Abstract
The present study aimed to identify genes and microRNAs (miRNAs or miRs) that were abnormally expressed in the vastus lateralis muscle of patients with acute exacerbations of chronic obstructive pulmonary disease (AECOPD). The gene expression profile of GSE10828 was downloaded from the Gene Expression Omnibus database, and this dataset was comprised of 4 samples from patients with AECOPD and 5 samples from patients with stable COPD. Differentially expressed genes (DEGs) were screened using the Limma package in R. A protein-protein interaction (PPI) network of DEGs was built based on the STRING database. Module analysis of the PPI network was performed using the ClusterONE plugin and functional analysis of DEGs was conducted using DAVID. Additionally, key miRNAs were enriched using gene set enrichment analysis (GSEA) software and a miR-gene regulatory network was constructed using Cytoscape software. In total, 166 up- and 129 downregulated DEGs associated with muscle weakness in AECOPD were screened. Among them, NCL, GOT1, TMOD1, TSPO, SOD2, NCL and PA2G4 were observed in the modules consisting of upregulated or downregulated genes. The upregulated DEGs in modules (including KLF6 and XRCC5) were enriched in GO terms associated with immune system development, whereas the downregulated DEGs were enriched in GO terms associated with cell death and muscle contraction. Additionally, 39 key AECOPD-related miRNAs were also predicted, including miR-1, miR-9 and miR-23a, miR-16 and miR-15a. In conclusion, DEGs (NCL, GOT1, SOD2, KLF6, XRCC5, TSPO and TMOD1) and miRNAs (such as miR-1, miR-9 and miR-23a) may be associated with the loss of muscle force in patients during an acute exacerbation of COPD which also may act as therapeutic targets in the treatment of AECOPD.
Keywords: loss of muscle force, acute exacerbation of chronic obstructive pulmonary disease, differentially expressed genes, miRNAs, miR-gene regulatory network
Introduction
Chronic obstructive pulmonary disease (COPD), characterized by a fixed obstruction of the airway caused by emphysema, chronic bronchitis, or both, is a common, growing public health problem that is responsible for a huge economic health burden worldwide (1). Stable COPD may lead to declines in lung functions, such as airflow obstruction, airway function decline, and respiratory muscle fatigue, which impair patient quality of life; although acute exacerbations of COPD (AECOPD) have various definitions, they are commonly characterized by worsened dyspnea and increased volumes of phlegm and phlegm purulence, usually accompanied by hypoxemia and worsened hypercapnia (2,3). Small airway lesions (due to chronic bronchiolitis) and destruction of the alveolar walls (emphysema) are the two major features of AECOPD (4). Based on available data in 2010, COPD is one of the 6 leading causes of death, and AECOPD is associated with high morbidity (5). To date, there are no methods for preventing AECOPD, and current medical therapies for AECOPD mainly involve bronchial relaxation and the use of glucocorticoids and antibiotics, which, however, are always associated with side effects leading to unsatisfactory prognosis (6).
Smoking, malnutrition, depression and drug addiction are the risk factors affecting the quality of life for patients with AECOPD (7). Furthermore, skeletal muscle dysfunction is the most severe complication of COPD, and progression is associated with oxidative stress, skeletal muscle fiber types, systemic inflammation and mitochondrial dysfunction (1), bringing great detrimental effects to patients. In fact, skeletal muscle depletion has been adopted as a predictor of mortality in patients with COPD (8). In COPD patients, a lack of antioxidant capacity and glutathione (GSH) indicated that oxidative stress is associated with skeletal muscle dysfunction (9). Furthermore, bacterial infection is another major cause of AECOPD (10). The increasing levels of systemic inflammatory factors, such as tumor necrosis factor α (TNF-α), interleukin (IL)-6 and IL-8, may inhibit muscle shrinkage and the protein degradation of skeletal muscle, thus leading to muscle atrophy (11). Mitochondrial dysfunction, in terms of sharply increased transmembrane potential and reduced mitochondrial density, is one of the factors associated with abnormal skeletal muscle in COPD (9). Thus, further studies are warranted in order to explore the mechanism underlying the pathogenesis of AECOPD.
Previous studies have identified a number of microRNAs (miRNAs or miRs) that may have significant regulatory functions in the progression of COPD, such as miR-223, miR-1274a and miR-15b in lung tissue (12), let-7c and miR-125b (13), as well as serum miR-20a, miR-28-3p, miR-34c-5p, miR-100 and miR-7 (14). Furthermore, the expression of miR-1 in quadriceps may be responsible for muscle dysfunction in COPD (15). To the best of our knowledge, however, there have been no reports on miRNAs contributing to skeletal muscle weakness in patients with AECOPD.
In order to identify the key genes and miRNAs that may be responsible for the loss of muscle force during an acute exacerbation in COPD patients, we downloaded a gene expression profile dataset GSE10828 (16) from the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/geo) (17), which were collected from muscle samples of patients with AECOPD or stable COPD. The authors submitting this dataset identified the differentially expressed genes (DEGs) that may be associated with loss of muscle force during AECOPD, and performed gene ontology (GO) enrichment analysis (16). In the present study, which is based on the identified DEGs, we constructed a protein-protein interaction (PPI) network and conducted subsequent module analysis in order to identify genes that play critical roles in the progression of AECOPD; we also built a miR-gene regulatory network. This study aimed to deepen our understanding of the molecular mechanisms underlying the loss of muscle force in AECOPD.
Materials and methods
Affymetrix microarray data
Only one microarray dataset (GSE10828) (16) was found to be associated with AECOPD in the GEO database of NCBI (17). The annotation platform of this dataset is the GPL2891 platform (GE Healthcare/Amersham Biosciences CodeLink™ UniSet Human 20K I Bioarray, Chalfont, UK). This dataset were collected from vastus lateralis samples from 4 male patients with acute COPD and 5 male patients with stable COPD. No significant differences were found in the basic characteristics between the patients with AECOPD and stable COPD with regard to age, body mass index (BMI), forced expiratory volume in the first second (FEV1), FEV1/forced vital capacity (FVC), arterial oxygen and carbon dioxide tension (PaO2 and PaCO2), and maximal inspiratory mouth pressure (PImax), apart from C-reactive protein (CRP) levels at admission and lower quadriceps force (16).
Data preprocessing and identification of DEGs
Firstly, the microarray data in .CEL format were converted into expression measures using the GEOquery package, a package for retrieving gene expression data sets in R/Bioconductor (Bioconductor version: Release 3.1, http://www.bioconductor.org/packages/release/bioc/html/GEOquery.html) (18). The Robust Multi-array Analysis (RMA) method was used to preprocess the downloaded raw data by background adjustment, quintile normalization and summarization (19). Subsequently, the differential expression values of genes between the acute COPD samples and the stable COPD samples were calculated by t-test using the Linear Models for Microarray Data (LIMMA) package (R/Bioconductor version: Release 3.1, http://www.bioconductor.org/packages/release/bioc/html/limma.html) (20). Multiple testing correction was conducted by the Bayesian method (21). Only genes with a false discovery rate (FDR) <0.01 and |log2 Fold-Change (FC)| >1.5 were identified as DEGs. Finally, the pheatmap package of R (http://cran.r-project.org/web/packages/pheatmap/index.html) was used for hierarchical clustering (22) of DEGs based on Euclidean distance (23), and the result was visualized using heat maps.
GO analysis
GO enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool (version 6.7, http://david.abcc.ncifcrf.gov/) (24) with a p-value <0.05 as threshold.
Construction of PPI network and functional analysis of significant modules
The Search Tool for the Retrieval of Interacting Genes (STRING) (version 10, http://string-db.org/) database was used to predict the interactions between the encoded proteins of these up- and downregulated DEGs, respectively (25). Only PPI pairs with a combination score >0.4 were included. Cytoscape software (version 3.2.1, http://cytoscape.org/), an open source software platform for integrating bimolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework (26), was used to visualize the resulting PPIs. The top five genes with the highest connection degrees were considered to be most closely associated with AECOPD. In addition, Cluster ONE plugin (27) was used to select the significant modules of the up- and downregulated genes in the PPI network, respectively. The top three modules according to p-values were selected for further analysis.
In addition, the biological processes of DEGs in the resulting networks were functionally analyzed using DAVID (version 6.7, http://david.abcc.ncifcrf.gov/) (24) with a p-value <0.05.
Enrichment analysis of key miRNAs and regulatory network construction of key miRNAs
We used the gene set enrichment analysis (GSEA) software (version 2.0.14, http://www.broadinstitute.org/gsea/index.jsp), based on the whole genome expression profile (28), to predict the miRNAs associated with AECOPD based on the microarray data. A p-value <0.01 was selected as threshold.
In addition, we used Cytoscape software to construct a miRNA-gene regulatory network with the miRNAs and their target DEGs. A miR-DEG regulatory network is a complicated biological system in which an miRNA functions in a variety of intracellular biological processes, such as gene regulation, cell signaling transduction, protein-protein interactions and metabolism, by regulating molecules at a transcriptional level. This provides a deeper understanding of the biological mechanisms involved (29).
Results
Screening of DEGs and hierarchical clustering analysis
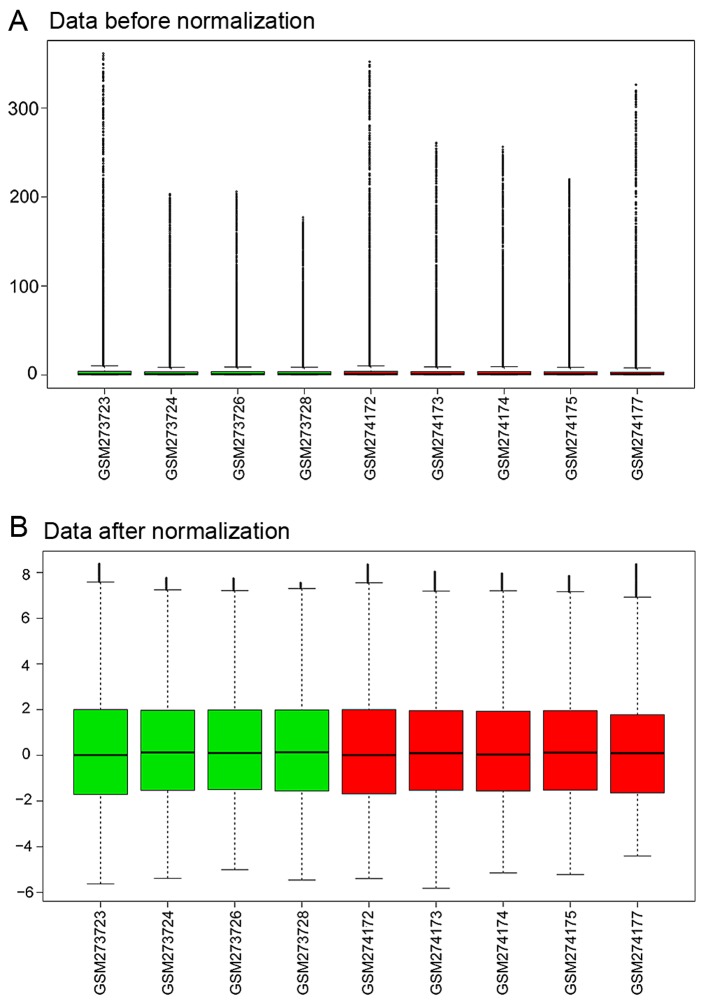
The expression profiling data were preprocessed using the R GEOquery package and were normalized by the RMA method in R (Fig. 1).
Figure 1.
Cascade figure of the normalization expression value. (A) Data before normalization. (B) Data after normalization. Green represents samples from patients with acute exacerbations of chronic obstructive pulmonary disease (AECOPD); red represents samples from patients with stable COPD.
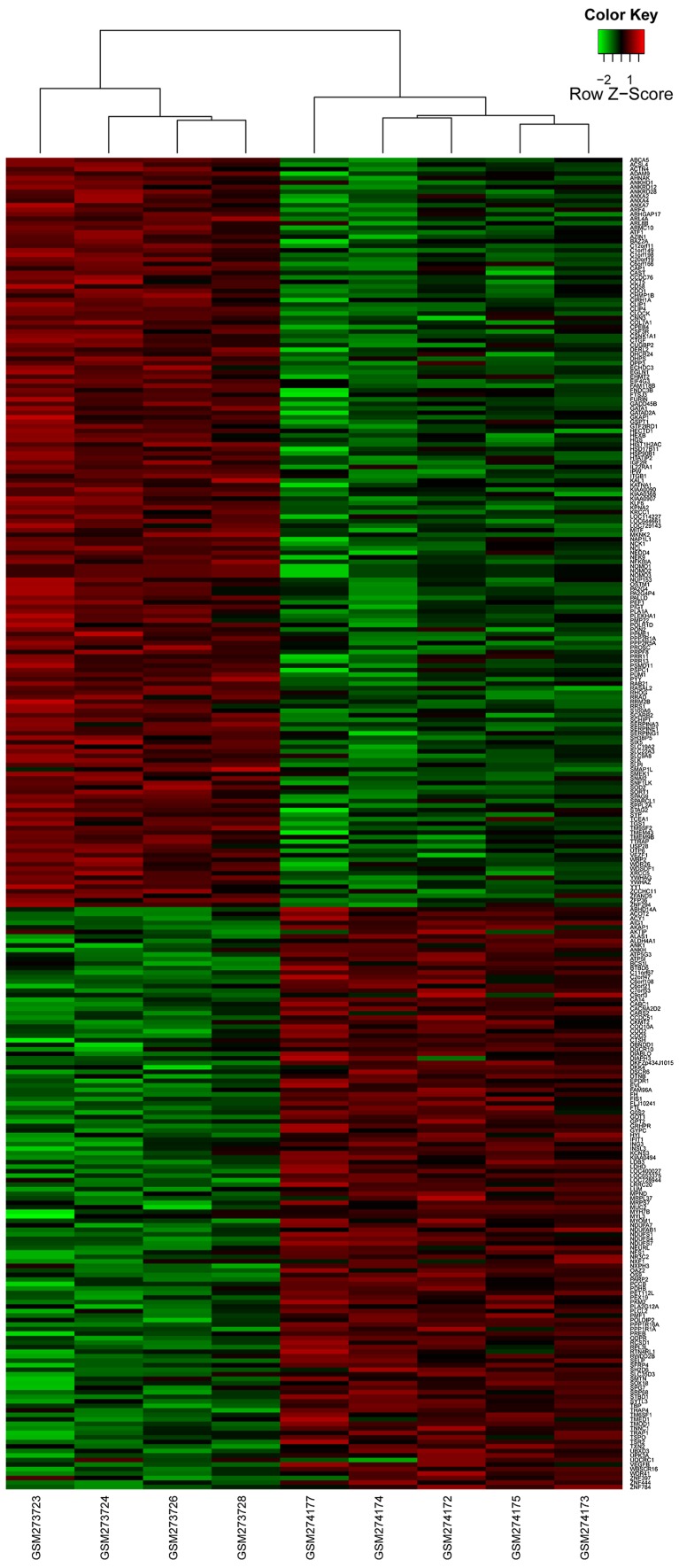
With the cut-offs of FDR <0.01 and |log2 FC| >1.5, a total of 295 DEGs were identified between the human AECOPD and stable COPD controls, including 166 upregulated genes and 129 downregulated genes (Fig. 2).
Figure 2.
Hierarchical clustering heat maps of differentially expressed genes (DEGs). The gradient color from green to red represents the expression level [acute chronic obstructive pulmonary disease (COPD) sample and stable COPD samples changes from upregulation to downregulation].
Functional annotation analysis of DEGs
According to the GO functional annotation, the top 10 enriched biological process terms included cellular respiration, generation of precursor metabolites and energy, and respiratory electron transport chain; the top 10 molecular function terms included cytoskeletal protein binding, serine-type endopeptidase inhibitor activity, protein complex binding and NADH dehydrogenase activity. Translocator protein (TSPO), superoxide dismutase 2 (SOD2) and proligeration associated protein (PA2G4) were the genes enriched in the significant functions (Table I).
Table I.
The top 10 enriched GO terms in BP and MF categories.
| Name | Term | Count | p-value | Genes |
|---|---|---|---|---|
| BP | GO:0015980 - energy derivation by oxidation of organic compounds | 11 | 7.94E-05 | NDUFS7, NDUFS4, GOT1, UQCRC1, PPP1R1A, NDUFA7, NDUFAB1, NDUFS1, PDHB, FH, SOD2 |
| BP | GO:0045333 - cellular respiration | 9 | 1.26E-04 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1, PDHB, FH, SOD2 |
| BP | GO:0006091 - generation of precursor metabolites and energy | 15 | 3.67E-04 | UQCRC1, TXN2, NDUFA7, NDUFAB1, ATP5G3, PDHB, SOD2, NDUFS7, GOT1, NDUFS4, PKM2, PPP1R1A, ATP5I, NDUFS1, FH |
| BP | GO:0022904 - respiratory electron transport chain | 7 | 4.50E-04 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1, SOD2 |
| BP | GO:0006119 - oxidative phosphorylation | 8 | 8.01E-04 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1 |
| BP | GO:0007005 - mitochondrion organization | 9 | 0.001355 | NDUFS7, FIS1, SPG7, TSPO, YWHAZ, NDUFS4, BCS1L, RRM2B, SOD2 |
| BP | GO:0042773 - ATP synthesis coupled electron transport | 6 | 0.001691 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1 |
| BP | GO:0042775 - mitochondrial ATP synthesis coupled electron transport | 6 | 0.001691 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1 |
| BP | GO:0022900 - electron transport chain | 8 | 0.001937 | NDUFS7, NDUFS4, UQCRC1, TXN2, NDUFA7, NDUFAB1, NDUFS1, SOD2 |
| BP | GO:0006120 - mitochondrial electron transport, NADH to ubiquinone | 5 | 0.003914 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 |
| MF | GO:0004857 - enzyme inhibitor activity | 16 | 1.89E-05 | CAST, PPME1, SERPING1, AZIN1, FURIN, ANXA4, ANXA2, OAZ2, SH3BP5, YWHAG, COL7A1, PPP1R1A, KAL1, SERPINE1, SERPINA3, SLPI |
| MF | GO:0008092 - cytoskeletal protein binding | 19 | 7.97E-04 | ACTN4, CNN3, TNNC1, MYL3, DIAPH3, EVL, PALLD, ANXA2, LOC729143, SPAG9, ANK1, SMTN,KATNA1, NCK1, CLIP1, ARL8B, CAP1, MYH7B, TMOD1 |
| MF | GO:0004867 - serine-type endopeptidase inhibitor activity | 7 | 0.002949 | COL7A1, KAL1, SERPINE1, SERPINA3, SLPI, SERPING1, FURIN |
| MF | GO:0019899 - enzyme binding | 18 | 0.003008 | PPME1, DIAPH3, NFKBIA, FURIN, ANXA2, SPAG9, YWHAG, PA2G4, ANK1, NEDD4, IGF2R, SERPINE1, SORT1, SYTL3, AKAP1, KPNA2, PA2G4P4, DHCR24, ADAM9 |
| MF | GO:0032403 - protein complex binding | 10 | 0.003358 | SYP, INSL3, YWHAZ, UQCRC1, ACTN4, TXN2, CTGF, NFKBIA, ITGB1, ADAM9 |
| MF | GO:0050136 - NADH dehydrogenase (quinone) activity | 5 | 0.004217 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 |
| MF | GO:0008137 - NADH dehydrogenase (ubiquinone) activity | 5 | 0.004217 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 |
| MF | GO:0003954 - NADH dehydrogenase activity | 5 | 0.004217 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 |
| MF | GO:0016655 - oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor | 5 | 0.006741 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 |
| MF | GO:0004866 - endopeptidase inhibitor activity | 8 | 0.007169 | CAST, COL7A1, KAL1, SERPINE1, SERPINA3, SLPI, SERPING1, FURIN |
GO, gene ontology; BP, biological process; MF, molecular function.
PPI network
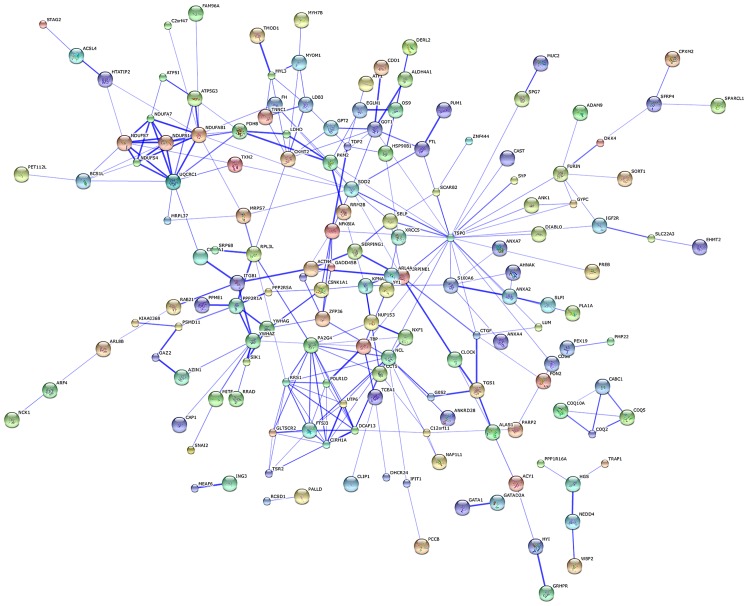
The protein-protein network of the identified DEGs was mapped using the software STRING in order to predict the protein interactions. By integrating these correlations, interaction networks between the target genes and their interactive genes were constructed (Fig. 3). In the core of the PPI network, genes belonged to more than one module. TSPO, nucleolin (NCL), NADH dehydrogenase (ubiquinone) 1, α/β subcomplex (NDUFAB1), PA2G4 and SOD2 were the top 5 protein nodes with the highest connection degrees (Table II and Fig. 3).
Figure 3.
Protein-protein interaction (PPI) network of differentially expressed genes (DEGs).
Table II.
Genes with the top 5 node degrees in PPI network.
| Gene | Degree | Log2FC | p-value |
|---|---|---|---|
| TSPO | 25 | −2.73239 | 0.007907 |
| NCL | 14 | 2.021786 | 0.001702 |
| NDUFAB1 | 13 | −27.2233 | 0.006066 |
| PA2G4 | 12 | 3.607543 | 0.006317 |
| SOD2 | 11 | 21.28352 | 0.005172 |
Degree, degree of node in PPI network; Log2FC, logarithms of amplitude changes for differentially expressed gene expression values; PPI, protein-protein interaction
Module analysis and functional analysis of DEGs in PPI network
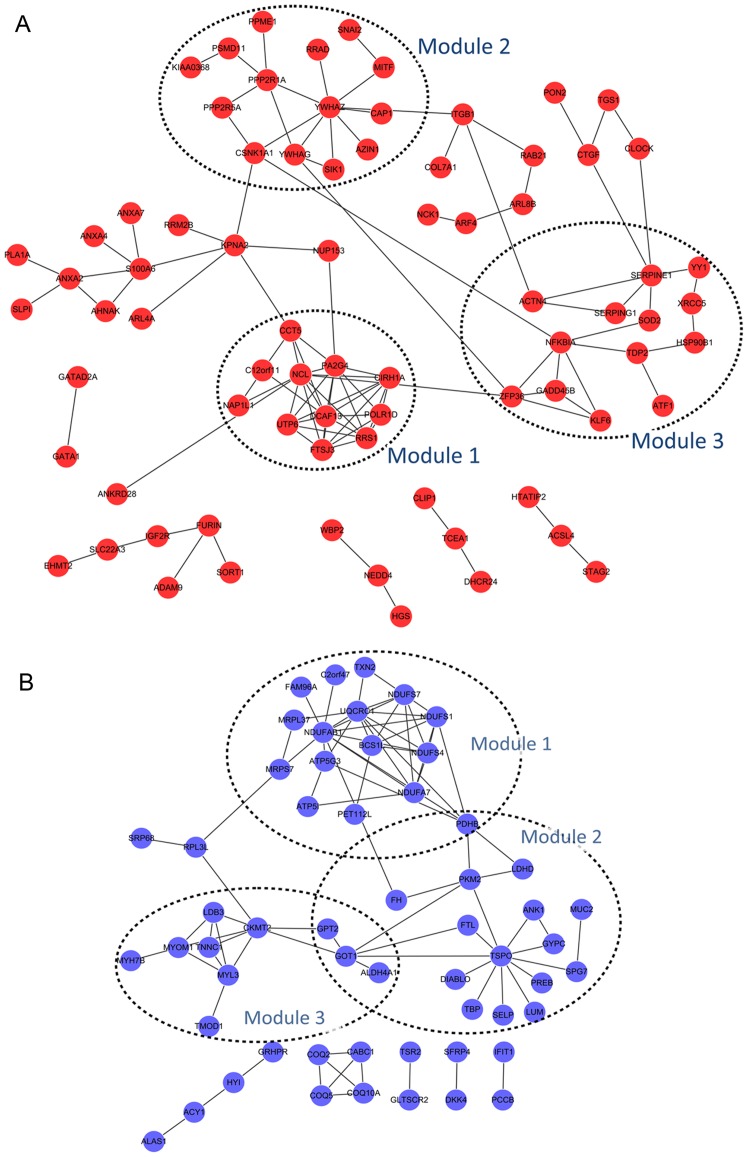
For the upregulated DEGs, three significant modules were obtained: module 1 consisting of 11 nodes (including NCL) and 33 edges (p-value=3.017E-5), module 2 of 14 nodes and 16 edges (p-value=3.115E-5), and module 3 of 13 nodes and 17 edges (p-value=3.194E-5) (Fig. 4A). Moreover, 3 significant modules were also obtained for the downregulated DEGs: module 1 consisting of 16 nodes and 35 edges (p-value=1.418E-6), module 2 of 18 nodes and 21 edges (p-value=4.592E-6), and module 3 of 10 nodes and 15 edges (p-value=3.642E-4). Pyruvate dehydrogenase (lipoamide) beta (PDHB) was commonly detected in module 1 and 2; glutamic pyruvate transaminase (alanine aminotransferase) 2 (GPT2), glutamic-oxaloacetic transaminase 1 (GOT1) and aldehyde dehydrogenase 4 family member A1 (ALDH4A1) were common in module 2 and 3 (Fig. 4B).
Figure 4.
Significant modules of differentially expressed genes (DEGs) in the protein-protein interaction (PPI) network. (A) Three modules for the upregulated DEGs, (B) three modules for the downregulated DEGs. Red nodes stand for the upregulated DEGs while blue nodes represent downregulated DEGs. Edges stand for the protein interaction and dot circle stand for the modules.
In addition, GO terms of the DEGs in the significant modules are presented in Table III. Functions of upregulated DEGs in module 1 were in the ribosome biogenesis. No GO term of the upregulated DEGs in module 2 was detected. Upregulated genes in module 3, such as X-ray repair complementing defective repair in Chinese hamster cells 5 (XRCC5) and Kruppel-like factor 6 (KLF6), were enriched in GO terms including hemopoietic or lymphoid organ development and immune system development (Table III-A). GO functions of downregulated DEGs in module 1 were in generation of precursor metabolites and energy, whereas that in module 2 was in cell death, and that in module 3 were in muscle contraction and muscle system process (Table III-B).
Table III.
GO terms of the DEGs in significant modules.
| Module | Term | Count | p-value | Genes |
|---|---|---|---|---|
| A, GO terms of the upregulated DEGs in the significant modules | ||||
|
| ||||
| Module 1 | Cluster 1 enrichment score: 4.308352643161865 | |||
| GO:0042254 - ribosome biogenesis | 5 | 7.66E-07 | DCAF13, PA2G4, UTP6, RRS1, FTSJ3 | |
| GO:0022613 - ribonucleoprotein complex biogenesis | 5 | 3.63E-06 | DCAF13, PA2G4, UTP6, RRS1, FTSJ3 | |
| GO:0006364 - rRNA processing | 4 | 2.48E-05 | DCAF13, PA2G4, UTP6, FTSJ3 | |
| GO:0016072 - rRNA metabolic process | 4 | 2.82E-05 | DCAF13, PA2G4, UTP6, FTSJ3 | |
| GO:0034470 - ncRNA processing | 4 | 2.05E-04 | DCAF13, PA2G4, UTP6, FTSJ3 | |
| GO:0034660 - ncRNA metabolic process | 4 | 3.78E-04 | DCAF13, PA2G4, UTP6, FTSJ3 | |
| GO:0006396 - RNA processing | 4 | 4.60E-03 | DCAF13, PA2G4, UTP6, FTSJ3 | |
| Module 2 | None | |||
| Module 3 | Cluster 1 enrichment score: 2.0090957996722376 | |||
| GO:0043066 - negative regulation of apoptosis | 4 | 2.51E-03 | XRCC5, HSP90B1, NFKBIA, SOD2 | |
| GO:0043069 - negative regulation of programmed cell death | 4 | 2.61E-03 | XRCC5, HSP90B1, NFKBIA, SOD2 | |
| GO:0060548 - negative regulation of cell death | 4 | 2.63E-03 | XRCC5, HSP90B1, NFKBIA, SOD2 | |
| GO:0042981 - regulation of apoptosis | 5 | 2.92E-03 | XRCC5, HSP90B1, ACTN4, NFKBIA, SOD2 | |
| GO:0043067 - regulation of programmed cell death | 5 | 3.03E-03 | XRCC5, HSP90B1, ACTN4, NFKBIA, SOD2 | |
| GO:0010941 - regulation of cell death | 5 | 3.07E-03 | XRCC5, HSP90B1, ACTN4, NFKBIA, SOD2 | |
| GO:0001666 - response to hypoxia | 3 | 5.05E-03 | HSP90B1, ACTN4, SOD2 | |
| GO:0070482 - response to oxygen levels | 3 | 5.58E-03 | HSP90B1, ACTN4, SOD2 | |
| GO:0006916 - anti-apoptosis | 3 | 1.16E-02 | HSP90B1, NFKBIA, SOD2 | |
| GO:0042592 - homeostatic process | 4 | 2.01E-02 | XRCC5, HSP90B1, SERPINE1, SOD2 | |
| Cluster 2 enrichment score: 1.7537673375612617 | ||||
| GO:0030097 - hemopoiesis | 3 | 1.50E-02 | XRCC5, KLF6, SOD2 | |
| GO:0048534 - hemopoietic or lymphoid organ development | 3 | 1.81E-02 | XRCC5, KLF6, SOD2 | |
| GO:0002520 - immune system development | 3 | 2.02E-02 | XRCC5, KLF6, SOD2 | |
| Cluster 3 enrichment score: 1.1482146347041753 | ||||
| GO:0051252 - regulation of RNA metabolic process | 6 | 9.85E-03 | ZFP36, KLF6, YY1, NFKBIA, ATF1, SOD2 | |
| GO:0006357 - regulation of transcription from RNA polymerase II promoter | 4 | 1.84E-02 | KLF6, YY1, NFKBIA, SOD2 | |
| GO:0006355 - regulation of transcription, DNA-dependent | 5 | 4.53E-02 | KLF6, YY1, NFKBIA, ATF1, SOD2 | |
|
| ||||
| B, GO terms of the downregulated DEGs in the significant modules | ||||
|
| ||||
| Module 1 | Cluster 1 enrichment score: 8.207305207832107 | |||
| GO:0006091 - generation of precursor metabolites and energy | 10 | 1.12E-12 | NDUFS7, NDUFS4, UQCRC1, TXN2, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1, PDHB | |
| GO:0006119 - oxidative phosphorylation | 8 | 1.39E-12 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1 | |
| GO:0045333 - cellular respiration | 7 | 1.92E-10 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1, PDHB | |
| GO:0022900 - electron transport chain | 7 | 5.13E-10 | NDUFS7, NDUFS4, UQCRC1, TXN2, NDUFA7, NDUFAB1, NDUFS1 | |
| GO:0042773 - ATP synthesis coupled electron transport | 6 | 1.27E-09 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1 | |
| GO:0042775 - mitochondrial ATP synthesis coupled electron transport | 6 | 1.27E-09 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1 | |
| GO:0015980 - energy derivation by oxidation of organic compounds | 7 | 2.11E-09 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1, PDHB | |
| GO:0022904 - respiratory electron transport chain | 6 | 2.53E-09 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, NDUFS1 | |
| GO:0006120 - mitochondrial electron transport, NADH to ubiquinone | 5 | 5.62E-08 | NDUFS7, NDUFS4, NDUFA7, NDUFAB1, NDUFS1 | |
| GO:0055114 - oxidation reduction | 8 | 6.79E-07 | NDUFS7, NDUFS4, UQCRC1, TXN2, NDUFA7, NDUFAB1, NDUFS1, PDHB | |
| GO:0016310 - phosphorylation | 8 | 3.09E-06 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1 | |
| GO:0006793 - phosphorus metabolic process | 8 | 1.14E-05 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1 | |
| GO:0006796 - phosphate metabolic process | 8 | 1.14E-05 | NDUFS7, NDUFS4, UQCRC1, NDUFA7, NDUFAB1, ATP5I, ATP5G3, NDUFS1 | |
| Cluster 2 enrichment score: 2.1705698016603754 | ||||
| GO:0010257 - NADH dehydrogenase complex assembly | 3 | 3.82E-05 | NDUFS7, NDUFS4, BCS1L | |
| GO:0032981 - mitochondrial respiratory chain complex I assembly | 3 | 3.82E-05 | NDUFS7, NDUFS4, BCS1L | |
| GO:0033108 - mitochondrial respiratory chain complex assembly | 3 | 5.60E-05 | NDUFS7, NDUFS4, BCS1L | |
| GO:0007005 - mitochondrion organization | 3 | 7.49E-03 | NDUFS7, NDUFS4, BCS1L | |
| GO:0043623 - cellular protein complex assembly | 3 | 1.02E-02 | NDUFS7, NDUFS4, BCS1L | |
| GO:0034622 - cellular macromolecular complex assembly | 3 | 3.62E-02 | NDUFS7, NDUFS4, BCS1L | |
| GO:0034621 - cellular macromolecular complex subunit organization | 3 | 4.47E-02 | NDUFS7, NDUFS4, BCS1L | |
| Cluster 3 enrichment score: 2.16913552816325 | ||||
| GO:0046034 - ATP metabolic process | 3 | 4.40E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009205 - purine ribonucleoside triphosphate metabolic process | 3 | 5.44E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009199 - ribonucleoside triphosphate metabolic process | 3 | 5.53E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009144 - purine nucleoside triphosphate metabolic process | 3 | 5.90E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009141 - nucleoside triphosphate metabolic process | 3 | 6.77E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009150 - purine ribonucleotide metabolic process | 3 | 7.49E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0009259 - ribonucleotide metabolic process | 3 | 8.46E-03 | ATP5I, ATP5G3, NDUFS1 | |
| GO:0006163 - purine nucleotide metabolic process | 3 | 1.33E-02 | ATP5I, ATP5G3, NDUFS1 | |
| Module 2 | Cluster 1 enrichment score: 2.0431078400255807 | |||
| GO:0008219 - cell death | 6 | 1.12E-03 | MUC2, SPG7, TSPO, PKM2, DIABLO, TBP | |
| GO:0016265 - death | 6 | 1.15E-03 | MUC2, SPG7, TSPO, PKM2, DIABLO, TBP | |
| GO:0012501 - programmed cell death | 4 | 3.31E-02 | MUC2, TSPO, PKM2, DIABLO | |
| GO:0006915 - apoptosis | 3 | 1.58E-01 | MUC2, TSPO, DIABLO | |
| Module 3 | Cluster 1 enrichment score: 4.256164391089315 | |||
| GO:0006936 - muscle contraction | 4 | 2.77E-05 | MYL3, TNNC1, CKMT2, MYOM1 | |
| GO:0003012 - muscle system process | 4 | 3.66E-05 | MYL3, TNNC1, CKMT2, MYOM1 | |
| GO:0006941 - striated muscle contraction | 3 | 1.68E-04 | MYL3, TNNC1, MYOM1 | |
GO, gene ontology; DEGs, differentially expressed genes.
Enrichment analysis of key miRNAs
In total, we identified 39 AECOPD-associated miRNAs using GSEA software at a p-value <0.01 (Table IV).
Table IV.
Enrichment analysis of key miRNAs.
| Name | Basic groups | Size | ES | NES | NOM | FDR |
|---|---|---|---|---|---|---|
| miR-23a, miR-23b | AATGTGA | 343 | 0.278676 | 1.438706 | 0 | 0.089588 |
| miR-103, miR-107 | ATGCTGC | 177 | 0.335453 | 1.486206 | 0 | 0.088895 |
| miR-221, miR-222 | ATGTAGC | 108 | 0.336992 | 1.396569 | 0 | 0.094994 |
| miR-320 | CAGCTTT | 213 | 0.343332 | 1.552349 | 0 | 0.078454 |
| miR-520f | AAGCACT | 194 | 0.344786 | 1.56579 | 0 | 0.091075 |
| miR-183 | GTGCCAT | 152 | 0.355742 | 1.541346 | 0 | 0.073964 |
| miR-524 | CTTTGTA | 365 | 0.35657 | 1.562035 | 0 | 0.084759 |
| miR-493 | ATGTACA | 266 | 0.384502 | 1.549554 | 0 | 0.074808 |
| miR-494 | ATGTTTC | 128 | 0.386803 | 1.631935 | 0 | 0.135062 |
| miR-498 | GCTTGAA | 92 | 0.397854 | 1.605433 | 0 | 0.115428 |
| miR-1, miR-206 | ACATTCC | 253 | 0.398441 | 1.603156 | 0 | 0.108254 |
| miR-323 | TAATGTG | 131 | 0.400318 | 1.56248 | 0 | 0.087979 |
| miR-373 | TTTTGAG | 194 | 0.406584 | 1.664687 | 0 | 0.125706 |
| miR-485-3p | TGTATGA | 128 | 0.41496 | 1.611675 | 0 | 0.123895 |
| miR-9 | TAGCTTT | 190 | 0.419432 | 1.627284 | 0 | 0.120835 |
| miR-17-3p | ACTGCAG | 87 | 0.421458 | 1.635642 | 0 | 0.150641 |
| miR-409-3p | AACATTC | 120 | 0.421607 | 1.671052 | 0 | 0.156583 |
| miR-422b, miR-422a | AAGTCCA | 56 | 0.428607 | 1.58016 | 0 | 0.109384 |
| miR-200a | GTAAGAT | 44 | 0.440217 | 1.63578 | 0 | 0.179569 |
| miR-518a-2 | TTTGCAG | 169 | 0.441104 | 1.684444 | 0 | 0.204788 |
| miR-202 | ATAGGAA | 84 | 0.445265 | 1.574246 | 0 | 0.100272 |
| miR-410 | GTTATAT | 76 | 0.466728 | 1.711476 | 0 | 0.295102 |
| miR-217 | ATGCAGT | 95 | 0.477771 | 1.610814 | 0.009328 | 0.113216 |
| miR-15a, miR-16, miR-15a, miR-195 | ||||||
| miR-424, miR-497 | TGCTGCT | 499 | 0.293906 | 1.427648 | 0.009452 | 0.09335 |
| miR-126 | TAATAAT | 179 | 0.379129 | 1.572559 | 0.009524 | 0.09167 |
| miR-186 | ATTCTTT | 234 | 0.390894 | 1.593438 | 0.009785 | 0.099291 |
| miR-182 | TTGCCAA | 274 | 0.355565 | 1.520188 | 0.00994 | 0.085925 |
| miR-519e | GGCACTT | 105 | 0.382867 | 1.579575 | 0.00994 | 0.104196 |
| miR-527 | CTTTGCA | 192 | 0.316954 | 1.487434 | 0.00996 | 0.089759 |
Basic groups: the targets of miRNAs; size, the number of target genes regulated by miRNA; ES, the integral enrichment of miRNAs; NES, standardization of the enrichment integration; NOM, standardization of p-value; FDR, false discovery rate.
miRNA regulatory network
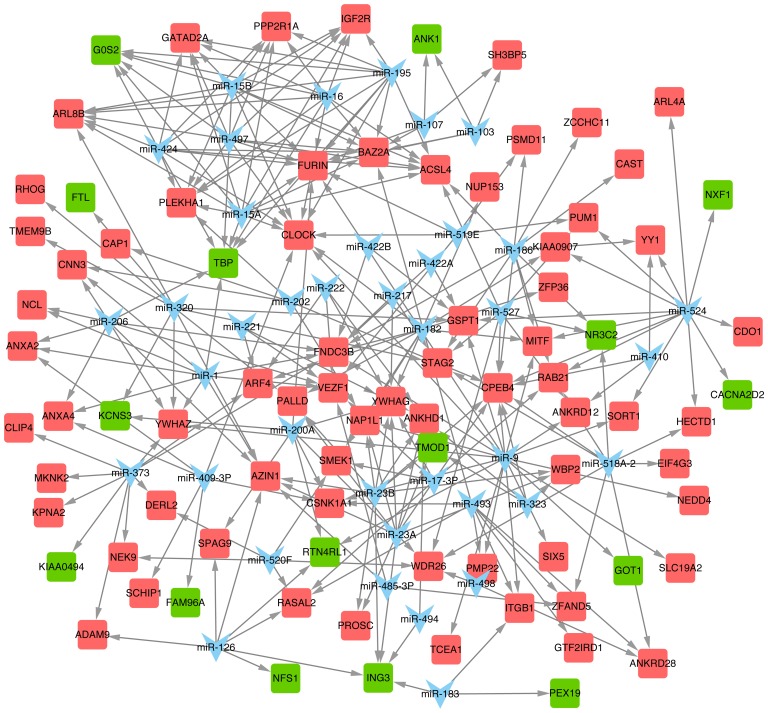
Thirty-nine predicted miRNAs and their target DEGS were constructed in an miR-DEG regulatory network (Fig. 5). miR-9, miR-524, miR-23a, miR-15a and miR-16 were the top 5 miRNAs with the most target DEGs (Table V), and their target DEGs included tropomodulin 1 (TMOD1), GOT1, NR3C2 and CPEB4 among others.
Figure 5.
The miRNA regulatory network. Blue represents miRNA; red represents upregulated differentially expressed genes (DEGs); green represents downregulated DEGs.
Table V.
miRNAs with the top 5 degrees in regulatory network.
| miRNA | Degree | miRNA | Degree |
|---|---|---|---|
| miR-9 | 20 | miR-15A | 11 |
| miR-524 | 13 | miR-15B | 11 |
| miR-23a | 10 | miR-16 | 11 |
| miR-23b | 10 | miR-182 | 11 |
| miR-320 | 10 | miR-186 | 11 |
| miR-373 | 10 | miR-195 | 11 |
| miR-1 | 9 | miR-424 | 11 |
| miR-206 | 9 | miR-497 | 11 |
| miR-518a-2 | 9 |
Discussion
Since AECOPD are one of the leading causes of death, there is an urgent need to investigate the mechanism underlying AECOPD and to develop an effective preventative strategy. Microarray-based studies have been performed to analyze the pathogenesis of AECOPD and to identify the AECOPD-associated genes in peripheral blood mononuclear cells (30) and skeletal muscle (16). However, no research investigating AECOPD-associated miRNAs has been reported, to the best of our knowledge. Hence, the present study was performed in order to predict AECOPD-associated mRNAs and miRNAs that may be responsible for the loss of muscle force, and to discuss the molecular mechanisms underlying the loss of muscle force during AECOPD.
NCL was observed in the Module 1 of the upregulated genes. It encodes a eukaryotic nucleolar phosphoprotein that is involved in the synthesis and maturation of ribosomes, which is mainly located in dense, fibrillar regions of the nucleolus. Nucleolin is one of the three components consisting of a D4Z4 repeat (31), in which the number variation is frequently detected in facioscapulohumeral muscular dystrophy (32). Thus, it can be inferred that NCL may also play a role in the loss of muscle force in AECOPD since it is associated with muscular function. Furthermore, NCL was also predicted to be regulated by miR-1 in the present study. miR-1 and miR-206, another miRNA also observed to regulate the differential gene expression herein, promote myotube formation (33). Another study reported the reduced expression of miR-1 in the quadriceps of patients with COPD, suggesting that miR-1 downregulation may contribute to COPD-associated skeletal muscle dysfunction (34), and they further observed an inverse correlation between miR-1 and Akt phosphorylation levels or HDAC4 protein levels in patients. Thus, it is likely that NCL may be downregulated during the AECOPD due to the downregulation of miR-1. SOD2 was another upregulated gene observed in module 3. An imbalance of the oxidation-antioxidant system in the body represents the principal cause of AECOPD (35). SOD2 (Mn-SOD) is a key enzyme that prevents cells from damage by eliminating the endogenous free radicals in the body (36), and increased expression was found in patients with AECOPD in the present study. Considering that samples were taken from patients with an exacerbation on day 4 of hospitalization, the antioxidant system may be activated by upregulating SOD2 in patients with AECOPD. However, this hypothesis requires further careful consideration. Additionally, Togliatto et al have reported that unacylated ghrelin (UnAG) induced skeletal muscle regeneration following hindlimb ischemia and was mediated by SOD2 (37). SOD2 may also play similar roles in muscle dysfunction during AECOPD, which suggests that SOD2 may be used as a therapeutic target in AECOPD.
In addition, XRCC5 and KLF6 were also found in module 3 of the upregulated DEGs. Both genes are involved in immune system development according to GO analysis, suggesting that the two genes may be important in AECOPD. KLF6 is a member of the Kruppel-like family of transcription factors that functions as a tumor suppressor (38). Mgbemena et al have proven that KLF6 regulated the apoptosis of lung cells through iNOS expression during respiratory syncytial virus infection (39). In addition, KLF6 may also be involved in cell atrophy during an acute exacerbation. On the other hand, XRCC5 is an ATP-dependent DNA helicase II or DNA repair protein (40). The role of XRCC5 in COPD has not been fully elucidated. However, previous findings have revealed that DNA damage or lack of DNA repair regulated the immune response to the tissue destruction in COPD (41). Therefore, XRCC5 may be a novel target for protecting against AECOPD.
TSPO displayed downregulated expression in patients with AECOPD, and this was observed in module 2. This gene encodes a protein transformation-related 18-kDa protein that assists in the recognition of the mitochondrial proteins prior to intracellular transportation (42). Otherwise, dysfunction of mitochondria caused by permeability transition would lead to the apoptosis of muscle cells, which plays a principal role in the progression of COPD (43). Thus, the downregulation of TSPO expression observed suggests that it may have an important role in the loss of muscle force occurring in AECOPD.
TMOD1 was another key downregulated gene, observed in module 3. Tropomodulin is a binding protein of tropomyosin, existing in the muscle cells, and is extracted from erythrocytes. It is necessary for many key biological functions including cell migration, differentiation and muscle contraction (44). The most important cause of the progression of AECOPD is oxygen deficiency resulting from several factors, such as the transformation of pulmonary blood vessel structures, manifested by hyperplasia and hypertrophy of pulmonary arterial muscle cells, leading to the incrassation of membranes and fibroblast proliferation (45). In the present study, TMOD1 was predicted to be regulated by miR-23a. It has been reported that the upregulation of miR-23a inhibits the development of B cells (46).
GOT1, displaying downregulated expression in AECOPD, was found in both modules 2 and 3 of the downregulated genes. GOT1 encodes glutamic-oxaloacetic transaminase 1, which is a cytoplasmic form of glutamic-oxaloacetic transaminase that is involved in amino acid metabolism. De Palma et al have also reported the decreased expression of GOT1 in patients with Ullrich congenital muscular dystrophy compared to controls (47). However, there have been no reports regarding the role of GOT1 in AECOPD, to the best of our knowledge. However, it has been reported as a putative target gene of miR-9 by Thulin et al (48), which is consistent with our predictions. miR-9 is known to play a key role in the activation of monocytes and/or macrophages during inflammatory responses (49). More importantly, it is also reported to play a role in Huntington's disease, a type of motor neuron disease (50). Furthermore, the abnormal expression of miR-9 alters motor neuron subtype differentiation as well as columnar development of spinal cords in chick embryos (51). Although the involvement of miR-9 in AECOPD has not been reported to date, to the best of our knowledge, it is possible that this miRNA may play a role in the loss of muscle force in AECOPD, and GOT1 may also be involved this process.
Finally, another two miRNAs, miR-15a and miR-16, were also found overexpressed in AECOPD patients in the present study. The pathway of Wnt signaling is known to be a promising target for mediating the development of COPD (52). A knockout of Wnt2 gene would induce lung hypoplasia and pulmonary hemorrhage caused by the abnormal muscle cells (53). Notably, miR-15a and miR-16-1 are reported to inhibit Wnt signaling (54). Thus, we hypothesized that both miR-15a and miR-16 play key roles in preventing the progression of AECOPD.
In conclusion, the present study identified some DEGs, such as NCL, GOT1, SOD2, KLF6, XRCC5, TSPO and TMOD1, and several miRNAs (e.g., miR-1, miR-9 and miR-23a) which may be associated with the pathomechanism of AECOPD. Among them, SOD2, KLF6 and XRCC5 may be involved in AECOPD due to infection via immune system development. The present study provides in-depth knowledge of the pathogenesis underlying the loss of muscle force during an acute exacerbation of COPD, despite using non-experimental methods. Since the public microarray data used in this study comes from a small sample size - 4 male patients with acute COPD and 5 male patients with stable COPD - it is necessary to validate our findings using experimental methods in a Chinese population with a larger sample size.
References
- 1.Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 2.Uzun S, Djamin R, Hoogsteden H, Aerts J, van der Eerden M. Acute exacerbations of chronic obstructive pulmonary disease. In: Kayembe JM, editor. Oncogenesis, Inflammatory and Parasitic Tropical Diseases of the Lung. InTech; 2013. pp. 77–98. Chapter 4. http://dx.doi.org/10.5772/54867. [Google Scholar]
- 3.Carrillo A, Ferrer M, Gonzalez-Diaz G, Lopez-Martinez A, Llamas N, Alcazar M, Capilla L, Torres A. Noninvasive ventilation in acute hypercapnic respiratory failure caused by obesity hypoventilation syndrome and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186:1279–1285. doi: 10.1164/rccm.201206-1101OC. [DOI] [PubMed] [Google Scholar]
- 4.Kim V, Rogers TJ, Criner GJ. New concepts in the pathobiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5:478–485. doi: 10.1513/pats.200802-014ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodhead M, Blasi F, Ewig S, Garau J, Huchon G, Ieven M, Ortqvist A, Schaberg T, Torres A, van der Heijden G, et al. Joint Taskforce of the European Respiratory Society and European Society for Clinical Microbiology and Infectious Diseases Guidelines for the management of adult lower respiratory tract infections - full version. Clin Microbiol Infect. 2011;17(Suppl 6):E1–E59. doi: 10.1111/j.1469-0691.2011.03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coventry PA, Gemmell I, Todd CJ. Psychosocial risk factors for hospital readmission in COPD patients on early discharge services: a cohort study. BMC Pulm Med. 2011;11:49. doi: 10.1186/1471-2466-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swallow EB, Reyes D, Hopkinson NS, Man WD, Porcher R, Cetti EJ, Moore AJ, Moxham J, Polkey MI. Quadriceps strength predicts mortality in patients with moderate to severe chronic obstructive pulmonary disease. Thorax. 2007;62:115–120. doi: 10.1136/thx.2006.062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabinovich RA, Bastos R, Ardite E, Llinàs L, Orozco-Levi M, Gea J, Vilaró J, Barberà JA, Rodríguez-Roisin R, Fernández-Checa JC, Roca J. Mitochondrial dysfunction in COPD patients with low body mass index. Eur Respir J. 2007;29:643–650. doi: 10.1183/09031936.00086306. [DOI] [PubMed] [Google Scholar]
- 10.Perotin JM, Dury S, Renois F, Deslee G, Wolak A, Duval V, De Champs C, Lebargy F, Andreoletti L. Detection of multiple viral and bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a pilot prospective study. J Med Virol. 2013;85:866–873. doi: 10.1002/jmv.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remels AH, Gosker HR, van der Velden J, Langen RC, Schols AM. Systemic inflammation and skeletal muscle dysfunction in chronic obstructive pulmonary disease: state of the art and novel insights in regulation of muscle plasticity. Clin Chest Med. 2007;28:537–552. vi. doi: 10.1016/j.ccm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Ezzie ME, Crawford M, Cho JH, Orellana R, Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67:122–131. doi: 10.1136/thoraxjnl-2011-200089. [DOI] [PubMed] [Google Scholar]
- 13.Van Pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, Vandesompele J, Brusselle GG. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 14.Akbas F, Coskunpinar E, Aynaci E, Oltulu YM, Yildiz P. Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp Lung Res. 2012;38:286–294. doi: 10.3109/01902148.2012.689088. [DOI] [PubMed] [Google Scholar]
- 15.Lewis A, Riddoch-Contreras J, Natanek SA, Donaldson A, Man WD, Moxham J, Hopkinson NS, Polkey MI, Kemp PR. Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD. Thorax. 2012;67:26–34. doi: 10.1136/thoraxjnl-2011-200309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crul T, Testelmans D, Spruit MA, Troosters T, Gosselink R, Geeraerts I, Decramer M, Gayan-Ramirez G. Gene expression profiling in vastus lateralis muscle during an acute exacerbation of COPD. Cell Physiol Biochem. 2010;25:491–500. doi: 10.1159/000303054. [DOI] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: Mining tens of millions of expression profiles - database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2013;14:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Šmídl V, Quinn A. Signals and Communication Technology. Springer Berlin; Heidelberg: 2006. The variational Bayes method in signal processing. [Google Scholar]
- 22.Szekely GJ, Rizzo ML. Hierarchical clustering via joint between-within distances: extending Ward's minimum variance method. J Classif. 2005;22:151–183. doi: 10.1007/s00357-005-0012-9. [DOI] [Google Scholar]
- 23.Mukherjee S, Chen Z, Gangopadhyay A. A privacy-preserving technique for Euclidean distance-based mining algorithms using Fourier-related transforms. VLDB J. 2006;15:293–315. doi: 10.1007/s00778-006-0010-5. [DOI] [Google Scholar]
- 24.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 25.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8 - a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita Y, Hayashida K, Nagai M, Inoue S, Matsumoto H, Okabe N, Reiprich TH, Sarazin CL, Takizawa M. Suzaku observation of the Ophiuchus galaxy cluster: one of the hottest cool core clusters. Publ Astron Soc Jpn Nihon Tenmon Gakkai. 2008;60:1133–1142. doi: 10.1093/pasj/60.5.1133. [DOI] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaisier CL, Pan M, Baliga NS. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012;22:2302–2314. doi: 10.1101/gr.133991.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Sun X, Chen C, Bai C, Wang X. Dynamic gene expressions of peripheral blood mononuclear cells in patients with acute exacerbation of chronic obstructive pulmonary disease: a preliminary study. Crit Care. 2014;18:508. doi: 10.1186/s13054-014-0508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/S0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 32.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 33.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;23:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig-Vilanova E, Aguiló R, Rodríguez-Fuster A, Martínez-Llorens J, Gea J, Barreiro E. Epigenetic mechanisms in respiratory muscle dysfunction of patients with chronic obstructive pulmonary disease. PLoS One. 2014;9:e111514. doi: 10.1371/journal.pone.0111514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barreiro E, de la Puente B, Minguella J, Corominas JM, Serrano S, Hussain SN, Gea J. Oxidative stress and respiratory muscle dysfunction in severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2005;171:1116–1124. doi: 10.1164/rccm.200407-887OC. [DOI] [PubMed] [Google Scholar]
- 36.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/S0891-5849(02)00905-X. [DOI] [PubMed] [Google Scholar]
- 37.Togliatto G, Trombetta A, Dentelli P, Cotogni P, Rosso A, Tschöp MH, Granata R, Ghigo E, Brizzi MF. Unacylated ghrelin promotes skeletal muscle regeneration following hindlimb ischemia via SOD-2-mediated miR-221/222 expression. J Am Heart Assoc. 2013;2:e000376. doi: 10.1161/JAHA.113.000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther. 2013;345:419–429. doi: 10.1124/jpet.113.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mgbemena V, Segovia J, Chang TH, Bose S. KLF6 and iNOS regulates apoptosis during respiratory syncytial virus infection. Cell Immunol. 2013;283:1–7. doi: 10.1016/j.cellimm.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 41.Brody JS, Spira A. State of the art. Chronic obstructive pulmonary disease, inflammation, and lung cancer. Proc Am Thorac Soc. 2006;3:535–537. doi: 10.1513/pats.200603-089MS. [DOI] [PubMed] [Google Scholar]
- 42.Becker T, Vögtle FN, Stojanovski D, Meisinger C. Sorting and assembly of mitochondrial outer membrane proteins. Biochim Biophys Acta. 2008;1777:557–563. doi: 10.1016/j.bbabio.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 43.Baraldo S, Turato G, Badin C, Bazzan E, Beghé B, Zuin R, Calabrese F, Casoni G, Maestrelli P, Papi A, et al. Neutrophilic infiltration within the airway smooth muscle in patients with COPD. Thorax. 2004;59:308–312. doi: 10.1136/thx.2003.012146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J Biol Chem. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 45.Peinado VI, Pizarro S, Barberà JA. Pulmonary vascular involvement in COPD. Chest. 2008;134:808–814. doi: 10.1378/chest.08-0820. [DOI] [PubMed] [Google Scholar]
- 46.Kong KY1, Owens KS, Rogers JH, Mullenix J, Velu CS, Grimes HL, Dahl R. miR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629–640. e621. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Palma S, Capitanio D, Vasso M, Braghetta P, Scotton C, Bonaldo P, Lochmüller H, Muntoni F, Ferlini A, Gelfi C. Muscle proteomics reveals novel insights into the pathophysiological mechanisms of collagen VI myopathies. J Proteome Res. 2014;13:5022–5030. doi: 10.1021/pr500675e. [DOI] [PubMed] [Google Scholar]
- 48.Thulin P, Wei T, Werngren O, Cheung L, Fisher RM, Grandér D, Corcoran M, Ehrenborg E. MicroRNA-9 regulates the expression of peroxisome proliferator-activated receptor δ in human monocytes during the inflammatory response. Int J Mol Med. 2013;31:1003–1010. doi: 10.3892/ijmm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuter S, Beckert H, Taube C. Take the Wnt out of the inflammatory sails: modulatory effects of Wnt in airway diseases. Lab Invest. 2015;2:177–185. doi: 10.1038/labinvest.2015.143. [DOI] [PubMed] [Google Scholar]
- 53.Goss AM, Tian Y, Cheng L, Yang J, Zhou D, Cohen ED, Morrisey EE. Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression. Dev Biol. 2011;356:541–552. doi: 10.1016/j.ydbio.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CH, Dixon RA, Ke LY, Willerson JT. Vascular progenitor cells in diabetes mellitus: roles of Wnt signaling and negatively charged low-density lipoprotein. Circ Res. 2009;9:1038–1040. doi: 10.1161/CIRCRESAHA.109.198051. [DOI] [PubMed] [Google Scholar]