Abstract
Background
Pituitary stem/progenitor cells give rise to all of the endocrine cell types within the pituitary gland and are necessary for both development and gland homeostasis. Recent studies have identified several key factors that characterize the progenitor cell population. However, little is known about the factors that regulate progenitor cell differentiation and maintenance. Therefore, it is crucial to identify novel factors that help elucidate mechanisms of progenitor cell function in the developing pituitary. Our studies are the first to characterize the expression of Grainyhead-like 2 (GRHL2), a transcription factor known to regulate progenitor cell plasticity, in the developing pituitary.
Results
Our studies show GRHL2 expression is highest in the embryonic and early postnatal pituitary and is localized in pituitary progenitor cells. We demonstrate GRHL2 expression is changed in Notch2 cKO and Prop1 df/df mice, mouse models that display progenitor cell number defects. In addition, our studies indicate a potential relationship between Notch signaling and GRHL2 expression in the developing pituitary.
Conclusions
Taken together, our results indicate GRHL2 as a novel progenitor cell maker in the developing pituitary that may contribute to progenitor cell function and maintenance.
Key Terms: Grhl2, pituitary development, progenitor, Notch signaling
Introduction
The pituitary gland regulates vast physiological functions such as growth, metabolism, stress response and reproduction by releasing hormones from endocrine cells. The development of these different hormone producing cell types require putative pituitary stem cells, referred to as progenitor cells, to interpret both intrinsic and extrinsic cues to regulate cell fate choice. Pituitary progenitors express proteins required for stem cell maintenance such as HMG box transcription factors SOX2 and SOX9 and remain highly proliferative during embryonic and postnatal pituitary development (Fauquier et al. 2008; Rizzoti et al. 2013). Recent studies have demonstrated that these cells are capable of differentiating into all of the endocrine cells of the pituitary during embryonic and postnatal development (Fauquier et al. 2008; Andoniadou et al. 2013). In addition, it has been demonstrated that pituitary progenitor cells in adult mice are capable of differentiating into endocrine cells when challenged with physiological demands such as target organ ablation (Nolan et al. 2004; Rizzoti et al. 2013). These studies demonstrate that the progenitor cell population functions as the precursor cells to each of the six distinct endocrine cells of the pituitary gland. However, the regulatory factors that influence progenitor cell maintenance and differentiation during development remain elusive. It is therefore important to identify novel molecules within the progenitor cell niche that may contribute to progenitor cell function.
The Notch signaling pathway is associated with progenitor maintenance in many organs including the developing pituitary. Notch signaling it most often thought to exert self-renewal processes in progenitor cells through transcriptional activation of its downstream canonical targets Hes1 and Hey1, which have been shown to transcriptionally repress pro-differentiation genes (Ishibashi et al. 1994; Ishibashi et al. 1995; Madsen et al. 2000; Weber et al. 2014). In addition, Notch has been shown to directly regulate the expression of genes essential for progenitor maintenance and function such as Sox2 and Sox9 (Li et al. 2012). Several Notch signaling genes including, Notch2, Notch3, Dll1, Hes1 and Hey1 are expressed in Rathke’s Pouch, in progenitor cells lining the marginal zone during postnatal development and the in adult pituitary. Manipulation of the Notch signaling pathway through genetic mouse models has demonstrated the importance of Notch signaling in maintaining progenitor cell population and cell fate selection in the developing pituitary (Nantie et al. 2014; Raetzman et al. 2004; Raetzman et al. 2007; Zhu et al. 2006). Studies from our lab have shown that Notch2 conditional knockout (cKO) mice display a misplacement and progressive loss of the progenitor cell population as well as decreased proliferation during postnatal pituitary development (Nantie et al. 2014). These data demonstrate that Notch signaling is essential for maintaining the correct number of progenitor cells in the pituitary. In addition, the Notch2 cKO mice can be used as a model for decreased progenitor cell number in the pituitary.
Another important factor in controlling pituitary progenitors in both mice and humans is PROP1, a pituitary specific homeodomain transcription factor. The importance of PROP1 during pituitary development is demonstrated by the fact that mutations in PROP1 are the most identified cause of combined pituitary hormone deficiency (CPHD), accounting for approximately 50% of familial cases (Cogan et al. 1998; Ward et al. 2005). In Ames dwarf mouse (Prop1df/df) pituitaries, a hypomorphic mutation of Prop1 leads to an inability of progenitor cells to migrate from the periluminal zone. This results in an abundance of progenitors during early gland development at the expense of differentiated cells of the PIT1 lineage: somatotropes, lactotropes and thyrotropes (Ward et al. 2005; Ward et al. 2006; Pérez Millán et al. 2016). The expansion of the progenitor cells is correlated with an increase in the Notch target Hey1 (Mortensen et al. 2011). Interestingly, in both the RBPJ and Notch2 conditional knockout models, reduced Notch signaling results in decreased Prop1 expression (Nantie et al. 2014; Zhu et al. 2006). These data indicate that Notch signaling and PROP1 both control progenitor maintenance and cell specification but do so through distinct mechanisms. Taken together, these studies indicate a major role for Notch signaling and PROP1 in coordinating pituitary progenitor cell fate, suggesting that they would be useful models to identify factors important in progenitor cells.
In the current study, we have used the models discussed above to identify the transcription factor Grainyhead-like 2 (GRHL2) as a novel progenitor cell marker in the developing pituitary. While our studies are the first to characterize GRHL2 expression in pituitary gland development, in other tissues it has been identified as a marker of epithelial progenitor cells where it has been shown to regulate cellular proliferation, differentiation and migration (Chen et al. 2010; Chen et al. 2016; Saaket Varma et al. 2012; Gao et al. 2015). Of particular interest, studies have demonstrated increased expression of GRHL2 resulted in increased proliferation, blockade of differentiation and increased cellular life span of human keratinocytes (Chen et al. 2012). In addition, GRHL2 has been shown to play a crucial role in embryonic development demonstrated by the fact that Grhl2-null mice and N-ethyl-N-nitrosourea (ENU) induced Grhl2 mutant mice die embryonically due to defects in neural tube closure (Werth et al. 2010; Pyrgaki et al. 2011). In particular, these defects in tissue development in Grhl2-deficient mice are suggested to be a result of decreased expression of cellular adhesion molecules such as E-cadherin (Ecad) and Claudin 4 (CLDN4), both of which have been shown to be direct transcriptional targets of GRHL2 (Werth et al. 2010; Aue et al. 2015). Due to its ability to regulate cellular processes such as cell-to-cell interactions, cell junctions and replicative potential, all of which are known to affect progenitor cell maintenance, we sought to characterize GRHL2 expression in the pituitary. Our studies demonstrate that GRHL2 expression is most robust in the embryonic and early postnatal pituitary and is predominately localized to pituitary progenitor cells. Our studies also show a significant decrease in Grhl2 expression in Notch2 cKO pituitaries and after chemical inhibition of Notch signaling via treatment. In addition, Grhl2 expression is increased in Prop1 mutants at a time when the Notch target Hey1 is increased. These studies are the first to characterize GRHL2 as a progenitor cell marker in the developing pituitary and correlate its expression with Notch signaling.
Results
Characterization of GRHL2 in the embryonic, postnatal and adult pituitary
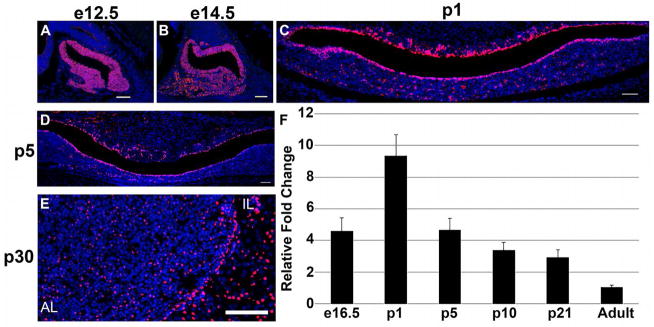
Expression of GRHL2 in the developing mouse pituitary has yet to be characterized. Therefore we examined the spatial and temporal expression patterns of GRHL2 in the embryonic, postnatal and adult pituitary. At e12.5 and e14.5, the vast majority of cells in Rathke’s pouch are GRHL2-immunopositive (Figure 1A and B). By postnatal day 1 (p1) GRHL2 expression is restricted to the marginal cells lining the lumen with a few positive cells scattered in the dorsal intermediate lobe (IL) and anterior lobe (AL) parenchyma (Figure 1C). At p5 there is a decrease in GRHL2 expression, with the most apparent reduction in the cleft cells of the intermediate lobe (IL) (Figure 1D). At p30, GRHL2 expression is maintained in the cleft cells but overall expression is reduced compared to the embryonic and postnatal pituitary (Figure 1E). In addition, we used qRT-PCR to determine relative mRNA levels of Grhl2 at e16.5, p1, p5, p10, and p21, compared to adult pituitaries. This data parallels the protein analysis of GRHL2 expression in that it shows Grhl2 mRNA is expressed throughout pituitary gland development. In addition, this data indicates that Grhl2 expression is highest during the early postnatal time period with a peak at p1, compared to adult pituitaries (Figure 1F). Taken together, this data shows GRHL2 is maintained throughout pituitary maturation and appears to peak during postnatal development.
Figure 1.
GRHL2 is expressed in the embryonic, postnatal and adult pituitary. GRHL2 is expressed throughout the pituitary at e12.5 (A) and is highly expressed in the cells lining the cleft as well as the intermediate (IL) and anterior lobe (AL) of the pituitary at e12.5 (A), e14.5 (B) and p1 (C). GRHL2 expression appears to be reduced at p5 (D). GRHL2 expression persists in the IL and AL lobes of p30 pituitaries. Grhl2 mRNA is detected in the embryonic, postnatal and in adult pituitaries. Ghrl2 expression peaks at p1 when compared to adult levels (F). Scale bar = 50 μm. n=3–4 (immunohistochemistry). n=3–4 (qRT-PCR).
GRHL2 is predominantly present in the non-hormone producing cells of the developing pituitary
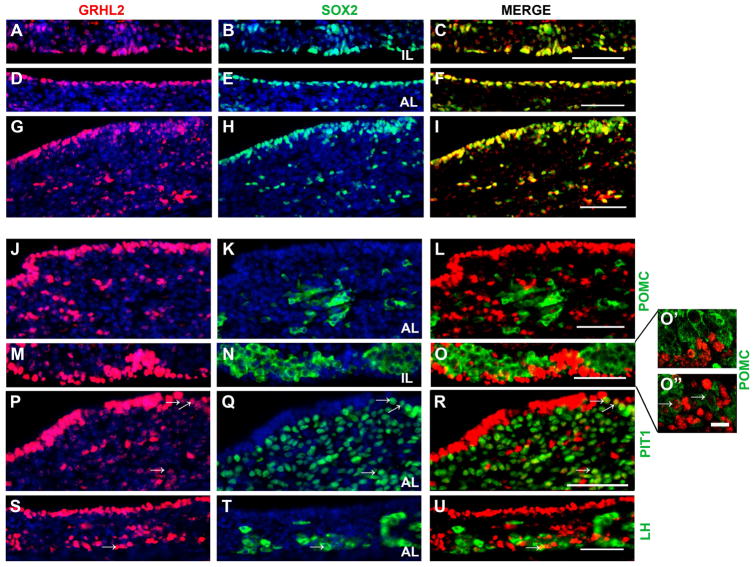
The localization of GRHL2 in cells that line the lumen of anterior (AL) and intermediate lobe (IL) of the pituitary strongly suggests GRHL2 is present in the progenitor cell population. However, GRHL2 is also expressed in the AL parenchyma and in cells outside of the marginal zone of the IL (Figure 2A, D, G, J, M P and S). These areas of the pituitary have been shown to contain a small number of progenitors but are predominately populated with differentiated endocrine cells. We used immunohistochemistry to determine if GRHL2 is expressed in both the progenitor and hormone producing cells in the AL and IL at p1. SOX2, a progenitor cell maker, is expressed predominantly in the cleft cells surrounding the lumen and in a few scatted cells within the anterior lobe parenchyma (Figure 2B, E and H). GRHL2 expression is detected in nearly all of the SOX2 positive cells of both the AL and IL (Figure 2C, F and I). POMC is expressed in corticotrope cells in the AL and in melanotropes in the IL (Figure 2K and N, respectively). GRHL2 does not appear to be expressed in corticotrope cells (Figure 2L). In the IL, cells that have high expression of GRHL2 did not co-express POMC (Figure 2O and 2O′). However, we did observe colocalization of GRHL2 and POMC in small number of cells that weakly express GRHL2 (Figure 2O″, arrows). To determine if GRHL2 expression was detected in somatotropes, thyrotropes and lactotropes we used immunohistochemistry of PIT1, a transcription factor that is necessary for the differentiation of each of these cell types (Figure 2Q). In cells that show a strong expression of GRHL2 we did not observe colocalization with PIT1. In contrast, we did observe colocalization of GRHL2 and PIT1 in cells that appeared to have reduced but not absent expression of GRHL2 (Figure 2R, arrows). Expression of GRHL2 in gonadotrope cells was determined by colocalization with luteinizing hormone (LHβ) (Figure 2T). The vast majority of GRHL2 positive cells were not found to be LHβ positive however, a doubled labeled cell was occasionally detected (Figure 2U, arrows). Taken together, this data indicates that GRHL2 is predominately expressed in the progenitor cell population in the developing pituitary.
Figure 2.
GRHL2 is predominantly expressed in progenitor cells in the postnatal pituitary. At p1 GRHL2 positive cells are mostly restricted to the intermediate lobe (IL) (A and M) and anterior lobe (AL) cleft cells (D and S) and a few GRHL2 positive cells are detected in the AL parenchyma (G, J and P). SOX2 is expressed in the cleft cells of the IL (B) and AL (E) cells of wild type pituitaries. A few SOX2 positive cells are detected in the AL parenchyma (H). GRHL2 is detected in the vast majority of SOX2 positive cells in both the IL (C) and AL (F and I). POMC is expressed in the AL parenchyma (K). GRHL2 expression is absent from POMC positive cells in the AL parenchyma (L). POMC is also expressed in dorsal IL (N). The majority of cells that express GRHL2 in the IL are not POMC positive (O and O′). A small subset of POMC positive cells appear to weakly express GRHL2 in the dorsal IL (O″, arrows). PIT1 is highly expressed in the anterior lobe parenchyma (Q). GRHL2 is absent from the majority of PIT1 positive cells but was detected in a small population of cells that weakly express GRHL2 (R). LH positive cells are confined to the AL parenchyma (T). Occasionally, cells double-labeled with GRHL2 and LHβ were detected in the AL but the majority of LHβ positive cells do not co-express GRHL2 (U). Arrows indicate double-labeled cells. Scale bar = 50 μm (A–U). Scale bar = 5 μm (O′ and O″). n=3–4.
GRHL2 expression is increased in the in Ames Dwarf (Prop1df/df) postnatal pituitary
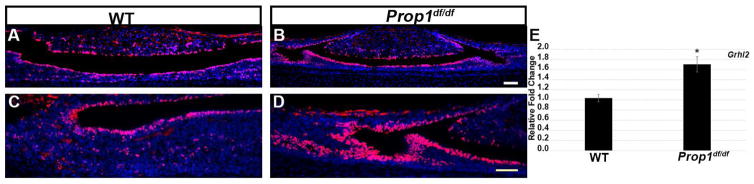
To further demonstrate that GRHL2 correlates with the presence of the progenitor cell population we examined its expression in Ames dwarf (Prop1df/df) mouse postnatal pituitaries by immunohistochemistry and qRT-PCR. Prop1df/df mice show an expansion of the luminal pituitary progenitor cell population due to an inhibition of these cells to differentiate into the endocrine cell types (Ward et al. 2005; Ward et al. 2006; Pérez Millán et al. 2016). We therefore used this mouse as a model for increased pituitary progenitor cell number. In control mice at p5, GRHL2 expression is present in the luminal cells of the anterior and intermediate lobes with a few cells scattered in the anterior lobe parenchyma (Figure 3A and C). A similar pattern of localization is detected in the Ames dwarf mouse pituitary. In addition, the number of GRHL2 positive cells surrounding the AL lumen appears to increase in Ames dwarf mice (Figure 3B and 3D). Coincidentally, we observe a significant increase in Grhl2 mRNA expression in Ames dwarf mice compared to controls (Figure 3E). These findings show GRHL2 protein and mRNA expression correlates with progenitor cell number in the developing pituitary.
Figure 3.
GRHL2 expression is increased in Prop1df/df mice at p5. GRHL2 expression is mainly detected in the cleft cells of the anterior lobe (AL) and intermediate lobe (IL) cells of control pituitaries (A and C). In Prop1df/df pituitaries, the number of GRHL2 positive cells appears to be increased in the AL cleft (B and D). Grhl2 mRNA levels are significantly increased in Prop1df/df mice pituitaries compared to controls (E). *, P < 0.05. n=3 (immunohistochemistry). n=3–4 (qRT-PCR).
GRHL2 expression is reduced in the Notch2 cKO mice
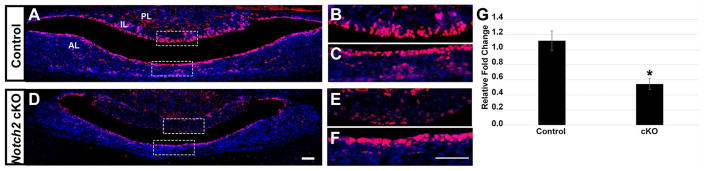
In contrast to Ames dwarf mice, the progenitor cell population is progressively lost in Notch2 cKO mice. Therefore, we sought to examine changes in GRHL2 expression in this model of decreased progenitor cell number. In control mice at p1, GRHL2 expression is robustly expressed in cleft cells of both the AL and IL and is present in a few scattered cells in the AL parenchyma (Figure 4A, B, and C). In contrast, GRHL2 expression appears to be significantly decreased in cells lining the marginal zone of Notch2 cKO mice (Figure 4D). Notably, the cleft cells of the intermediate lobe appear to have the most marked reduction in GRHL2 expression (Figure 4E) while the AL cleft cells appear to be unaffected (Figure 4F). These findings are further supported by the significant decrease in Grhl2 mRNA levels in the Notch2 cKO mice compared to controls at p1 (Figure 4G). These data indicate that loss of progenitor cells in the Notch2 cKO mice results in decreased expression of GRHL2 during early postnatal development.
Figure 4.
Loss of Notch2 results in decreased expression of GRHL2 in p1 pituitaries. GRHL2 is expressed in the anterior (AL) and intermediate (IL) lobe cleft cells and in cells scattered throughout the AL parenchyma of control pituitaries (A). Boxed region of IL is shown in (B) and AL in (C). In the Notch2 cKO mice, GRHL2 expression is drastically decreased in the cleft cells of the IL (D, boxed region shown in E). GRHL2 expression appears to be unaffected in anterior lobe cells (D, boxed region shown in F). A significant decrease in mRNA levels of Grhl2 is observed in the Notch2 cKO pituitaries compared to controls. *, P < 0.05. Scale bar = 50 μm. n=3 (immunohistochemistry). n=7 (qRT-PCR).
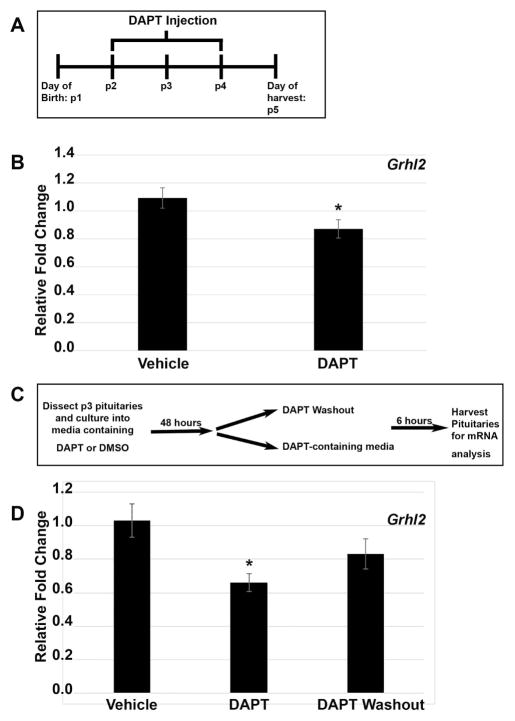
Postnatal Notch inhibition results in decreased Grhl2 expression
Loss of GRHL2 in Notch2 cKO may simply be due to a reduction in progenitor cell numbers. However, this observation may indicate that Grhl2 expression is regulated by Notch signaling. To determine if an acute reduction in Notch signaling during the postnatal period was sufficient to decrease Grhl2 expression, mice were injected with N-[N-(3,5- difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester (DAPT), a gamma secretase inhibitor that blocks Notch signaling, during the early postnatal period following the dosing paradigm in Figure 5A. A significant reduction in Grhl2 mRNA levels was observed in mice dosed with DAPT compared to vehicle-treated controls, suggesting Grhl2 as a Notch regulated gene (Figure 5B). To determine if the potential regulation of Grhl2 by Notch signaling is direct or indirect, we used an explant culture system to examine effects of DAPT treatment at the level of the pituitary. Pituitaries were treated as depicted in Figure 5C. Quantifications of Grhl2 mRNA levels in each of the conditions show Grhl2 mRNA levels are reduced after DAPT treatment and recover to control levels after 6 hours of DAPT washout (Figure 5D). These data suggest that the decrease in Grhl2 we observe in the Notch2 cKO mice is due to inherent changes at the level of the pituitary. Furthermore, the increase in Grhl2 mRNA levels within 6 hours of DAPT removal further suggests Grhl2 as a potential direct Notch target, as this same pattern is observed with Hes1, Hey1 and Prop1 (Nantie et al. 2014).
Figure 5.
Grhl2 is significantly decreased in vivo and in vitro after treatment with DAPT during postnatal pituitary development. Schematic representing dosing paradigm for mice treated with DAPT, a gamma secretase inhibitor that is known to block Notch signaling (A). A significant decrease is observed in Grhl2 mRNA levels after in vivo treatment with DAPT (B). Schematic showing paradigm for pituitary explant culture treatment with DAPT (C). A significant decrease is observed in Grhl2 mRNA levels after in vitro DAPT treatment compared to vehicle treated controls. Grhl2 mRNA levels recover after DAPT is removed from the culture media (D). *, P < 0.05. n= 5 to 8 (qRT-PCR in vivo), and n= 4 to 8 (qRT-PCR in vitro).
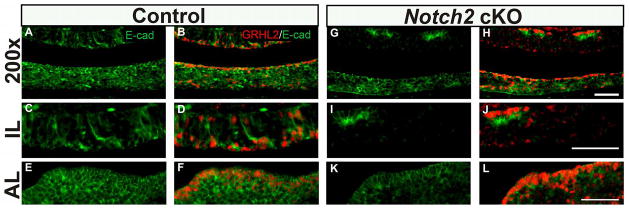
Downstream Targets of GRHL2, E-cadherin and PCNA are decreased in the Notch2 cKO mice
It is well established in other tissues that GRHL2 is a suppressor of epithelial differentiation (Chen et al. 2012; Tanimizu, Kobayashi, et al. 2014). GRHL2 controls this process by directly regulating the expression of genes necessary for maintaining cellular proliferative capacity. Included in the GRHL2 regulatory network are genes responsible for cellular adhesion such as E-Cadherin (E-cad), as well as genes that direct cellular proliferation such as Proliferating Cell Nuclear Antigen (PCNA) (Chen et al. 2010; Werth et al. 2010). We therefore sought to characterize expression of these downstream targets in the postnatal pituitary by immunohistochemistry. Furthermore, because we observe a reduction of GRHL2 expression in Notch2 cKO mice pituitaries we analyzed E-cad and PCNA expression in these mice. In control mice, E-cad is highly expressed in both the intermediate and anterior lobes of the developing pituitary (Figure 6A, C and E). GRHL2 strongly colocalizes in cells that express E-cad, which is most apparent in the luminal cells of both IL and AL. However, we did observe cells that expressed E-cad but did not appear to be GRHL2 positive (Figure 6B, D and F). In Notch2 cKO pituitaries, we observe a dramatic reduction in E-cad expression specifically in the IL with only a few positive cells remaining (Figure 6G and I). Interestingly, the few cells that maintain E-cad expression in the IL also express GRHL2 (Figure 6H and J). In contrast, the expression of E-cadherin and GRHL2 appear to be relatively unaffected in anterior lobe of cKO pituitaries (Figure 6K and L).
Figure 6.
Notch2 cKO mice have decreased E-cadherin expression at p1. In control mice, E-cadherin expression is detected throughout the anterior pituitary but appears to be concentrated in the cleft cells of the intermediate lobe (IL) (A and C) and anterior lobe (AL) (A and E). In control mice, GRHL2 immunopositive cells appear to co-express E-cadherin in the pituitary cleft cells of the IL (B and D) andAL (B and F). Colocalization of E-cadherin and GRHL2 is also detected AL parenchyma (F). In contrast, in cKO mice, E-cadherin expression is decreased in the cleft cells of IL (G and I). Colocalization of E-cadherin and GRHL2 is lost in the cleft cells of IL (J). A few cells show E-cadherin and GRHL2 co-localization in dorsal IL (H and J). Expression of E-cadherin appears unaffected in the AL (G and K). No change in colocalization of E-cadherin and GRHL2 is observed in the AL cleft and AL parenchyma (H and L). Scale bar = 50 μm. n=3–4.
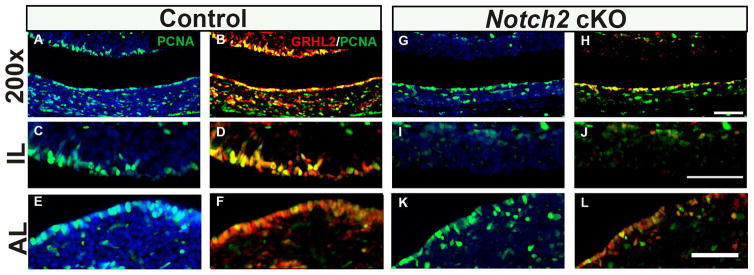
In control mice, PCNA expression closely mimics that of other cell proliferation markers such as Ki67, with concentrated expression in the luminal cells and appreciable levels throughout the AL and IL (Figure 7A, C and E). As expected, PCNA colocalizes with GRHL2 expressing cells particularly in luminal cells of both the AL and IL, where we almost never observed a GRHL2 positive cell that was not also positive for PCNA (Figure 7B, D and F). In cKO mice, there appears to be a reduction in PCNA expression specifically in IL cleft cells where we detect very few PCNA-positive cells (Figure 7G and I). Although PCNA and GRHL2 expression is drastically reduced in the IL cleft, a small number of positive cells remain in the dorsal IL that co-express PCNA and GRHL2 (Figure 7H and J). No obvious changes in expression of PCNA are detected in the AL marginal cells of Notch2 cKO mice compared to control (Figure 7K). Conversely, colocalization of GRHL2 and PCNA appears to be unaffected in the AL (Figure 7L). Taken together, these data indicate known GRHL2 targets E-cad and PCNA are highly expressed in the postnatal pituitary. In addition, we demonstrate that these targets are reduced in the Notch2 cKO mice pituitaries suggesting that the loss of Notch and subsequent loss of GRHL2 expression correlates with changes in these downstream genes.
Figure 7.
Notch2 cKO pituitaries have decreased PCNA expression at p1. In control animals, PCNA is detected in the intermediate lobe (IL) (A and C) and anterior lobe (AL) cleft cells (A and E) and in cells scattered in the dorsal IL and AL parenchyma (A and E). The vast majority of PCNA immunopositive cells co-express GRHL2 in the cleft of both the IL (B and D) and AL (B and F). In the dorsal IL (B) and AL parenchyma (F), a large number of cells are double-labeled and small subset of cells that express only GRHL2 or PCNA are detected. In contrast, in cKO mice the number of PCNA positive cells appears to be decreased specifically in the IL cleft cells (G and I). The colocalization of PCNA and GRHL2 is not detected in the IL cleft cells of cKO mice (H and J). Expression of PCNA in the AL cleft and AL parenchyma appears unaffected (G and K). Colocalization of PCNA and GRHL2 does not appear to be changed in the AL cleft and AL parenchyma (H and L). Scale bar = 50 μm. n=4.
Discussion
The stem/progenitor cell population of the pituitary gland has been identified as the precursor cells that give rise to all of the hormone producing cell types. This cell population is necessary for embryonic and postnatal expansion of the gland and also plays a critical role in the physiological response of the adult. While recent advances have been made in identifying these cells relatively little is known about the factors that regulate the progenitor cell niche in the developing pituitary. Our studies characterize the expression of GRHL2 in the pituitary and identify this transcription factor as a novel pituitary progenitor cell marker.
The pituitary progenitor cells are characterized by the expression of hallmark stem cell factors such as the high-mobility group (HMG) box transcription factor, SOX2. Recent studies, both in vitro and in vivo studies have demonstrated that SOX2 expressing progenitors differentiate into all of the different endocrine cell types of the anterior pituitary (Fauquier et al. 2008). Additional factors have also been identified as progenitor cell markers, including SOX9, OCT4, GRFa2, S100 and PROP1 (Chen et al. 2005; Fauquier et al. 2008; Garcia-Lavandeira et al. 2009; Yoshida et al. 2009). Interestingly, these factors are not ubiquitously expressed in all pituitary progenitors, indicating a heterogeneous population of progenitor cells whose functional differences have yet to be elucidated. More recently, in vivo studies have shown that PROP1 expressing cells also have the ability to differentiate into all of the hormone-producing cells types of the anterior pituitary (Davis et al. 2016). These studies demonstrate that SOX2 and PROP1 expressing progenitors serve as the precursor cells to the differentiated pituitary cell types, however loss of function studies for both of these factors does not eliminate differentiation of all the pituitary lineages (Ward et al. 2005; Ward et al. 2006; Jayakody et al. 2012). Taken together, these studies indicate that other pituitary progenitor factors must play a role in pituitary development. Therefore, identification of novel progenitor cell markers such as GRHL2 will provide new insight into the regulation and maintenance of pituitary progenitor cells.
GRHL2 expression marks the progenitor cell population in the developing pituitary. The spatial and temporal pattern of GRHL2 in the pituitary closely mimics the expression pattern of other progenitor cell markers including SOX2, SOX9, Notch2 and Prop1 (Garcia-Lavandeira et al. 2009; Yoshida et al. 2011; Ward et al. 2005; Nantie et al. 2014). In agreement with this observation, we show that GRHL2 is expressed almost exclusively in SOX2 positive cells and is either absent or minimally detected in the majority of the endocrine cell types of the anterior pituitary. This finding is consistent with localization patterns of GRHL2 in other tissues including the liver, where GRHL2 is detected in epithelial cells that express stemness markers such as SOX9 (Tanimizu et al. 2013). Interestingly, at e14.5 GRHL2 can be detected in both Rathke’s Pouch (RP) and in the anterior lobe (AL). At this period in development, the AL contains differentiated cell types and a large number of cells that have been designated as non-cycling precursor cells that are thought to be transitioning into fully differentiated endocrine cell types (Bilodeau et al. 2009). A similar phenomena can be seen during postnatal pituitary development in which we observe a low level of GRHL2 expression in cells directly outside of the progenitor cell niche that are positive for lineage commitment makers such as PIT1. A subset of PIT1 positive cells also express the progenitor cell marker PROP1, and are also suggested to be cells transitioning from a progenitor cell state to terminally differentiated cell (Yoshida et al. 2009; Yoshida et al. 2013). In addition, it has been shown that a population of terminally differentiated cells are mitotic during postnatal pituitary expansion (Carbajo-Perez & Watanabe 1990; Taniguchi et al. 2002). Therefore, cells that co-express both GRHL2 and terminal differentiation makers, as was observed in a minority of melanotropes and gonadotropes, could possibly be indicative of differentiated cells that display mitotic activity. Together, these observations indicate that GRHL2 expression may mark both a progenitor cell population and transitional cells.
We further demonstrate GRHL2 as a progenitor cell marker by correlating its expression with changes in progenitor cell number in the Prop1df/df and Notch2 cKO pituitaries. PROP1, a pituitary specific transcription factor expressed in pituitary progenitor cells, coordinates endocrine cell differentiation by regulating the expression of PIT1, which is necessary for expression of GH, PRL, and TSHβ. Prop1df/df mice have an expansion of progenitor cells in the marginal zone (MZ) of the pituitary due to a failure of epithelial to mesenchymal transition and the subsequent inability of these cells to further differentiate (Ward et al. 2006; Pérez Millán et al. 2016). Therefore, we used Prop1df/df mice as a model for progenitor cell increase during early pituitary development. We show that expression of GRHL2 is significantly increased in the MZ of Prop1df/df pituitaries at postnatal day 5 (p5), indicating that an increase in progenitor cell number is associated with an increase in GRHL2 expression. In contrast, the loss of Notch2 during pituitary development is associated with a decrease in the progenitor cell pool. Therefore, this model was used to examine changes in GRHL2 expression in pituitaries with a decreased number of progenitor cells. We show that loss of Notch2 results in decreased expression of GRHL2 at p1. In particular, the most drastic reduction in GRHL2 expression was seen in IL progenitor cells. Similar observations were reported for other progenitor cell markers SOX2 and SOX9 in Notch2 cKO mice (Nantie et al. 2014). Taken together, these data demonstrate that modulations in the progenitor cell population can be detected by the changes in GRHL2 expression.
It can be postulated that the reduction in GRHL2 expression in the Notch2 cKO is solely a consequence of progenitor cell loss. Alternatively, this reduction could imply Notch signaling as novel regulator of GRHL2 expression in the developing pituitary. We therefore sought to assess if Grhl2 could be regulated by Notch. Our data shows that pituitaries exposed acutely in vivo and in vitro to DAPT, a gamma secretase inhibitor, display a significant reduction in mRNA levels of Grhl2. This data has several implications about Notch regulation of Grhl2 expression. First, this suggests that the decrease is GRHL2 expression we observe in the Notch2 cKO mice is not merely a consequence of the long term loss of Notch. In addition, our in vitro data implies that the reduction of Grhl2 in Notch2 cKO mice is a direct effect on the pituitary and not due to extrinsic factors. Furthermore, in vitro Grhl2 mRNA levels recover after 6 hours of DAPT washout, a pattern that is observed with direct downstream Notch targets including Hes1 and Hey1 (Weng et al. 2006; Chadwick et al. 2009; Nantie et al. 2014). Therefore, these data may suggest that Grhl2 is directly regulated by Notch signaling. GRHL2 has not been previously been shown to be regulated by Notch signaling in mammalian cells. However, a recent study demonstrated the drosophila homologue, Grainyhead (Grh) is directly regulated by Notch signaling in neuroblasts (Zacharioudaki et al. 2016). Furthermore, studies in other mammalian tissues have eluded to a potential relationship between Notch signaling and GRHL2 expression. For example, several studies have demonstrated that active Notch signaling promotes the differentiation of cholangiocytes from hepatoblasts during liver development (Tanimizu & Miyajima 2004; Wang et al. 2014). Interestingly, GRHL2 in addition to other well established Notch targets including Hes1, Hey1, and Sox9 were identified as cholangiocyte specific markers (Tanimizu, Nishikawa, et al. 2014), suggesting Notch regulation of GRHL2 occurs in the liver. In addition, in lung epithelium GRHL2 has been suggested to regulate expression of Notch3 and Notch1 (Gao et al. 2015). This may indicate that there is a reciprocal relationship between Notch and GRHL2 regulation. However, further studies are necessary to determine if Grhl2 is in fact a direct Notch target in the pituitary.
Another link between the Notch signaling pathway and GRHL2 is that they appear to regulate many of the same cellular pathways. Both Notch and GRHL2 are thought to be important in stem cell maintenance and regulation of differentiation. A potential relationship between the two has been established in drosophila studies, identifying that GRH works in synergy with Notch to regulate target-gene expression (Furriols et al. 2001). In support of this observation, a recent study identified GRHL2 and the essential Notch co-activator protein, RBPJ, as a part of the OCT4 interactome, a transcription factor critical for the maintenance of stem cell pluripotency. These studies indicate a large number of transcription factors work in concert to regulate cellular plasticity and self-renewal (van den Berg et al. 2010). This implies that GRHL2 and Notch may work synergistically to regulate genes necessary for progenitor cell maintenance in the pituitary.
While the function of GRHL2 in the pituitary has yet to be elucidated, its function is well established in other tissues. GRHL2 has been shown to regulate genes that are important in maintaining epithelial integrity and regulation of cellular differentiation. In particular, GRHL2 is a direct transcriptional regulator of Cdh1 (E-cadherin), a cellular adhesion molecule, during embryonic development and in epithelial cells of several different tissues including the liver, lung and gut (S. Varma et al. 2012; Werth et al. 2010). In human keratinocytes, GRHL2 also been shown to directly regulate the expression of cellular proliferation proteins such as Ki67 and PCNA (Chen et al. 2010; Chen et al. 2012). Our studies demonstrate that GRHL2 expression is strongly associated with both E-cadherin and PCNA expression in the postnatal pituitary. In addition, Notch2 cKO mice show a drastic reduction in E-cadherin and PCNA expression in the in the cleft cells of the IL, a region in which GRHL2 expression is also reduced. The bias of IL expression disruption correlates with selective stem cell marker loss in the IL of Notch2 cKO mice at the same age (Nantie et al. 2014). These data may indicate that GRHL2 functions to regulate cellular proliferation and cellular adhesion in the developing pituitary. Furthermore, E-cadherin expression is also important for stem cell adhesion and is a necessary factor for the maintenance of the stem-cell niche in other tissues (Chen et al. 2013). Therefore, it could be postulated that expression of GRHL2 is important in the maintenance of the progenitor cell niche in the developing pituitary. However, the reduced expression of known GRHL2 targets we observe in Notch2 cKO mice could be due to expression changes in other regulators of these genes. Notch signaling directly regulates transcription of cell cycle genes including Ccnd1, Ccnd3 and Cdnk1 (Cohen et al. 2010; Georgia et al. 2006; Joshi et al. 2009). Therefore, the decrease in the cellular proliferation genes in Notch2 cKO mice could be a direct consequence of reduced Notch signaling. While our data provided evidence to a potential function of GRHL2 in the pituitary, future studies are necessary to elucidate the role of GRHL2 in the developing pituitary.
Experimental Procedures
Mice
For GRHL2 protein analysis, wildtype mice of mixed genetic background were used. CD-1 mice (Charles River) from a breeding colony maintained in our lab were used for the Grhl2 mRNA analysis and the in vivo and vitro DAPT treatment studies.
Notch2 cKO mice were generated as previously described (Nantie et al. 2014). Briefly, Notch2fl/fl mice, obtained from Jackson Laboratory (Bar Harbor) were bred to Foxg1+/cre, also obtained from Jackson Laboratory. Notch2+/fl; Foxg1+/cre and Notch2+/fl; Foxg1+/+ littermates were then used to generate cKO animals. For genotyping, tail biopsies were obtained and DNA was extracted via HotSHOT (Truett et al. 2000). Genotyping for Cre and Notch2 alleles was determined by PCR using as previously described using published primers (Hébert & McConnell 2000; McCright et al. 2006).
Ames Prop1+/df mice on the C57BL/6J background, provided by Dr. Sally Camper (University of Michigan, Ann Arbor, Michigan), were bred together to generate Prop1+/+ and Prop1df/df mice. For genotyping, tail biopsies were obtained and DNA was extracted via salt-out method. Genotyping PCR for Prop1df mutant allele was performed as previously described using published primers (Himes & Raetzman 2009).
DAPT injections
CD1 mice were injected starting at p2, daily for 3 days with 100 mg/kg DAPT (N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-Butyl Ester, Gamma Secretase Inhibitor IX (Millipore, Billerica) diluted in Dimethyl Sulfoxide (DMSO, Sigma-Aldrich) or DMSO alone. Pituitaries were collected 24 hours after last injection.
Pituitary Explant Culture and DAPT Washout
p3 pituitaries were dissected from CD-1 mice and cultured in a 96 well plate. Culture media consists of DMEM/F-12 media (Cellgro) supplemented with 10% fetal bovine serum (Hyclone) and 1% Penicillin Streptomycin (Fisher). For DAPT treatment, 10μM DAPT in DMSO or an equal amount of DMSO was added to cell culture media at the start of the culture. After 24 hours, media was spiked with DAPT or DMSO and pituitaries remained in culture for an additional 24 hours. For the washout assay, after 48 hours in culture, medium was removed, pituitaries were rinsed and media containing either, DMSO or 10μM DAPT dissolved in DMSO was added. Pituitaries were harvested after 6 hours.
Immunohistochemistry
Processing of pituitaries for the immunohistological analysis at e14.5, e16.5, p1 and p5, in Notch2 cKO mice and in Ames Dwarf mice was performed as previously described (Nantie et al. 2014). Briefly, whole e14.5 and e16.5 embryos and heads of postnatal mice were fixed in 3.7% formaldehyde and embedded in paraffin for sectioning. Samples were deparaffinized, rehydrated and boiled in a 10 mM citrate solution. Anti-GRHL2 antibody was purchased from Sigma Aldrich and was previously validated by the Human Protein Atlas (HPA) project (Uhlén et al. 2015). For Immunohistological detection of GRHL2 (1:500–1:1000) tyramide signal amplification was used (TSA) (Perkin Elmer) and slides were blocked with TNB blocking solution containing 0.1 M Tris–HCl, 0.15 M NaCl, and 0.5% TSA Blocking Reagent diluted in sterile ddH2O (pH 7.5). Slides were incubated with biotin-conjugated rabbit secondary antibody (1:200; Jackson ImmunoResearch) diluted in the TNB blocking solution. This was followed by incubation with streptavidin-HRP (1:100) and incubation with Cyanine-3 within the TSA Kit (Perkin Elmer) according to the manufacturer’s protocol. For p30 animals, pituitaries were fixed in 3.7% formaldehyde, cyroprotected in a 30% sucrose/PBS solution, then frozen in Optimal Cutting Temperature compound (Electron Microscopy Sciences). Frozen sections, were thawed for 10 minutes, then fixed with 3.7% formaldehyde. Immunohistological detection of GRHL2 was performed using TSA as described above.
For double-stains, p1 heads were fixed using a Zinc fixative (BD Pharmigen). Slides were first stained with GRHL2 using TSA as described above. Slides were then blocked with a blocking solution (3% bovine serum albumin (Jackson ImmunoResearch) and 0.5% Triton X-100 (Sigma Aldrich) diluted in PBS), 1% Peroxidase-conjugated streptavidin (Jackson ImmunoResearch) and 1% Donkey Anti-Rabbit IgG (Jackson ImmunoResearch). Following incubation with the blocking solution, slides were incubated with the following primary antibodies, SOX2 (1:1000; Millipore), PIT1 (1:1000; kind gift of Dr. Simon Rhodes), adrenocorticotropic hormone (ACTH) (1:100; DAKO), LHβ (1:100; Dr. A. F. Parlow and the National Hormone and Pituitary Program, University of California, Los Angeles), PCNA (1:200; Cell Signaling), and E-cadherin (1:100; Cell Signaling). These antibodies were diluted in the blocking solution described previously. Slides were then incubated with either biotin-conjugated rabbit secondary antibody (SOX2, Pit1, POMC, LHβ and E-cadherin) or biotin-conjugated mouse secondary antibody (PCNA) (1:200; Jackson ImmunoResearch). This was followed by incubation with streptavidin-conjugated Cy2 fluorphore (1:200; Jackson ImmunoResearch). As a control, slides without each primary antibody were included for each experiment to ensure staining specificity. All slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1:1000; Life Technologies). Slides were visualized at 200x and 400x with a Leica DM2560 microscope or at 630x using a using a Leica DMI4000B confocal microscope and processed as previously described (Nantie et al. 2014).
Quantitative RT-PCR (qRT-pCR)
RNA was processed as previously described (Nantie et al. 2014). Briefly, an RNAqueous micro kit (Ambion) was used to isolate RNA from individual pituitaries and total amount of RNA obtained from each pituitary was converted to cDNA with ProtoScript M-Mulv First Strand cDNA Synthesis Kit (New England Biolabs). For adults, 0.5ug of RNA was converted to cDNA. The following primer sequences were used; Grhl2 forward: GAA AGC CAC AAA GCA TCA GGA C, Grhl2 reverse: AGA CAG CAC AGC GAC ATG GAAG. Gapdh levels were used to normalized data, using the following primers; Gapdh forward: GGT GAG GCC GGT GCT GAG TAT G, Gapdh Reverse: GAC CCG TTT GGC TCC ACC CTT C. The standard comparative Δcycle threshold value method was used for data analysis as previously described (Goldberg et al. 2011). Statistical significance was determined by Student’s t test.
Acknowledgments
Grant support: NIH R01 DK076647 and NIH T32 ES007326
We thank Dan Getz for technical assistance. The work was supported by National Institute of Health Grants R01 DK076647 (LTR) and T32 ES007326 (WE).
References
- Andoniadou CL, et al. Sox2(+) stem/progenitor cells in the adult mouse pituitary support organ homeostasis and have tumor-inducing potential. Cell stem cell. 2013;13(4):433–45. doi: 10.1016/j.stem.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Aue A, et al. A Grainyhead-Like 2/Ovo-Like 2 Pathway Regulates Renal Epithelial Barrier Function and Lumen Expansion. Journal of the American Society of Nephrology. 2015;26(11):2704–2715. doi: 10.1681/ASN.2014080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DLC, et al. An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J. Distinct Developmental Roles of Cell Cycle Inhibitors p57Kip2 and p27Kip1 Distinguish Pituitary Progenitor Cell Cycle Exit from Cell Cycle Reentry of Differentiated Cells. Molecular and Cellular Biology. 2009;29(7):1895–1908. doi: 10.1128/MCB.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo-Perez E, Watanabe YG. Cellular proliferation in the anterior pituitary of the rat during the postnatal period. Cell and tissue research. 1990;261(2):333–338. doi: 10.1007/BF00318674. [DOI] [PubMed] [Google Scholar]
- Chadwick N, et al. Identification of novel Notch target genes in T cell leukaemia. Molecular cancer. 2009;8:35. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. The adult pituitary contains a cell population displaying stem/progenitor cell and early embryonic characteristics. Endocrinology. 2005;146(9):3985–98. doi: 10.1210/en.2005-0185. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140(2):255–65. doi: 10.1242/dev.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell death & disease. 2012;3(12):e450. doi: 10.1038/cddis.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. The Journal of biological chemistry. 2010;285(52):40852–63. doi: 10.1074/jbc.M110.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 2016;37(5):500–10. doi: 10.1093/carcin/bgw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogan JD, et al. The PROP1 2-base pair deletion is a common cause of combined pituitary hormone deficiency. The Journal of clinical endocrinology and metabolism. 1998;83(9):3346–3349. doi: 10.1210/jcem.83.9.5142. [DOI] [PubMed] [Google Scholar]
- Cohen B, et al. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast cancer research and treatment. 2010;123(1):113–124. doi: 10.1007/s10549-009-0621-9. [DOI] [PubMed] [Google Scholar]
- Davis SW, et al. All Hormone-Producing Cell Types of the Pituitary Intermediate and Anterior Lobes Derive From Prop1-Expressing Progenitors. Endocrinology. 2016;157(4):1385–96. doi: 10.1210/en.2015-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquier T, et al. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proceedings of the National Academy of Sciences. 2008;105(8):2907–2912. doi: 10.1073/pnas.0707886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M, et al. A model Notch response element detects Suppressor of Hairless–dependent molecular switch. Current Biology. 2001;11(1):60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gao X, et al. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. The Journal of Cell Biology. 2015;211(3):669–682. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lavandeira M, et al. A GRFa2/Prop1/Stem (GPS) Cell Niche in the Pituitary J. A. L. Calbet, ed. PLoS ONE. 2009;4(3):e4815. doi: 10.1371/journal.pone.0004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S, et al. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Developmental biology. 2006;298(1):22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Goldberg LB, Aujla PK, Raetzman LT. Persistent expression of activated Notch inhibits corticotrope and melanotrope differentiation and results in dysfunction of the HPA axis. Developmental biology. 2011;358(1):23–32. doi: 10.1016/j.ydbio.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) Locus Mediates loxP Recombination in the Telencephalon and Other Developing Head Structures. Developmental biology. 2000;222(2):296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Himes AD, Raetzman LT. Premature differentiation and aberrant movement of pituitary cells lacking both Hes1 and Prop1. Developmental biology. 2009;325(1):151–161. doi: 10.1016/j.ydbio.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, et al. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. The EMBO journal. 1994;13(8):1799–805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, et al. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes & development. 1995;9(24):3136–48. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jayakody SA, et al. SOX2 regulates the hypothalamic-pituitary axis at multiple levels. The Journal of clinical investigation. 2012;122(10):3635–46. doi: 10.1172/JCI64311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113(8):1689–98. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem cells. 2012;30(4):741–52. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen OD, et al. Control of endodermal endocrine development by Hes-1. Nature Genetics. 2000;24(1):36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44(1):29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- Mortensen AH, et al. Candidate genes for panhypopituitarism identified by gene expression profiling. Physiological genomics. 2011;43(19):1105–16. doi: 10.1152/physiolgenomics.00080.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantie LB, et al. Notch signaling in postnatal pituitary expansion: proliferation, progenitors, and cell specification. Molecular endocrinology. 2014;28(5):731–744. doi: 10.1210/me.2013-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan LA, Thomas CK, Levy A. Pituitary mitosis and apoptotic responsiveness following adrenalectomy are independent of hypothalamic paraventricular nucleus CRH input. Journal of Endocrinology. 2004;181(3):521–529. doi: 10.1677/joe.0.1810521. [DOI] [PubMed] [Google Scholar]
- Pérez Millán MI, et al. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. In: Rossant J, editor. eLife. Vol. 5. 2016. p. e14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Developmental Biology. 2011;353(1):38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetzman L, et al. Developmental regulation of Notch signaling genes in the embryonic pituitary: Prop1 deficiency affects Notch2 expression. Developmental Biology. 2004;265(2):329–340. doi: 10.1016/j.ydbio.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Raetzman LT, Cai JX, Camper SA. Hes1 is required for pituitary growth and melanotrope specification. Developmental biology. 2007;304(2):455–466. doi: 10.1016/j.ydbio.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoti K, Akiyama H, Lovell-Badge R. Mobilized adult pituitary stem cells contribute to endocrine regeneration in response to physiological demand. Cell stem cell. 2013;13(4):419–432. doi: 10.1016/j.stem.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, et al. Proliferation and differentiation of rat anterior pituitary cells. Anatomy and Embryology. 2002;206(1–2):1–11. doi: 10.1007/s00429-002-0271-8. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Kobayashi S, et al. Downregulation of miR122 by grainyhead-like 2 restricts the hepatocytic differentiation potential of adult liver progenitor cells. Development. 2014;141(23):4448–56. doi: 10.1242/dev.113654. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, et al. Hepatic biliary epithelial cells acquire epithelial integrity but lose plasticity to differentiate into hepatocytes in vitro during development. Journal of cell science. 2013;126(Pt 22):5239–46. doi: 10.1242/jcs.133082. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa Y, et al. Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM-) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. The Journal of biological chemistry. 2014;289(11):7589–98. doi: 10.1074/jbc.M113.517243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Miyajima A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. Journal of cell science. 2004;117(Pt 15):3165–74. doi: 10.1242/jcs.01169. [DOI] [PubMed] [Google Scholar]
- Truett GE, et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) BioTechniques. 2000;29(1):52, 54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Varma S, et al. The Transcription Factors Grainyhead-like 2 and NK2-Homeobox 1 Form a Regulatory Loop That Coordinates Lung Epithelial Cell Morphogenesis and Differentiation. Journal of Biological Chemistry. 2012;287(44):37282–37295. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S, et al. The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. The Journal of biological chemistry. 2012;287(44):37282–95. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, et al. Notch inhibition promotes fetal liver stem/progenitor cells differentiation into hepatocytes via the inhibition of HNF-1β. Cell and tissue research. 2014;357(1):173–84. doi: 10.1007/s00441-014-1825-9. [DOI] [PubMed] [Google Scholar]
- Ward RD, et al. Cell proliferation and vascularization in mouse models of pituitary hormone deficiency. Molecular endocrinology. 2006;20(6):1378–90. doi: 10.1210/me.2005-0409. [DOI] [PubMed] [Google Scholar]
- Ward RD, et al. Role of PROP1 in Pituitary Gland Growth. Molecular Endocrinology. 2005;19(3):698–710. doi: 10.1210/me.2004-0341. [DOI] [PubMed] [Google Scholar]
- Weber D, Wiese C, Gessler M. Chapter Eight – Hey bHLH Transcription Factors. Current Topics in Developmental Biology. 2014:285–315. doi: 10.1016/B978-0-12-405943-6.00008-7. [DOI] [PubMed] [Google Scholar]
- Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & development. 2006;20(15):2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137(22):3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- Yoshida S, et al. PROP1 coexists with SOX2 and induces PIT1-commitment cells. Biochemical and biophysical research communications. 2009;385(1):11–5. doi: 10.1016/j.bbrc.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Yoshida S, et al. Rapid transition of NESTIN-expressing dividing cells from PROP1-positive to PIT1-positive advances prenatal pituitary development. Journal of neuroendocrinology. 2013;25(9):779–91. doi: 10.1111/jne.12077. [DOI] [PubMed] [Google Scholar]
- Yoshida S, et al. Significant quantitative and qualitative transition in pituitary stem/progenitor cells occurs during the postnatal development of the rat anterior pituitary. Journal of neuroendocrinology. 2011;23(10):933–43. doi: 10.1111/j.1365-2826.2011.02198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudaki E, et al. Genes implicated in stem cell identity and temporal programme are directly targeted by Notch in neuroblast tumours. Development. 2016;143(2):219–31. doi: 10.1242/dev.126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, et al. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes & development. 2006;20(19):2739–2753. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]