Abstract
Myocardial ischemia/reperfusion (I/R) injury may cause the apoptosis of cardiomyocytes as well as cardiac fibrosis, which is characterized as the transdifferentiation of fibroblasts to myofibroblasts and collagen deposition. MicroRNAs (miRNAs or miRs) have been demonstrated to be involved in myocardial I/R injury. However, the underlying molecular mechanism remains largely unclear. In the present study, mouse cardiomyocyte M6200 cells were treated with hypoxia/reoxygenation (H/R). Our data indicated that H/R treatment led to cell apoptosis, the increased expression of fibrosis-related proteins, namely collagen I, II, III, and fibronectin, as well as the downregulation of miR-142-3p in M6200 cells. Overexpression of miR-142-3p suppressed the H/R-induced apoptosis and fibrosis of M6200 cells. Bioinformatics analysis and a Dual-Luciferase reporter assay further identified high mobility group box 1 (HMGB1) as a direct target gene of miR-142-3p, and miR-142-3p negatively regulated the protein level of HMGB1 in M6200 cells. Furthermore, knockdown of HMGB1 enhanced cell proliferation whereas it inhibited the apoptosis and fibrosis of M6200 cells. In addition, TGF-β1/Smad3 signaling was suggested to be involved in the miR-142-3p/HMGB1-mediated apoptosis and fibrosis of M6200 cells treated with H/R. Taken together, the findings of the present study demonstrate that miR-142-3p inhibits H/R-induced apoptosis and fibrosis of cardiomyocytes, partly at least, by the direct inhibition of HMGB1 expression. Therefore, these findings have increased our understanding of the pathogenesis of H/R-induced myocardial injury.
Keywords: hypoxia/reoxygenation, cardiomyocyte, apoptosis, fibrosis, microRNA-142-3p, high mobility group box 1
Introduction
Ischemia/reperfusion (I/R) injury causes the apoptosis of cardiomyocytes and cardiac fibrosis, which is characterized as the transdifferentiation of fibroblasts to myofibroblasts and collagen deposition (1–3). Attenuating the apoptosis of cardiomyocytes and cardiac fibrosis effectively reduces the extent of myocardial I/R injury and the degree of heart failure (4,5). Therefore, revealing the underlying molecular mechanism responsible for these effects is important for the development of therapeutic strategies for myocardial I/R injury.
MicroRNAs (miRNAs or miRs), a class of non-coding RNAs 18–25 nucleotides in length, directly bind to the 3′-untranslational region (3′UTR) of their target miRs, which may cause further degradation of miR or inhibit protein translation (6). Through inhibition of the expression of their target genes at the post-transcriptional level, miRs have been demonstrated to play a key role in the regulation of a variety of cellular processes, such as cell proliferation, differentiation and apoptosis, as well as fibrosis (7,8). miR-142-3p has previously been reported to be associated with cardiovascular diseases (9–11). For example, miR-142-3p was upregulated in the peripheral blood mononuclear cells derived from chronic heart failure patients affected by non-ischemic dilated cardiomyopathy (12). Kee et al reported that miR-142-5p inhibited the proliferation of vascular smooth muscle cells by directly targeting B cell translocation gene 3 (13). However, whether miR-142-3p plays a role in hypoxia/reoxygenation (H/R)-induced apoptosis of cardiomyocytes and cardiac fibrosis has never previously been studied, to the best of our knowledge.
High-mobility group box 1 (HMGB1) is a non-histone, nuclear DNA binding protein that belongs to the HMGB superfamily. HMGB1 has been found to participate in the organization of DNA and regulate gene transcription, playing a role in several cellular processes, including inflammation, cell differentiation and tumor cell migration (14,15). Moreover, HMGB1 has been implicated in I/R-induced myocardial injury (16,17). The protein expression of HMGB1 is significantly increased in cardiac tissues following I/R injury, and the inhibition of HMGB1 effectively attenuates the extent of the tissue injury and improves cardiac performance (16,17). Therefore, HMGB1 is a promising therapeutic target for the treatment of myocardial I/R injury. In addition, transforming growth factor-β1 (TGF-β1)/Smad3 signaling has been found to be involved in HMGB1-mediated pulmonary fibrosis (18). However, the regulatory mechanism of HMGB1 expression during myocardial I/R injury remains largely unknown.
In the present study, we aimed to reveal the regulatory mechanism of miR-142-3p in H/R-induced apoptosis of cardiomyocytes and cardiac fibrosis. Moreover, we also studied the association between miR-142-3p and HMGB1 in H/R-treated cardiomyocytes, as well as the downstream signaling pathway.
Materials and methods
Cell culture and H/R treatment
The mouse cardiomyocyte line M6200 was purchased from ScienCell Research Laboratories (San Diego, CA, USA). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS; both from Life Technologies, Carlsbad, CA, USA) at 37°C in a humidified incubator containing 5% CO2. For H/R treatment, the M6200 cells were cultured under hypoxic conditions for 24 h, followed by reoxygenation for 1 h. Subsequently, M6200 cells were harvested for analysis.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. TaqMan MicroRNA Reverse Transcription kit (Life Technologies) was then used to synthesize cDNA, according to the manufacturer's instructions. SYBR-Green Universal qPCR Master Mix (Bio-Rad, Hercules, CA, USA) was then used to perform qPCR on an ABI 7500 thermocycler (Life Technologies) in a total volume of 20 µl reaction system, including 10 µl 2X SYBR-Green qPCR Mix, 1 µl primer (10 µmol/l), 1 µl cDNA and 7 µl H2O. The thermal cycling condition was set as 95°C for 3 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The following primer sequences were used: HMGB1 sense, GCTGACAAGGCTCGTTATGAA and antisense, CCTTTGATTTTGGGGCGGTA; fibronectin sense, 5′-TCTGTGCCTCCTATCTATGTGC-3′ and antisense, 5′-GAGGGACCACGACAACTCTTC-3′; COL1A1 sense, GCTCCTCTTAGGGGCCACT and antisense, ATTGGGGACCCTTAGGCCAT; COL1A2 sense, TCGTGCCTAGCAACATGCC and antisense, CCATAGCTGAACTGAAAACCACC; COL2A1 sense, GGGTCACAGAGGTTACCCAG and antisense, ACCAGGGGAACCACTCT CAC; COL3A1 sense, CTGTAACATGGAAACTGGGGAAA and antisense, CCATAGCTGAACTGAAAACCACC; COL4A1 sense, TCCGGGAGAGATTGGTTTCC and antisense, CTGGCCTATAAGCCCTGGT; and GAPDH sense, AGGTCGGTGTGAACGGATTTG and antisense, GGGGTCGTTGATGGCAACA. GAPDH was used as the internal control. Gene expression was determined by 2−ΔΔCt, where ΔCt = (Ctgene − CtGAPDH) and Ct is the threshold cycle.
Western blot analysis
The cells were solubilized in cold RIPA Lysis and Extraction buffer (Life Technologies), and 50 µg protein was separated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto a polyvinylidene difluoride membrane (Pierce Chemical Co., Rockford, IL, USA), which was incubated with PBS containing 5% milk overnight at 4°C; and then with rabbit anti-cleaved caspase-3 (1:100; ab32042), mouse Bcl-2 (1:100; ab692), mouse anti-fibronectin (1:200; ab2413), mouse anti-collagen I (1:100; ab90395), mouse anti-collagen II (1:100; ab3092), mouse anti-collagen III (1:100; ab7778), mouse anti-collagen IV (1:50; ab6311), mouse anti-HMGB1 (1:200; ab11354), mouse anti-TGF-β1 (1:50; ab64715), mouse anti-Smad3 (1:100; ab75512), mouse anti-GAPDH (1:200; ab8245) monoclonal antibodies and rabbit anti-phosphorylated (p-)Smad3 (1:200; ab63403) polyclonal antibody (all from Abcam, Cambridge, MA, USA) at room temperature for 3 h; and then with rabbit anti-mouse monoclonal IgG (1:5,000; ab190475; unconjugated) or goat anti-rabbit monoclonal IgG (1:10,000; ab190492; unconjugated) secondary antibodies at room temperature for 1 h, followed by chemiluminescence for visualization with an ECL kit (Pierce Chemical Co.). The relative protein expression was analyzed by Image-Pro Plus 6.0 software, represented as the density ratio versus GAPDH.
Cell proliferation assay
M6200 cells were plated into a 96-well plate, and cultured at 37°C with 5% CO2 for 12, 24, 48 or 72 h. Subsequently, 20 µl MTT (5 mg/ml; Life Technologies) was added. Following incubation at 37°C for 4 h, 150 µl dimethylsulphoxide (DMSO) was added. After incubation at room temperature for 10 min, the production of formazan was detected by determining the optical density (OD) at 570 nm, using a Multiskan FC enzyme immunoassay analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Cell apoptosis assay
An Annexin V apoptosis detection kit (Life Technologies) was used for apoptosis detection. Following H/R treatment, the M6200 cells were washed with cold phosphate buffer saline (PBS; Life Technologies) and resuspended with 500 µl binding buffer. Subsequently, 5 µl propidium iodide (PI) and 5 µl Annexin V-FITC were added and mixed. The cells were then incubated at room temperature in darkness for 30 min. Thereafter, the apoptosis of M6200 cells was analyzed using a BD C6 flow cytometer (BD Biosciences, San Jose, CA, USA).
Transfection
Lipofectamine 2000 (Life Technologies) was used to perform cell transfection according to the manufacturer's instructions. Briefly, miR-142-3p mimic, miR-142-3p inhibitor, HMGB1 siRNA and Lipofectamine 2000 were diluted with serum-free medium, respectively. The diluted Lipofectamine 2000 was added into the diluted miR-142-3p mimic, miR-142-3p inhibitor, or HMGB1 siRNA plasmid, respectively, and incubated for 20 min at room temperature; and then added into the M6200 cell suspension. The M6200 cells were then incubated at 37°C with 5% CO2 for 6 h. Subsequently, the medium in each well was replaced by DMEM with 10% FBS, and cultured for 48 h prior to the following analyses.
Bioinformatics prediction and Dual-Luciferase reporter assay
The TargetScan online software (http://www.targetscan.org/vert_60/) was used to analyze the putative target genes of miR-142-3p. For the luciferase reporter experiments, the wild-type (WT) of the 3′UTR segment of the HMGB1 gene containing the miR-142-3p binding sequences was amplified by PCR from human genomic DNA. A Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA) was used to construct the mutant type (MT) of HMGB1 3′UTR lacking complementarity with miR-142-3p seed sequence, in accordance with the manufacturer's instructions. Subsequently, the WT or MT of HMGB1 3′UTR was cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA), downstream of the Renilla luciferase gene, respectively (Amspring, Changsha, China). The M6200 cells were then transfected with the WT or MT HMGB1-3′UTR-psiCHECK-2 combined with miR-142-3p mimic or miR negative control (NC) mimic, respectively, using Lipofectamine 2000 according to the manufacturer's instructions. Following transfection for 48 h, the M6200 cells were lysed with a 1X passive lysis buffer, and lucif-erase activity was measured using the Dual-Luciferase reporter assay system (Promega) on an LMax multiwell Luminometer (Molecular Devices, Sunnyvale, CA, USA), in accordance with the manufacturer's instructions.
Statistical analysis
All data in this study are presented as the means ± SD. GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA) was used to perform statistical analysis. Comparisons between groups were performed by one-way analysis of variance (ANOVA). A P-value <0.05 was considered to indicate a statistically significant difference.
Results
Treatment with H/R induces the apoptosis and fibrosis of cardiomyocytes as well as the inhibition of miR-142-3p expression
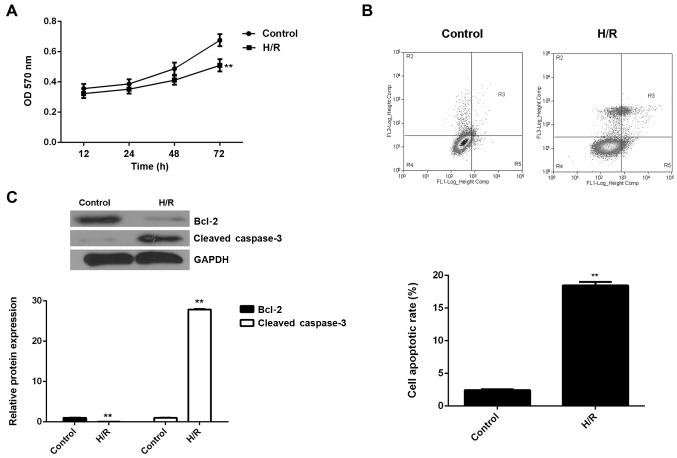
The M6200 cells were exposed to hypoxic conditions for 24 h, followed by reoxygenation for 1 h. An MTT assay was conducted to examine cell proliferation. As shown in Fig. 1A, H/R treatment led to a significant decrease in the proliferation of M6200 cells. We speculated that the downregulation of cell proliferation may be caused by the induction of cell apoptosis. Therefore, we subsequently performed flow cytometry in order to determine the level of apoptosis in M6200 cells treated with H/R. As shown in Fig. 1B, the apoptosis rate was significantly increased after H/R treatment. To further confirm these findings, western blot analysis was performed to evaluate the expression of apoptosis-related proteins. Our data showed that Bcl-2, a key inhibitor of cell apoptosis, was markedly downregulated, whereas cleaved caspase-3, a marker of cell apoptosis, was significantly upregulated in M6200 cells following H/R (Fig. 1C). Accordingly, H/R treatment inhibited the proliferation of M6200 cells probably through the induction of cell apoptosis.
Figure 1.
(A) MTT assay and (B) flow cytometry were conducted to examine the proliferation and apoptosis of M6200 cells, respectively, treated with hypoxia/reoxygenation (H/R). (C) Western blot analysis was used to examine the expression of apoptosis-related proteins in M6200 cells treated with H/R. GAPDH was used as an internal reference. Non-treated M6200 cells were used as a control. **P<0.01 vs. control.
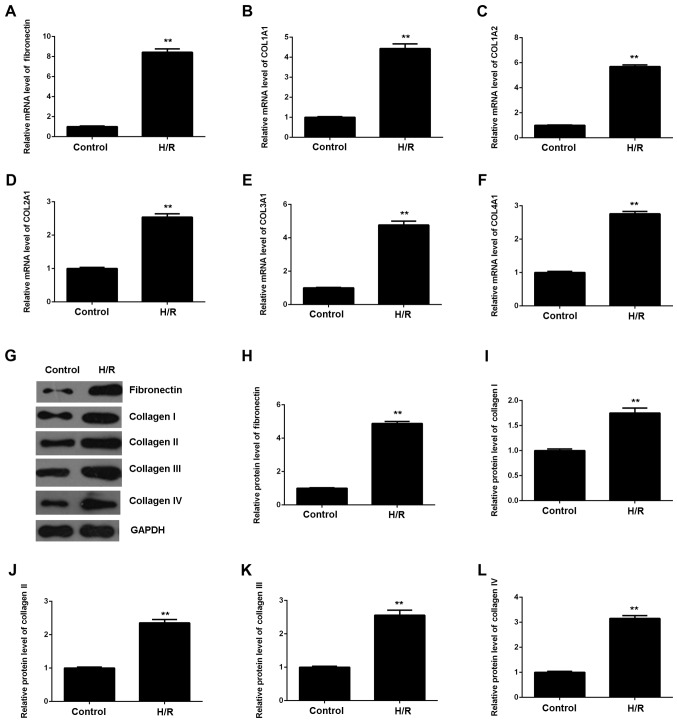
As myocardial I/R injury may also cause cardiac fibrosis, we then evaluated the expression of fibrosis-related proteins using RT-qPCR and western blot analysis. As shown in Fig. 2, the mRNA and protein expression of fibronectin and collagen I, II, III and IV were all significantly upregulated in M6200 cells following H/R treatment, when compared with the control group, respectively. Therefore, H/R treatment also induced the fibrosis of M6200 cardiomyocytes.
Figure 2.
(A–F) RT-qPCR and (G–L) western blot analysis of the mRNA and protein expression of fibronectin and collagen I, II, III and IV in M6200 cells treated with hypoxia/reoxygenation (H/R). GAPDH was used as an internal reference. Non-treated M6200 cells were used as a control. **P<0.01 vs. control.
Overexpression of miR-142-3p suppresses H/R-induced apoptosis and fibrosis of M6200 cells
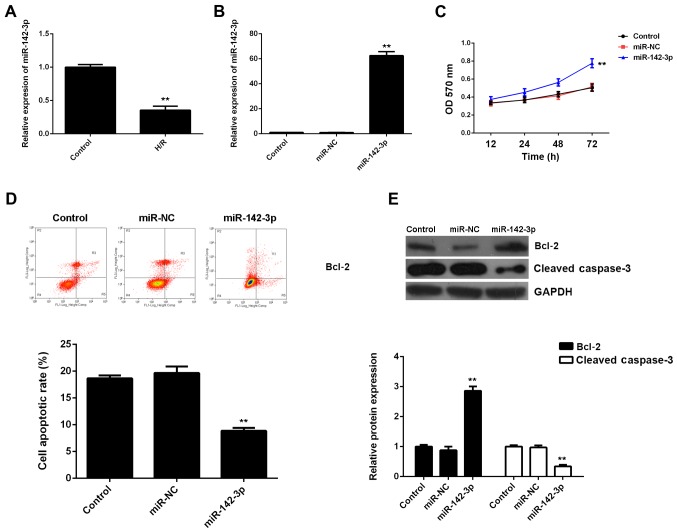
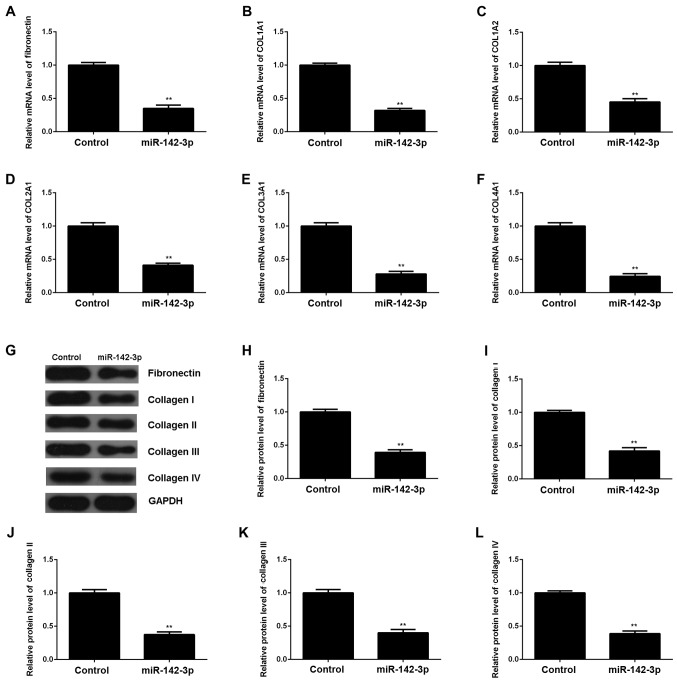
Subsequently, we evaluated the expression of miR-142-3p in M6200 cells treated with H/R. Our data showed that miR-142-3p was markedly downregulated following H/R treatment, when compared with the control group (Fig. 3A), suggesting that it may be involved in myocardial I/R injury. To further reveal the role of miR-142-3p in H/R-treated cardiomyocytes, the gain-of-function model of miR-142-3p was then conducted. Following transfection with miR-142-3p mimic, miR-142-3p expression was significantly increased compared with the control group (Fig. 3B). Thereafter, we found that cell proliferation was higher in miR-142-3p-overexpressing M6200 cells after H/R treatment when compared with that in the control group (Fig. 3C). Moreover, the apoptosis level was lower following the overexpression of miR-142-3p in M6200 cells treated with H/R (Fig. 3D), and this was accompanied by the increased expression of Bcl-2 and the decreased expression of cleaved caspase-3 (Fig. 3E). In addition, the upregulation of miR-142-3p significantly suppressed the mRNA and protein levels of fibrosis-related proteins (Fig. 4). Accordingly, the restoration of miR-142-3p expression inhibited the H/R-induced apoptosis and fibrosis of M6200 cells.
Figure 3.
(A) RT-qPCR of the mRNA expression of miR-142-3p in M6200 cells treated with hypoxia/reoxygenation (H/R). M6200 cells without H/R treatment were used as a control. **P<0.01 vs. control. (B) RT-qPCR of the mRNA expression of miR-142-3p in M6200 cells transfected with miR-negative control (NC) mimic or miR-142-3p mimic, respectively. Non-transfected M6200 cells were used as a control. **P<0.01 vs. control. (C) MTT assay and (D) flow cytometry were conducted to examine the proliferation and apoptosis of H/R-treated M6200 cells, respectively, with or without miR-142-3p overexpression. (E) Western blot analysis was used to examine the protein levels of Bcl-2 and cleaved caspase-3. (C–E) H/R-treated M6200 cells without any transfection were used as a control. **P<0.01 vs. control.
Figure 4.
(A–F) RT-qPCR and (G–L) western blot analysis of the mRNA and protein expression of fibronectin and collagen I, II, III and IV in hypoxia/reoxygenation (H/R)-treated M6200 cells with or without transfection with miR-142-3p mimic. H/R-treated M6200 cells, without transfection with miR-142-3p mimic were used as a control. **P<0.01 vs. control.
Identification of HMGB1 as a target gene of miR-142-3p and the negatively mediated expression of HMGB1 by miR-142-3p in M6200 cells
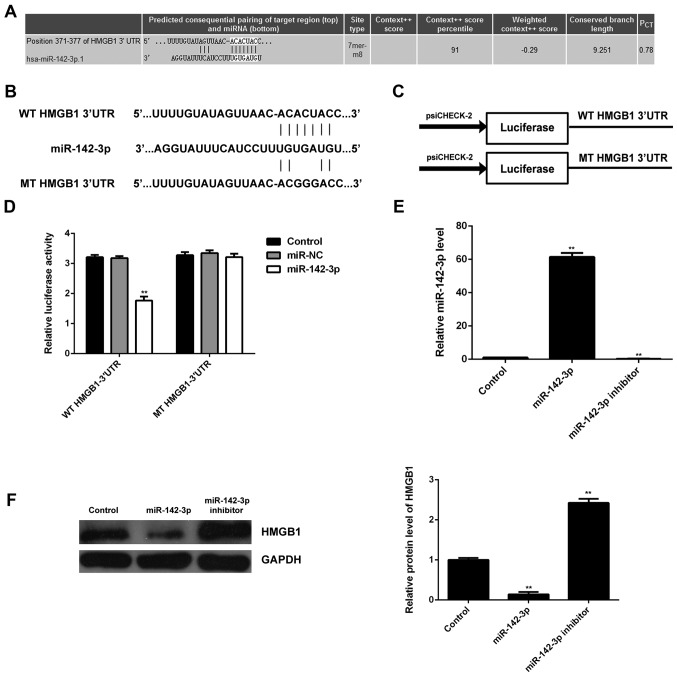
Bioinformatics analysis was performed in order to predict the target genes for miR- 142-3p. HMGB1 was predicted to be a candidate target of miR-142-3p (Fig. 5A). In order to confirm whether HMGB1 was indeed functionally targeted by miR-142-3p, the WT or MT of HMGB1 3′UTR was cloned into the psiCHECK-2 vector downstream of the Renilla luciferase gene, respectively (Fig. 5B and C). M6200 cells were then transfected with the WT or MT HMGB1-3′UTR-psiCHECK-2, combined with miR-142-3p mimic or miR-NC mimic, respectively, and the Dual-Luciferase reporter assay was then performed. Our data showed that the luciferase activity was significantly reduced in M6200 cells co-transfected with the WT HMGB1-3′UTR-psiCHECK-2 vector and miR-142-3p mimics, whereas luciferase activity was unchanged in the cells co-transfected with MT HMGB1-3′UTR-psiCHECK-2 vector and miR-142-3p mimics, when compared with the control group (Fig. 5D). Accordingly, HMGB1 was identified as a target gene of miR-142-3p.
Figure 5.
(A) Identification of high mobility group box 1 (HMGB1) as a candidate target of miR-142-3p. (B and C) The wild type (WT) or mutant type (MT) of HMGB1 3′UTR was cloned into the psiCHECK-2 vector downstream of the Renilla luciferase gene, respectively. (D) M6200 cells were transfected with the WT or MT HMGB1-3′UTR-psi-CHECK-2 reporter vectors, combined with miR-142-3p mimic or miR-negative control (NC) mimic, respectively, and luciferase activity was then determined. M6200 cells only transfected with the WT or MT HMGB1-3′UTR-psi-CHECK-2 were used as a control. **P<0.01 vs. control. (E) RT-qPCR of the expression of miR-142-3p in M6200 cells transfected with miR-142-3p mimic or inhibitor, respectively. (F) Western blot analysis was then used to examine the protein expression of HMGB1. Non-transfected M6200 cells were used as a control. **P<0.01 vs. control.
Subsequently, we examined the expression of HMGB1 in M6200 cells transfected with miR-142-3p mimic or inhibitor, respectively. As shown in Fig. 5E, transfection with miR-142-3p mimic enhanced the miR-142-3p level, whereas transfection with miR-142-3p inhibitor decreased the miR-142-3p level in M6200 cells when compared with that in the control group, respectively. Western blot analysis was performed to examine the protein level of HMGB1. Our data indicated that overexpression of miR-142-3p led to a significant decrease in the protein level of HMGB1, whereas knockdown of miR-142-3p upregulated HMGB1 protein expression in M6200 cells when compared with that in the control group, respectively (Fig. 5F). These data indicated that HMGB1 was negatively mediated by miR-142-3p at the post-transcriptional level in M6200 cells.
Knockdown of HMGB1 suppresses H/R-induced apoptosis and fibrosis of M6200 cells
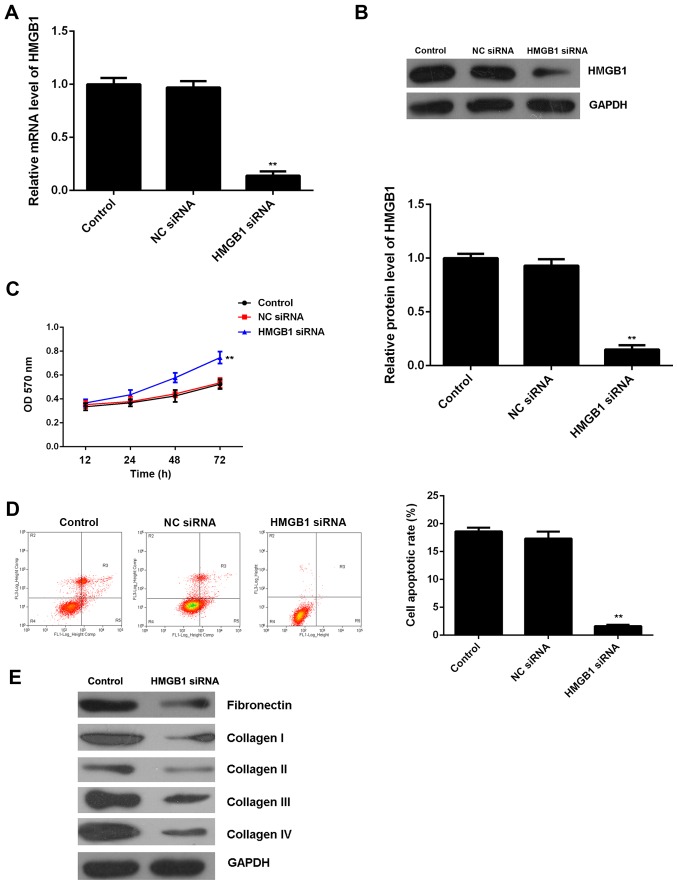
To further clarify whether HGMB1 was involved in the miR-142-3p-mediated proliferation, apoptosis and fibrosis of M6200 cells treated with H/R, HMGB1 siRNA was transfected into M6200 cells. Following transfection, both the mRNA and protein expression of HMGB1 were significantly reduced when compared with the control group, (Fig. 6A and B). Moreover, the cell proliferation was higher after knockdown of HMGB1 in the M6200 cells following H/R treatment, and this was accompanied by a lower level of cell apoptosis (Fig. 6C and D). Furthermore, we found that knockdown of HMGB1 also suppressed the protein levels of fibrosis-related proteins (Fig. 6E). Accordingly, these findings demonstrate that knockdown of HMGB1 suppresses H/R-induced apoptosis and fibrosis of M6200 cells, which is similar to the effects of miR-142-3p upregulation.
Figure 6.
(A) RT-qPCR and (B) western blot analysis of the mRNA and protein expression of high mobility group box 1 (HMGB1) in M6200 cells transfected with HMGB1 siRNA or negative control (NC) siRNA, respectively. (C) MTT assay and (D) flow cytometry were conducted to examine cell proliferation and apoptosis. Non-transfected M6200 cells were used as a control. **P<0.01 vs. control. (E) Western blot analysis of the levels of fibrosis-related proteins in M6200 cells with or without transfection of HMGB1 siRNA.
Involvement of TGF-β1/Smad3 signaling in the miR-142-3p/HMGB1-mediated apoptosis and fibrosis of M6200 cells treated with H/R
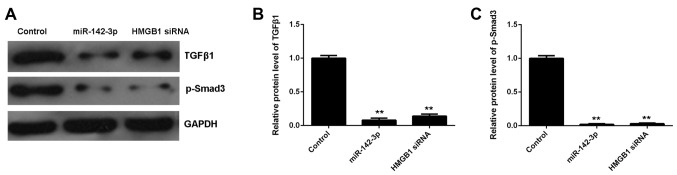
As the TGF-β1/Smad3 signaling pathway has been reported to be mediated by HMGB1 and involved in HMGB1-mediated apoptosis and fibrosis, we performed western blot analysis in order to measure the activity of this signaling pathway in M6200 cells in each group. Our data showed that the protein levels of TGF-β1 and p-Smad3 were significantly decreased in miR-142-3p-overexpressing M6200 cells treated with H/R, when compared with the control group (Fig. 7). As overexpression of miR-142-3p may inhibit the protein expression of HMGB1, we suggest that the TGF-β1/Smad3 signaling pathway is involved in the miR-142-3p/HMGB1-mediated apoptosis and fibrosis of M6200 cells treated with H/R.
Figure 7.
(A–C) Western blot analysis of the protein expression of transforming growth factor-β1 (TGF-β1) and phosphorylated (p-)Smad3 in hypoxia/reoxygenation (H/R)-treated M6200 cells transfected with miR-142-3p mimic or high mobility group box 1 (HMGB1) siRNA, respectively. Non-transfected M6200 cells treated with H/R were used as a control. **P<0.01 vs. control.
Discussion
Myocardial I/R injury may cause apoptosis of cardiomyocytes as well as cardiac fibrosis, which may further lead to heart failure. Therefore, understanding the underlying molecular mechanism is important for the development of effective therapeutic strategies for myocardial I/R injury. Herein, we examined the role of miR-142-3p in H/R-induced apoptosis and fibrosis of M6200 cardiomyocytes. Our data showed that treatment with H/R induced cell apoptosis and the upregulation of fibrosis-related proteins, namely fibronectin and collagen I, II, III and IV, which was accompanied by the downregulation of miR-142-3p in M6200 cells. Further investigation indicated that miR-142-3p suppressed H/R-induced apoptosis and fibrosis of M6200 cells partly at least, through the direct inhibition of HMGB1 expression in M6200 cells. Moreover, we found that the TGF-β1/Smad3 signaling pathway was involved in the miR-142-3p/HMGB1-mediated apoptosis and fibrosis of M6200 cells treated with H/R.
Previous studies have focused on the role of miR-142 in human cancers (19,20). For example, the serum level of miR-142-3p was found to be significantly decreased in patients with colorectal carcinoma, suggesting that miR-142-3p may serve as a novel, non-invasive biomarker in the diagnosis of colorectal cancer (21). On the contrary, miR-142-3p was markedly upregulated in the plasma of patients with head and neck squamous cell cancer, and its upregulation correlated with a poorer prognosis (22). Isobe et al reported that miR-142 enhanced the tumorigenicity of human breast cancer stem cells partly at least, by directly targeting the canonical Wnt signaling pathway and regulating the expression of miR-150 (23). In addition, the downregulation of miR-142-3p was found to enhance thyroid follicular tumorigenesis by enhancing the expression of ASH1L and MLL1 (24). Recently, miR-142-3p has been implicated in cardiovascular diseases (9). In the present study, we found that H/R treatment led to a significant decrease in the miR-142-3p level as well as inducing the apoptosis and fibrosis of M6200 cells. We speculated that the downregulation of miR-142-3p may play a role in H/R-induced apoptosis and fibrosis of M6200 cells. Therefore, M6200 cells were transfected with miR-142-3p mimic to upregulate its expression. Our data indicated that the restoration of miR-142-3p markedly suppressed the H/R-induced apoptosis and fibrosis of M6200 cells, which confirmed our speculation.
As miRs function through negative mediation of their target genes (25), we further focused on the investigation of the putative targets of miR-142-3p using bioinformatics analysis and a Dual-Luciferase reporter assay. We identified HMGB1 as a direct target gene of miR-142-3p and indicated that the expression of HMGB1 was negatively regulated by miR-142-3p in M6200 cells. HMGB1 can be produced by various types of cell such as immune cells, hepatocytes or endothelial cells, as well as cardiomyocytes, and has been found to play a critical role in cell death and survival (26–29). Moreover, HMGB1 is an important pro-inflammatory cytokine in cardiovascular diseases (30). The cardiac expression of HMGB1 is significantly increased in I/R injury, which makes it a potential therapeutic target (16). The inhibition of HMGB1 may protect against I/R-induced myocardial injury and improve cardiac performance (17). In our study, knockdown of HMGB1 markedly suppressed H/R-induced apoptosis and fibrosis in M6200 cells, similar to the effects of miR-142-3p overexpression. Based on these findings, we suggest that the suppressive role of miR-142-3p on the H/R-induced apoptosis and fibrosis of M6200 cells occurs partly at least through the direct inhibition of HMGB1 expression.
The TGF-β1/Smad3 signaling pathway plays an important role in tissue fibrosis through regulating the production of collagens as well as other extracellular matrix (ECM) components (33). Previous research has found that HMGB1 participates in the epithelial-to-mesenchymal transition (EMT) in pulmonary fibrosis through regulating the activity of TGF-β1/Smad3 signaling (18). Li et al found that HMGB1 enhanced TGF-β1 expression and triggered Smad2/3 phosphorylation, whereas TGF-β1 deficiency ameliorated HMGB1-mediated EMT with reduced p-Smad2/3 in A549 cells (18). In our study, we found that both miR-142-3p overexpression and HMGB1 knockdown inhibited the activity of the TGF-β1/Smad3 signaling pathway in M6200 cells treated with H/R. Based on these findings, we suggest that overexpression of miR-142-3p inhibits HMGB1 expression, which suppresses the activation of TGF-β1/Smad3 signaling, and subsequently suppresses ECM production and fibrosis in H/R-treated M6200 cells.
In conclusion, the present study demonstrates that miR-142-3p inhibits H/R-induced apoptosis and fibrosis of cardiomyocytes partly at least, by the direct inhibition of HMGB1 expression. Therefore, these findings have expanded our understanding of the pathogenesis of H/R-induced myocardial injury.
References
- 1.Yong KW, Li Y, Huang G, Lu TJ, Safwani WK, Pingguan-Murphy B, Xu F. Mechanoregulation of cardiac myofibroblast differentiation: implications for cardiac fibrosis and therapy. Am J Physiol Heart Circ Physiol. 2015;309:H532–H542. doi: 10.1152/ajpheart.00299.2015. [DOI] [PubMed] [Google Scholar]
- 2.Korkmaz-Icöz S, Lehner A, Li S, Vater A, Radovits T, Hegedűs P, Ruppert M, Brlecic P, Zorn M, Karck M, Szabó G. Mild Type 2 diabetes mellitus reduces the susceptibility of the heart to ischemia/reperfusion injury: identification of underlying gene expression changes. J Diabetes Res. 2015;2015:396414. doi: 10.1155/2015/396414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Y, Birnbaum GD, Perez-Polo JR, Nanhwan MK, Nylander S, Birnbaum Y. Ticagrelor protects the heart against reperfusion injury and improves remodeling after myocardial infarction. Arterioscler Thromb Vasc Biol. 2015;35:1805–1814. doi: 10.1161/ATVBAHA.115.305655. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Ji SY, Liu SZ, Jing R, Lou WJ. Cardioprotective effect of breviscapine: inhibition of apoptosis in H9c2 cardiomyocytes via the PI3K/Akt/eNOS pathway following simulated ischemia/reperfusion injury. Pharmazie. 2015;70:593–597. [PubMed] [Google Scholar]
- 5.Li R, Xiao J, Qing X, Xing J, Xia Y, Qi J, Liu X, Zhang S, Sheng X, Zhang X, Ji X. Sp1 mediates a therapeutic role of MiR-7a/b in angiotensin II-induced cardiac fibrosis via mechanism involving the TGF-β and MAPKs pathways in cardiac fibroblasts. PLoS One. 2015;10:e0125513. doi: 10.1371/journal.pone.0125513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Ji X, Wu B, Fan J, Han R, Luo C, Wang T, Yang J, Han L, Zhu B, Wei D, et al. The anti-fibrotic effects and mechanisms of microRNA-486-5p in pulmonary fibrosis. Sci Rep. 2015;5:14131. doi: 10.1038/srep14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Li W, Wang S, Wu Y, Li Z, Wang W, Liu R, Ou J, Zhang C, Wang S. MiR-142-3p attenuates the migration of CD4+ T cells through regulating actin cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans. PLoS One. 2014;9:e95514. doi: 10.1371/journal.pone.0095514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair N, Kumar S, Gongora E, Gupta S. Circulating miRNA as novel markers for diastolic dysfunction. Mol Cell Biochem. 2013;376:33–40. doi: 10.1007/s11010-012-1546-x. [DOI] [PubMed] [Google Scholar]
- 11.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 12.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, Melillo G, Rigolini R, Costa E, Crea F, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42:420–426. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 13.Kee HJ, Park S, Kwon JS, Choe N, Ahn Y, Kook H, Jeong MH. B cell translocation gene, a direct target of miR-142-5p, inhibits vascular smooth muscle cell proliferation by down-regulating cell cycle progression. FEBS Lett. 2013;587:2385–2392. doi: 10.1016/j.febslet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Gruber HE, Hoelscher GL, Bethea S, Ingram J, Cox M, Hanley EN., Jr High-mobility group box-1 gene, a potent proinflammatory mediators, is upregulated in more degenerated human discs in vivo and its receptor upregulated by TNF-α exposure in vitro. Exp Mol Pathol. 2015;98:427–430. doi: 10.1016/j.yexmp.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Liu Y, Varley P, Chang Y, He XX, Huang H, Tang D, Lotze MT, Lin J, Tsung A. High-mobility group box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res. 2015;75:1645–1656. doi: 10.1158/0008-5472.CAN-14-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diao H, Kang Z, Han F, Jiang W. Astilbin protects diabetic rat heart against ischemia-reperfusion injury via blockade of HMGB1-dependent NF-κB signaling pathway. Food Chem Toxicol. 2014;63:104–110. doi: 10.1016/j.fct.2013.10.045. [DOI] [PubMed] [Google Scholar]
- 17.Ding HS, Yang J, Chen P, Yang J, Bo SQ, Ding JW, Yu QQ. The HMGB1-TLR4 axis contributes to myocardial ischemia/reperfusion injury via regulation of cardiomyocyte apoptosis. Gene. 2013;527:389–393. doi: 10.1016/j.gene.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 18.Li LC, Li DL, Xu L, Mo XT, Cui WH, Zhao P, Zhou WC, Gao J, Li J. High-mobility group box 1 mediates epithelial-to-mesenchymal transition in pulmonary fibrosis involving transforming growth factor-β1/Smad2/3 signaling. J Pharmacol Exp Ther. 2015;354:302–309. doi: 10.1124/jpet.114.222372. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Shan WF, Jin TT, Wu GQ, Xiong XX, Jin HY, Zhu SM. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J Transl Med. 2014;12:279. doi: 10.1186/s12967-014-0279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chai S, Tong M, Ng KY, Kwan PS, Chan YP, Fung TM, Lee TK, Wong N, Xie D, Yuan YF, et al. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget. 2014;5:5725–5735. doi: 10.18632/oncotarget.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghanbari R, Mosakhani N, Asadi J, Nouraee N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S, Malekzadeh R. Downregulation of plasma miR-142-3p and miR-26a-5p in patients with colorectal carcinoma. Iran J Cancer Prev. 2015;8:e2329. doi: 10.17795/ijcp2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summerer I, Unger K, Braselmann H, Schuettrumpf L, Maihoefer C, Baumeister P, Kirchner T, Niyazi M, Sage E, Specht HM, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113:76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isobe T, Hisamori S, Hogan DJ, Zabala M, Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS, et al. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife. 2014;3:e01977. doi: 10.7554/eLife.01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colamaio M, Puca F, Ragozzino E, Gemei M, Decaussin-Petrucci M, Aiello C, Bastos AU, Federico A, Chiappetta G, Del Vecchio L, et al. miR-142-3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J Clin Endocrinol Metab. 2015;100:E59–E69. doi: 10.1210/jc.2014-2280. [DOI] [PubMed] [Google Scholar]
- 25.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Wang Z, Han G, Huang L, Jiang J, Li S. Ketamine attenuates high mobility group box-1-induced inflammatory responses in endothelial cells. J Surg Res. 2015;200:593–603. doi: 10.1016/j.jss.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 27.Lea JD, Clarke JI, McGuire N, Antoine DJ. Redox-dependent HMGB1 isoforms as pivotal co-ordinators of drug-induced liver injury: mechanistic biomarkers and therapeutic targets. Antioxid Redox Signal. 2016;24:652–665. doi: 10.1089/ars.2015.6406. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Yong Y, Li X, Hu Y, Wang J, Wang YQ, Song W, Chen WT, Xie J, Chen XM, et al. Vagal modulation of high mobility group box-1 protein mediates electroacupuncture-induced cardioprotection in ischemia-reperfusion injury. Sci Rep. 2015;5:15503. doi: 10.1038/srep15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh V, Roth S, Veltkamp R, Liesz A. HMGB1 as a key mediator of immune mechanisms in ischemic stroke. Antioxid Redox Signal. 2016;24:635–651. doi: 10.1089/ars.2015.6397. [DOI] [PubMed] [Google Scholar]
- 30.Cai J, Wen J, Bauer E, Zhong H, Yuan H, Chen AF. The role of HMGB1 in cardiovascular biology: danger signals. Antioxid Redox Signal. 2015;23:1351–1369. doi: 10.1089/ars.2015.6408. [DOI] [PubMed] [Google Scholar]
- 31.Lytle KA, Depner CM, Wong CP, Jump DB. Docosahexaenoic acid attenuates western diet-induced hepatic fibrosis in Ldlr−/− mice by targeting the TGFβ-Smad3 pathway. J Lipid Res. 2015;56:1936–1946. doi: 10.1194/jlr.M061275. [DOI] [PMC free article] [PubMed] [Google Scholar]