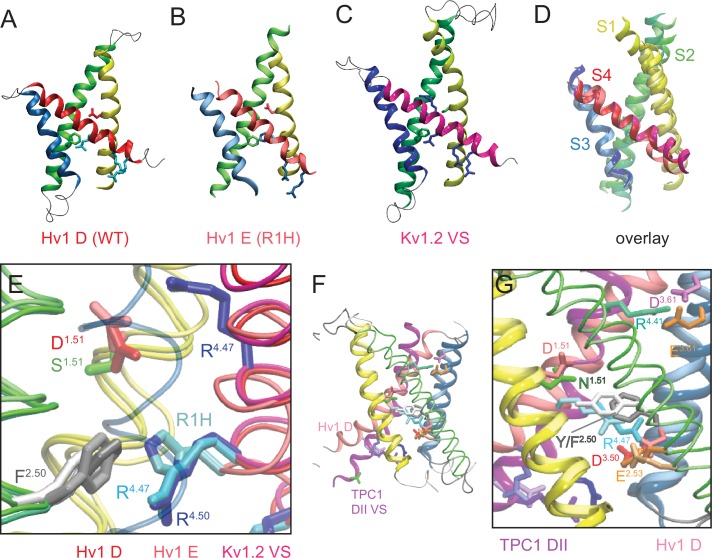
Figure 5. New Hv1 D (WT) and Hv1 E (R1H) VS domain resting-state model structures.
(A–D) Ribbon diagrams represent backbone structures in snapshots taken from MD simulations of the Hv1 D (WT) VS domain resting-state model structure (A), Hv1 E R1H mutant model structure (B), resting-state Kv1.2 VS domain Rosetta model structure (Pathak et al., 2007) that was used as the template for construction of Hv1 D (C). The model structures in A–C are overlain in D to illustrate their overall structural similarity. Transmembrane helical backbones in A–D are color coded: S1, yellow; S2, green; S3, blue; S4 red. Video 1 shows similar representations of Hv1 D, Hv1 E and Kv1.2 resting-state model structures rotated about the vertical axis. (E) An overlay of the Hv1 D, Hv1 E and Kv1.2 resting-state Rosetta VS domain model structures illustrates the relative positions of S1-S4 helical backbones (tubes colored as in A–D). Selected side chains (Hv1 D: D112/D1.51, red; F150/F2.50, light gray; R1/R205/R4.47, cyan; Hv1 E: D112/D1.51, light red; F150/F2.50, white; R1H/R205H, cyan/blue; Kv 1.2: S176/S1.51, green; F233/F2.50, dark gray; R1/R4.47 and R2/R4.50, blue) are shown in colored licorice. For clarity, only the S3 helix from Hv1 D (transparent blue tube) is shown. (F) The backbone structures of Hv1 D model and At TPC1 DII X-ray (pdb: 5EJ1) VS domains are overlain. S1, S3 and S4 helices are shown as ribbons and S2 helices are shown as tubes. Helical segments are colored as in A–D and loop regions are gray; lighter shades represent Hv1 D and darker shades represent TPC1. Selected side chains in Hv1 D/TPC1 (D/N1.51, red/green; F/Y2.50, gray/white; D/E3.61, magenta/orange) are shown in colored licorice. (G) A magnified view of the overlain Hv1 D and TPC1 structures illustrates the similar positions of selected side chains, which are shown in colored licorice (Hv1 D: D112/D1.51, pale red; F150/F2.50, light gray; E153/E2.53, pale orange; D174/D3.50, pale red; D185/D3.61, pale magenta; R1/R205/R4.47, pale cyan; TPC1: N443/N1.51, green; Y475/Y2.50, white; E478/E2.53, orange; D500/D3.50, red; E511/E3.61, orange; R531/R4.41, aqua; R537/R1/R4.47, cyan).