ml), and bFGF (20 ng/ml) (Sigma-Aldrich). For IR treatment, cells were treated with the indicated doses of 137Cs radiation. To analyze the role of ABL1 activity, the ABL inhibitors imatinib mesylate (LKT Laboratories Inc.) and ponatinib (APExBIO) were used at the indicated concentrations for 24 hours.
RNA interference
siRNAs were transfected into cells using INTERFERin transfection reagent (Polyplus-transfection) at concentrations of 10 nM or otherwise indicated; after 48 hours of transfection, the knockdown efficiency was evaluated by determining the protein levels in whole-cell lysates. The siRNA sequences used are listed in table S1.
Cell viability and colony-forming assays
Cells were plated at subconfluent density, and 24 hours later, they were irradiated (15 Gy) and kept in an incubator for an additional 24 to 72 hours at 37°C. For transfection with siRNAs or DNA plasmids, cells were plated 1 day before transfection, and 24 hours later, they were irradiated. CellTiter-Blue Cell Viability Assay (Promega) was used according to the manufacturer’s instructions. When indicated, cells were stained with crystal violet solution (0.1% crystal violet and 20% methanol) for 15 min at room temperature (RT), and the results were imaged. To perform a colony-forming assay, 25 and 50 cells were plated per well in six-well plates and irradiated with indicated doses of radiation. Cells were incubated for 14 days, and the visible colonies were counted after they were stained with crystal violet, as described above.
Plasmid construction, transfection, and primers
The pcDNA3-Tie2, containing full-length human Tie2 cDNA (complementary DNA), and pcDNA3.1-Tie2-myc plasmids have been previously described (36, 37). Mutagenesis was performed using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies), following the manufacturer’s recommended protocol. Cells were transfected using X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science), following the manufacturer’s protocol. The primers used are listed in table S2.
Reverse transcription PCR and quantitative real-time PCR analysis
Total RNA was extracted with TRIzol (Invitrogen), and 2 μg of RNA was reverse-transcribed to cDNA with High Capacity RNA-to-cDNA Kit (Applied Biosystems). Quantitative gene expression was measured using PowerUp SYBR Green Master Mix (Applied Biosystems) on ABI PRISM 7500 Fast (Applied Biosystems). Expression levels of target genes were normalized to β-actin mRNA levels. For reverse transcription polymerase chain reaction (PCR) expression, cDNA was subjected to a PCR cycle analyzer with a Kapa PCR master mixture (Kapa Biosystems) using the following PCR conditions: 95°C for 2 min, 95°C for 15 s, 64°C for 15 s, 72°C for 15 s with 32 cycles, and final extension at 72°C for 10 min and hold at 4°C. Primer sequences are provided in table S2.
Cell cycle analysis
Cells were fixed in ice-cold 95% ethanol for 30 min at 4°C, washed once with phosphate-buffered saline (PBS), and stained with propidium iodide (50 μg/ml) in PBS containing ribonuclease A (50 μg/ml). Further, the cells were incubated at 37°C for 20 min and analyzed on a Gallios 561 flow cytometer (Beckman Coulter) using Kaluza software (Beckman Coulter).
Flow cytometric analysis
Cells were harvested, washed three times with PBS containing 0.5% bovine serum albumin (BSA), and incubated with phycoerythrin (PE)–conjugated mouse anti-human TIE2 for 30 min on ice at 4°C in the dark. BSA (0.1%) was used as a negative control for treatment, and PE-conjugated mouse IgG was used as a negative control for the staining procedure. All flow cytometric experiments were performed using a BD FACSAria (BD Biosystems). The antibodies and work conditions used are listed in table S3.
Subcellular fractionation
Cells were washed with ice-cold PBS, harvested by scraping or trypsinization, and collected in a hypotonic lysis buffer [20 mM Hepes (pH 7.0), 10 mM KCl, 2 mM MgCl2, 0.5% NP-40, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and aprotinin (2 μg/ml)] for 10 min on ice. Cells were then homogenized by 20 strokes in a tight-fitting Dounce homogenizer (Kontes disposable pestle with microtubes, Fisher Scientific). After centrifugation at 4500 rpm for 5 min to sediment the nuclei, the supernatant was transferred to a new tube and centrifuged at 14,000 rpm for 20 min. The resulting supernatant was collected as a cytoplasmic fraction and transferred to a prechilled tube. The nuclear pellet was washed three times with hypotonic lysis buffer, resuspended, and periodically vortexed in nuclear extraction buffer [20 mM Hepes (pH 7.9), 400 mM NaCl, 1 mM EDTA (pH 8.0), 1 mM EGTA (pH 7.0), phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich), and a protease inhibitor cocktail (Sigma-Aldrich)] on ice for 30 min. After centrifugation at 14,000 rpm for 10 min at 4°C, the nuclear lysates were collected in prechilled tubes.
Isolation of chromatin fraction
Chromatin isolation by small-scale biochemical fractionation has been described previously (38). Cells were harvested and lysed with a cytoplasmic buffer, buffer A [10 mM Hepes (pH7.9), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 1 mM dithiothreitol (DTT)] containing a phosphatase and protease inhibitor cocktail, and Triton X-100 to a final concentration of 0.1% for 8 min on ice. The supernatant was separated by centrifugation at 14,000 rpm for 20 min at 4°C. The nuclear pellet was lysed with buffer B (3 mM EDTA, 0.2 mM EGTA, and 1 mM DTT) with phosphatase and protease inhibitor cocktail for 30 min on ice. The chromatin pellet obtained following centrifugation at 14,000 rpm for 10 min at 4°C was resuspended in an SDS sample buffer and boiled for 10 min at 90°C; the chromatin-associated proteins were analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting.
Isolation of the nucleosomes
Nucleosomes were isolated as previously described (39). Cells were lysed with a hypotonic lysis buffer [10 mM tris-Cl (pH 8.0), 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 1.5% Triton-X, 5 mM sodium butyrate, 1 mM Na3VO4, and a phosphatase inhibitor cocktail] with rotation at 4°C for 10 min. After centrifugation at 400g for 10 min at 4°C, the cell pellet was collected, washed three times with the hypotonic lysis buffer and once with a wash buffer [10 mM tris-Cl (pH 8.0), 1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 5 mM sodium butyrate, 1 mM Na3VO4, and a phosphatase inhibitor cocktail], resuspended in the wash buffer and 0.8 M H2SO4 (v/v: 1:1), and kept in a rotator at 4°C overnight. Acid-extracted nucleosomes were precipitated by centrifugation at 13,000 rpm for 10 min at 4°C, dried, and dissolved in water.
Immunofluorescence microscopy
Cells were seeded in chamber slides (Lab-Tek), and after the indicated treatments, they were washed with PBS, fixed with 4% paraformaldehyde, permeabilized for 30 min with 0.2% Triton X-100 in PBS, and blocked for 30 min in a blocking buffer (3% BSA and 2% horse serum) at RT. After incubation with the indicated primary antibodies in blocking buffer overnight at 4°C, the cells were incubated with secondary fluorescent antibodies for 60 min at RT. After final washing, we added Vectashield Mounting Medium with DAPI (Vector Laboratories). Images were captured using a confocal microscope (Olympus FluoView FV1000). To detect TIE2 and ANG1 in vivo, GSC-7-2 and GSC-20 cells were implanted in the brains of nude mice (day 0); the mice were treated with radiation (10 Gy) on day 30 (for GSC-20) and day 56 (for GSC-7-2). Mice were sacrificed 24 hours after IR, and their brains were extracted, formalin-fixed, and paraffin-embedded. Sections (5 μm) were deparaffinized and rehydrated. Antigen retrieval was performed using a prewarmed 10 mM citric acid (pH 6.0) buffer. The slides were incubated for 10 min in a pressure cooker, allowed to cool down, and blocked with 5% BSA. Primary antibodies were added overnight at 4°C, followed by biotinylated anti-rabbit for 1 hour. DyLight594-conjugated streptavidin was added for 30 min at RT. The signal was amplified using biotinylated anti-streptavidin for 30 min at RT, followed by the additional step of DyLight594-conjugated streptavidin for 15 min. Nuclear staining was performed with DAPI (Invitrogen), and images were captured with a confocal microscope (Olympus FluoView FV1000). All animal experimentation was approved by the Institutional Animal Care and Use Committee and performed at the veterinary facilities of The University of Texas MD Anderson Cancer Center in accordance with institutional guidelines.
Enzyme-linked immunosorbent assay
After irradiation, the concentrations of ANG1 and ANG2 in the conditioned medium were measured using the Human Angiopoietin-1 and Human Angiopoietin-2 Quantikine ELISA Kits (R&D Systems), respectively, according to the manufacturer’s instructions. To measure TIE2 protein concentration in the cytoplasm and nucleus, cells were treated for 30 min with different ligands [ANG1 (400 ng/ml), ANG2 (400 ng/ml), VEGF (100 ng/ml), EGF (20 ng/ml), and bFGF (20 ng/ml); Sigma-Aldrich]. Following subcellular fractionation, ELISA was performed with the Human Tie-2 Quantikine ELISA Kit (R&D Systems) according to the manufacturer’s instructions.
Lysate preparation, immunoprecipitation, and Western blot analysis
Cells were treated with ANG1 or radiation. After the indicated time periods at 37°C, the cells were incubated for 15 to 30 min at RT with 1 mM protein cross-linker dithiobis(succinimidyl propionate) (Lomant’s Reagent; Thermo Scientific) in PBS made freshly from a 0.1 M stock dissolved in dimethyl sulfoxide (40). Cells were then washed with PBS, collected with a cell scraper, and lysed with IPH buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.2% NP-40, 1 mM Na3VO4 containing phosphatase, and protease inhibitor cocktails]. After incubation on ice for 15 min, the lysates were subjected to sonication with 30× amplitude for 10 s for three cycles (total, 30 s) in a Qsonica sonicator (VWR). The supernatant was cleaned by centrifugation at 14,000 rpm for 10 min at 4°C to obtain total cell lysates. When nuclear extracts were needed, cells were lysed with cytoplasmic extraction buffer after protein cross-linking, as described before, and the nuclear pellet was lysed with IPH buffer following sonication. The lysate was subjected to immunoprecipitation with the specific antibody overnight at 4°C. The resulting mixture was incubated with protein A–agarose (for rabbit host) (Millipore) or protein G–agarose (for mouse host) (Millipore) for 30 min, washed, and separated by 4 to 20% SDS-PAGE. The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, and Western blot analysis was conducted with the indicated antibodies. Restore PLUS Western Blot Stripping Buffer (Thermo Scientific) was used to reblot the membranes as recommended by the manufacturer.
Isolation of DNA-bound proteins
Cells were irradiated and cross-linked with 1% formaldehyde for 10 min at RT. Cells were then washed with ice-cold PBS and scraped and collected by centrifugation. The nuclear extraction was prepared as described above. DNA was precipitated from the nuclear extract by adding 3 M sodium acetate (pH 5.2) and ice-cold ethanol at −80°C for 1 hour. The DNA was washed three times with ethanol and dried. The bound proteins were resolved by adding SDS sample buffer, separated by SDS-PAGE, and further processed for Western blotting as described above.
In vivo end-joining assays
Cells were exposed to a low dose of IR (1.5 Gy), and 2 hours later, they were transfected with the reporter plasmids for end-joining efficiency. For the NHEJ assay, the pEGFP-Pem1-Ad2 plasmid was used as reporter plasmid (16). The NHEJ plasmid was digested with Hind III or I-Sce I restriction enzyme for 12 to 24 hours, and the linearized plasmid was extracted from the gel using the QIAquick Gel Extraction Kit (Qiagen), following the manufacturer’s protocol. Circular pEGFP-Pem1-Ad2 (1.0 μg) or linearized pEGFP-Pem1-Ad2 (1.0 μg) was cotransfected with 0.5 μg of pDsRed2-C1 plasmid (Clontech) to determine transfection efficiency. pEGFP-C1 transfection (Clontech) was used as a control for FACS analysis. To overexpress Tie2 constructs, 0.5 μg of pcDNA3-EV, pcDNA3-Tie2 (wild type), pcDNA3-Tie2d585-586 (d585-586), and pcDNA3-Tie2SS582/86AA (SS582/86AA) with linearized pEGFP-Pem1-Ad2 (1.0 μg) were transfected into U251, HEK293, and HeLa cells. After 48 to 72 hours of transfection, the enhanced GFP (EGFP) signal was monitored by microscopy and images were obtained. Cells were harvested by trypsinization, resuspended in PBS, and analyzed by FACS analysis on a FACSCalibur instrument. The values of rejoined DNA were calculated as the ratio of green cells to red fluorescent cells. The efficiency of NHEJ was calculated relative to U251.P cells or EV-transfected cells. For siRNA-mediated knockdown in NHEJ assay, siRNAs (10 nM) were transfected 1 day after cell plating. Twenty-four hours later, linearized NHEJ plasmid was transfected and FACS analysis was performed after 48 hours. For the HR assay, the stable transfected HR reporter gene in U2OS cells, U2OS.DR-GFP, was transfected with I-Sce I restriction enzyme–expressing plasmid (pCBASceI, provided by G. Peng) (35), and FACS analyses were performed to analyze EGFP+ cells after successful digestion of the HR reporter in vivo with I-Sce I enzyme.
Plasmid rescue
After transfecting the linear NHEJ reporter plasmid into cells, the repaired NHEJ reporter plasmids were rescued. Total genomic DNA was isolated from U251.P, U251.EV, and U251.Tie2 cells using QIAamp DNA Blood Mini Kit (Qiagen). The DNA concentration was measured, and an equal amount of DNA was transformed into DH5α-competent Escherichia coli cells (Invitrogen). The transformed competent cells were plated onto LB medium containing kanamycin (50 μg/ml) to select the colonies that contained circularized NHEJ plasmid. Plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen), the frequency of NHEJ repair was determined by restriction digestion with Hind III, and the digestion pattern was evaluated by agarose gel electrophoresis.
Prediction of Tie2 NLS
The Tie2 NLS signal was identified using the PredictProtein Web site (www.predictprotein.org) (41).
In vitro kinase assay
In vitro kinase assays were conducted with 200 ng of purified glutathione S-transferase (GST)–tagged TIE2 active protein (SignalChem, no. T04-11G) or the His-tagged TIE2 with deleted kinase domain (Creative BioMart, no. TEK-153H) in kinase buffer [25 mM MOPS (pH 7.2), 12.5 mM β-glycerol phosphate, 20 mM MgCl2, 12.5 mM MnCl2, 2 mM EDTA (pH 8.0), 5 mM EGTA (pH 7.0), and 0.25 mM DTT] with the addition of 20 μM adenosine triphosphate (ATP) for cold reactions, and 10 μM ATP mixed with 10 μCi of [γ-32P]ATP for radioactive reactions. The substrates were added to the reaction mixture and incubated at 37°C for 30 min. The following substrates were used in this study: core histone for calf thymus (Roche, no. 10223565001), histone H3.3 (New England Biolabs, no. M2507S), and histone H4 (New England Biolabs, no. M2504S). The kinase reaction was stopped by the addition of 4× protein sample buffer (Invitrogen) and boiled for 5 min, followed by SDS-PAGE (16%) analyses. Phosphorylated histone proteins were detected by autoradiography or Western blotting. The poly (4:1 Glu, Tyr) peptide (SignalChem, no. P61-58) was used as universal substrate for protein tyrosine kinases.
Generation of H4pY51 antibody
Two histone H4 peptides were used to generate a phospho–histone H4 antibody: acetyl-GLI[pY]EETRGVL-Ahx-C-amide (phosphopeptide) and acetyl-GLIYEETRGVL-Ahx-C-amide (nonphosphopeptide). These sequences showed no homology with other proteins but histone H4 (42). H4pY51 antibodies were generated by 21st Century Biochemicals according to their standard protocols.
Phospho-tag gel analysis
SuperSep Phos-tag (50 μM) 12.5%, 13-well gels (Wako Pure Chemicals Industries Ltd.) were used to detect phosphorylated proteins by SDS-PAGE using an analogous Phos-tag complex with two manganese (II) ions, Mn2+–Phos-tag. To analyze the phosphoproteins, an in vitro kinase assay was performed, and the samples were then loaded onto a Phos-tag gel; the proteins were transferred to PVDF membranes according to the manufacturer’s protocol. Western blot analyses were conducted using specific antibodies. Alkaline phosphatase (Roche) was used to dephosphorylate the proteins after the in vitro kinase assay for 1 hour at 37°C.
MS analysis
Nucleosome proteins were prepared as described above. Nano–LC-MS/MS (liquid chromatography–MS/MS) was performed on a Thermo Finnigan LTQ Orbitrap Velos (Thermo Scientific), coupled with an Eksigent NanoLC Ultra nano-HPLC (AB SCIEX). The sample was injected onto a nano-trap column (100 μm inner diameter × 1 cm, C18 PepMap100) with an autosampler and then passed down a C18 reversed-phase home-packed column (SB-C18, Zorbax, 5 μm) (Agilent). The flow rate was 400 nl/min with 60-min LC gradient, where mobile phases are A [5% acetonitrile (ACN) and 0.1% formic acid (FA)] and B (100% ACN and 0.1% FA). Eluted peptides were sprayed through a charged emitter tip (PicoTip emitter, New Objective, 10 ± 1 μm) into the mass spectrometer. Parameters included the following: spray voltage at +2.0 kV, Fourier transform MS mode for MS acquisition of precursor ions (60,000 resolution); ion trap MS mode for subsequent MS/MS of top six precursors selected; and fragmentation accomplished via collision-induced dissociation. Proteome Discoverer version 1.2 (Thermo Scientific) was used for protein identification and modification analysis. Parameters included the following: selection of the enzyme as trypsin; maximum missed cleavages = 2; variable modifications include oxidation (M) and tyrosine phosphorylation; precursor ion tolerance was at 0.02 dalton; and MS/MS fragment tolerance was at 0.6 dalton. The significance of a peptide match was based on expectation values (<0.05).
Purification of H4 recombinant proteins
Full-length human histone H4 cDNA was cloned into the pET-32a(+) vector (Novagen, no. 69015-3) between Xho I and BamH I sites. Histone H4 wild-type and two other mutant (H4Y51F and H4Y88F) plasmids were transformed into BL21 (DE3)–competent cells (New England Biolabs), and proteins were expressed with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction at 18°C overnight and purified according to the manufacturer’s protocol (Novagen). Briefly, the cell pellet was lysed with lysis buffer [50 mM tris-Cl (pH 7.5), 200 mM NaCl, 1.0% NP-40, and 0.1% Na-deoxycholate with a protease inhibitor cocktail] on ice for 30 min, and the supernatant was collected by centrifugation at 13,000 rpm for 10 min at 4°C. His-tagged histone H4 proteins were precipitated using Ni-nitrilotriacetic acid resins (Sigma-Aldrich) with rotation at 4°C for 1 hour. The resins were washed three times with lysis buffer and eluted with lysis buffer containing 150 mM imidazole. Histone H4 protein expression was confirmed by colloidal blue staining (Invitrogen) in addition to Western blotting using antibodies against His-tag and H4.
Phosphotyrosine binding SH2 domain array
The cloning of the human SH2 domain library has been described previously (21). All SH2 domains were expressed as GST fusions in E. coli and purified on glutathione-Sepharose beads. The recombinant SH2 domains were arrayed onto nitrocellulose-coated glass slides (Oncyte Avid slides, Grace Bio-Labs) using a robotic pin arrayer as previously described (22). A list of the SH2 domains on this array is given in fig. S13. The fluorescent labeling of the biotinylated peptide probe and slide binding has also been previously described (22). Fluorescent signal was detected using a GeneTAC LSIV scanner (Genomic Solutions). The peptides used in this array [phosphopeptide: Biot-Ahx-KRISGLI(pY)EETRGVL-amide; nonphosphopeptide: Biot-Ahx-KRISGLIYEETRGVL-amide] were synthesized by 21st Century Biochemicals.
Supplementary Material
Acknowledgments
We thank A. M. Sutton (Department of Scientific Publications, MD Anderson Cancer Center, Houston, TX) for scientific editorial assistance and X. Fan and J. Gumin for technical assistance (Brain Tumor Program, MD Anderson Cancer Center, Houston, TX). We acknowledge F. Bussolino (University of Torino, Italy) for providing the pcDNA3-Tie2 plasmid; G. Peng (University of Texas MD Anderson Cancer Center, Houston, TX) for providing U2OS.DR-GFP cells and pCBASceI; J. Kwon (Ewha Womans University, Seoul, Republic of Korea) for providing Flag-H2AX– and Flag-H2AX S139A–expressing 293T cells; V. Gorbunova (University of Rochester, Rochester, NY) for providing pEGFP-Pem1-Ad2 plasmid; and W. A. Barton (Virginia Commonwealth University, Richmond, VA) for providing pcDNA3.1-Tie2-myc. Funding: This work was supported by NIH grants R01 NS069964 and P50 CA127001 and Cancer Center Support Grant P30CA01667 (Sequencing and Microarray, Flow Cytometry and Cellular Imaging, and Research Animal Support Facilities). The Protein Array Core was supported by the Cancer Prevention Research Institute of Texas (RP130432) and the Centre for Environmental and Molecular Carcinogenesis at MD Anderson Cancer Center. Author contributions: M.B.H., J.F., and C.G.-M. developed the original hypothesis, designed the study, analyzed the data, and wrote the manuscript. R.S. carried out in vitro experiments. D.G.J. contributed to the original hypothesis and assisted with study design and data interpretation. M.T.B. and S.S.C.L. carried out and interpreted the SH2 high-throughput array. Z.L. guided the in vitro kinase assays. F.F.L. and E.P.S. provided GSCs and guided the in vivo experiments. R.E. was involved with the in vivo experiments. K.R.G. and N.C.-S. carried out immunofluorescence studies and analyses. J.T. and M.-C.H. assisted with the study design and data interpretation. H.J. assisted with discussion of results. X.L. carried out and interpreted the mass spectrometry experiments. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1501290/DC1
Fig. S1. Additional analyses on TIE2 nuclear expression and glioma radioresistance.
Fig. S2. TIE2 nuclear translocation signal.
Fig. S3. ANG1 regulates TIE2 nuclear translocation.
Fig. S4. S582 and S586 may modulate TIE2 nuclear translocation.
Fig. S5. Additional analysis on nuclear TIE2 binding γH2AX complexes.
Fig. S6. Additional analysis of the role of nuclear TIE2 in the regulation of DNA repair via NHEJ (Hind III).
Fig. S7. Additional analysis of the role of nuclear TIE2 in the regulation of DNA repair via NHEJ (I-Sce I).
Fig. S8. Analysis of the relation of Tie2 expression and cell cycle.
Fig. S9. Kinase activity of TIE2 is required for TIE2-mediated NHEJ repair.
Fig. S10. Nuclear TIE2 is not involved in DNA repair via HR.
Fig. S11. Additional analyses on the TIE2-mediated phosphorylation of the conserved Tyr51 residue in H4.
Fig. S12. Additional analysis on TIE2/H4pY51 complexes in TIE2-negative cells.
Fig. S13. Additional analysis on H4Y51 reader.
Fig. S14. Additional analysis on TIE2/ABL1 complexes upon ANG1 or IR.
Fig. S15. Additional analyses on nuclear TIE2/DNA repair complexes.
Fig. S16. Additional analysis on TIE2/ABL1 axes in NHEJ repair.
Table S1. List of siRNA sequences used for this study.
Table S2. List of primer sequences.
Table S3. List of antibodies used for this study.
REFERENCES AND NOTES
- 1.Bao S., Wu Q., McLendon R. E., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., Rich J. N., Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hambardzumyan D., Squatrito M., Holland E. C., Radiation resistance and stem-like cells in brain tumors. Cancer Cell 10, 454–456 (2006). [DOI] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network , Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R. A., DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 8, 193–204 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Lee O.-H., Xu J., Fueyo J., Fuller G. N., Aldape K. D., Alonso M. M., Piao Y., Liu T.-J., Lang F. F., Bekele B. N., Gomez-Manzano C., Expression of the receptor tyrosine kinase Tie2 in neoplastic glial cells is associated with integrin β1-dependent adhesion to the extracellular matrix. Mol. Cancer Res. 4, 915–926 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Martin V., Xu J., Pabbisetty S. K., Alonso M. M., Liu D., Lee O.-H., Gumin J., Bhat K. P., Colman H., Lang F. F., Fueyo J., Gomez-Manzano C., Tie2-mediated multidrug resistance in malignant gliomas is associated with upregulation of ABC transporters. Oncogene 28, 2358–2363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S.-C., Nakajima Y., Yu Y.-L., Xia W., Chen C.-T., Yang C.-C., McIntush E. W., Li L.-Y., Hawke D. H., Kobayashi R., Hung M.-C., Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8, 1359–1368 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Huang H., Bhat A., Woodnutt G., Lappe R., Targeting the ANGPT-TIE2 pathway in malignancy. Nat. Rev. Cancer 10, 575–585 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Bogdanovic E., Nguyen V. P. K. H., Dumont D. J., Activation of Tie2 by angiopoietin-1 and angiopoietin-2 results in their release and receptor internalization. J. Cell Sci. 119, 3551–3560 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Lukas J., Lukas C., Bartek J., More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Keogh M.-C., Kim J.-A., Downey M., Fillingham J., Chowdhury D., Harrison J. C., Onishi M., Datta N., Galicia S., Emili A., Lieberman J., Shen X., Buratowski S., Haber J. E., Durocher D., Greenblatt J. F., Krogan N. J., A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439, 497–501 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Kwon S.-J., Park J.-H., Park E.-J., Lee S.-A., Lee H.-S., Kang S. W., Kwon J., ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34, 303–313 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lee H.-S., Park J.-H., Kim S.-J., Kwon S.-J., Kwon J., A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandyopadhyay D., Mandal M., Adam L., Mendelsohn J., Kumar R., Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J. Biol. Chem. 273, 1568–1573 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Lieber M. R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seluanov A., Mittelman D., Pereira-Smith O. M., Wilson J. H., Gorbunova V., DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 101, 7624–7629 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller K. M., Jackson S. P., Histone marks: Repairing DNA breaks within the context of chromatin. Biochem. Soc. Trans. 40, 370–376 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Williamson E. A., Wray J. W., Bansal P., Hromas R., Overview for the histone codes for DNA repair. Prog. Mol. Biol. Transl. Sci. 110, 207–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oubaha M., Gratton J.-P., Phosphorylation of endothelial nitric oxide synthase by atypical PKCζ contributes to angiopoietin-1-dependent inhibition of VEGF-induced endothelial permeability in vitro. Blood 114, 3343–3351 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Hu H.-T., Huang Y.-H., Chang Y.-A., Lee C.-K., Jiang M.-J., Wu L.-W., Tie2-R849W mutant in venous malformations chronically activates a functional STAT1 to modulate gene expression. J. Invest. Dermatol. 128, 2325–2333 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S., Li C., Roy P., Ho K., Songyang Z., Pawson T., Gao Y., Li S. S.-C., Defining the specificity space of the human Src homology 2 domain. Mol. Cell. Proteomics 7, 768–784 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Espejo A., Côté J., Bednarek A., Richard S., Bedford M. T., A protein-domain microarray identifies novel protein–protein interactions. Biochem. J. 367, 697–702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaidi A., Jackson S. P., KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 498, 70–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Greuber E. K., Smith-Pearson P., Wang J., Pendergast A. M., Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 13, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J. Y. J., The capable ABL: What is its biological function? Mol. Cell. Biol. 34, 1188–1197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearl L. H., Schierz A. C., Ward S. E., Al-Lazikani B., Pearl F. M. G., Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 15, 166–180 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Tjeertes J. V., Miller K. M., Jackson S. P., Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou R.-H., Wang Y.-N., Hsieh Y.-H., Li L.-Y., Xia W., Chang W.-C., Chang L.-C., Cheng C.-C., Lai C.-C., Hsu J. L., Chang W.-J., Chiang S.-Y., Lee H.-J., Liao H.-W., Chuang P.-H., Chen H.-Y., Wang H.-L., Kuo S.-C., Chen C.-H., Yu Y.-L., Hung M.-C., EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell 30, 224–237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao A., Li H., Shechter D., Ahn S. H., Fabrizio L. A., Erdjument-Bromage H., Ishibe-Murakami S., Wang B., Tempst P., Hofmann K., Patel D. J., Elledge S. J., Allis C. D., WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R. K., Kabbaj M.-H. M., Paik J., Gunjan A., Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11, 925–933 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan K., Fang B., Koomen J. M., Mahajan N. P., H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat. Struct. Mol. Biol. 19, 930–937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson M. A., Bannister A. J., Göttgens B., Foster S. D., Bartke T., Green A. R., Kouzarides T., JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461, 819–822 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basnet H., Su X. B., Tan Y., Meisenhelder J., Merkurjev D., Ohgi K. A., Hunter T., Pillus L., Rosenfeld M. G., Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 516, 267–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H., Gomez-Manzano C., Aoki H., Alonso M. M., Kondo S., McCormick F., Xu J., Kondo Y., Bekele B. N., Colman H., Lang F. F., Fueyo J., Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: Role of autophagic cell death. J. Natl. Cancer Inst. 99, 1410–1414 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Hu R., Wang E., Peng G., Dai H., Lin S.-Y., Zinc finger protein 668 interacts with Tip60 to promote H2AX acetylation after DNA damage. Cell Cycle 12, 2033–2041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audero E., Cascone I., Maniero F., Napione L., Arese M., Lanfrancone L., Bussolino F., Adaptor ShcA protein binds tyrosine kinase Tie2 receptor and regulates migration and sprouting but not survival of endothelial cells. J. Biol. Chem. 279, 13224–13233 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Seegar T. C. M., Eller B., Tzvetkova-Robev D., Kolev M. V., Henderson S. C., Nikolov D. B., Barton W. A., Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol. Cell 37, 643–655 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wysocka J., Reilly P. T., Herr W., Loss of HCF-1–chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21, 3820–3829 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Xu, S. Chen, B. Zhu, in Nucleosomes, Histones & Chromatin, Part A, vol. 512 of Methods in Enzymology, C. Wu, C. D. Allis, Eds. (Elsevier, San Diego, CA, ed. 1, 2012). [DOI] [PubMed] [Google Scholar]
- 40.Tamburini B. A., Carson J. J., Adkins M. W., Tyler J. K., Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot. Cell 4, 1583–1590 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cokol M., Nair R., Rost B., Finding nuclear localization signals. EMBO Rep. 1, 411–415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1501290/DC1
Fig. S1. Additional analyses on TIE2 nuclear expression and glioma radioresistance.
Fig. S2. TIE2 nuclear translocation signal.
Fig. S3. ANG1 regulates TIE2 nuclear translocation.
Fig. S4. S582 and S586 may modulate TIE2 nuclear translocation.
Fig. S5. Additional analysis on nuclear TIE2 binding γH2AX complexes.
Fig. S6. Additional analysis of the role of nuclear TIE2 in the regulation of DNA repair via NHEJ (Hind III).
Fig. S7. Additional analysis of the role of nuclear TIE2 in the regulation of DNA repair via NHEJ (I-Sce I).
Fig. S8. Analysis of the relation of Tie2 expression and cell cycle.
Fig. S9. Kinase activity of TIE2 is required for TIE2-mediated NHEJ repair.
Fig. S10. Nuclear TIE2 is not involved in DNA repair via HR.
Fig. S11. Additional analyses on the TIE2-mediated phosphorylation of the conserved Tyr51 residue in H4.
Fig. S12. Additional analysis on TIE2/H4pY51 complexes in TIE2-negative cells.
Fig. S13. Additional analysis on H4Y51 reader.
Fig. S14. Additional analysis on TIE2/ABL1 complexes upon ANG1 or IR.
Fig. S15. Additional analyses on nuclear TIE2/DNA repair complexes.
Fig. S16. Additional analysis on TIE2/ABL1 axes in NHEJ repair.
Table S1. List of siRNA sequences used for this study.
Table S2. List of primer sequences.
Table S3. List of antibodies used for this study.