Abstract
Rationale
We have previously shown that innate immunity is necessary for transdifferentiation of fibroblasts to endothelial cells. A major signaling molecule involved in innate immunity is inducible nitric oxide synthase (iNOS). Accordingly, we hypothesized that iNOS-generated nitric oxide (NO) might enhance transdifferentiation.
Objective
To elucidate the role of NO in epigenetic plasticity during transdifferentiation.
Methods and Results
We exposed the BJ fibroblasts to transdifferentiation formulation that included endothelial growth factors and innate immune activator polyinosinic:polycytidylic acid (PIC) to induce endothelial cells (iECs). Generation of iECs was associated with iNOS expression and NO elaboration. In absence of PIC, or in presence of antagonists of NFκB or iNOS activity, NO synthesis and iEC generation was reduced. Furthermore, genetic knockout (in MEFs) or siRNA knockdown (in BJ fibroblasts) of iNOS nearly abolished transdifferentiation, an effect which could be reversed by iNOS overexpression. Notably, PIC induced nuclear localization of iNOS, and its binding to, and nitrosylation of, the epigenetic modifier RING1A as assessed by immunostaining, Co-IP and mass spectrometry. Nitrosylation of RING1A reduced its binding to chromatin, and reduced global levels of repressive histone marker H3K27trimethylation. Overexpression of a mutant form of RING1A (C398A) lacking the nitrosylation site almost abrogated transdifferentiation.
Conclusions
Overall, our data indicates that during transdifferentiation, innate immune activation increases iNOS generation of NO to S-nitrosylate RING1A, a key member of the polycomb repressive complex. Nitrosylation of RING1A reduces its binding to chromatin and decreases H3K27trimethylation level. The release of epigenetic repression by nitrosylation of RING1A is critical for effective transdifferentiation.
Keywords: Innate immunity, epigenetics, endothelial cell, inducible nitric oxide synthase, transdifferentiation
Subject Terms: Cellular Reprogramming, Endothelial/Vascular Type/Nitric Oxide, Inflammation
INTRODUCTION
Transdifferentiation is the process by which a somatic cell undergoes a metamorphosis into a different type of somatic cell. This process of cellular transformation plays a role in malignancy, but it may also play a role in the physiological response to injury. For the purpose of regenerative medicine applications, several groups have converted somatic cells such as fibroblasts into neurons 1, cardiomyocytes 2 or endothelial cells (ECs) 3 by overexpressing specific lineage transcription factors. Transdifferentiation from fibroblasts to endothelial cells may be useful in cardiovascular regeneration. For example, an effective transdifferentiation strategy might modify the healing process by preventing formation of scar tissue, and facilitating the generation of vascularized tissues, in response to an injury.
Most investigators that are transdifferentiating cells for regenerative medicine applications have achieved their goal using viral vectors encoding transcriptional factors mediating cell lineage 4,7,5. By contrast, we have developed a non-viral strategy using small molecules and growth factors. This strategy was made possible by leveraging our observation that cellular transformations (either nuclear reprogramming to pluripotency, or transdifferentiation to another cell type) require innate immune activation 6, 7. Indeed, the viral vectors used in nuclear reprogramming studies are more than passive vehicles; by activating innate immunity they facilitate reprogramming7. We have shown that innate immune activation increases the expression of histone acetyltransferase (HAT) family members, and suppresses the expression of histone deactylase (HDAC) family members and DOT1L7. These changes in the balance of epigenetic modifiers would be expected to increase epigenetic plasticity.
It is well known that inducible nitric oxide synthase (iNOS) is a major effector of innate immune signaling 8. Activation of innate immunity leads to NFκB-mediated expression of iNOS 9, which S-nitrosylates proteins to affect their activity and cellular location 10. Accordingly, we hypothesized that innate immune activation of iNOS, and S-nitrosylation of epigenetic modifiers, might contribute to epigenetic plasticity required for transdifferentiation. Driven by this hypothesis, we now describe the discovery of a novel iNOS-dependent post-translational modification of the repressive epigenetic machinery to facilitate transdifferentiation.
METHODS
Chemicals and reagents
All chemicals were purchased from Sigma-Aldrich (St Louis, MO) unless otherwise stated. Other reagents included FBS, Taqman RT-PCR primers, Taqman master mix, 4-Amino-5-methylamino-2′, 7′-difluorofluorescein Diacetate (DAF-FM DA), RNAimax and DAPI (Life Technologies, Carlsbad, CA); polyinosinic:polycytidylic acid (PIC) and NFκB inhibitors Bay117082, celastrol and dexamethasone (Invivogen, San Diego, CA); antibody against the S-nitroso (SNO) moiety (Sigma-Aldrich); antibody against 3-nitrotyrosine (NT) (Cayman Chemical, Ann Arbor, Michigan); PE- antibody against human CD31 and APC- antibody against mouse CD144 (BD Biosciences, San Jose, CA); antibody against inducible nitric oxide synthase (iNOS), HRP conjugated goat anti mouse or rabbit antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); antibody against β-tubulin, Ring1A, H3K27 trimethylation (H3K27me3) and H3K9 dimethylation (H3K9me2; Abcam, Cambridge, UK); bone morphogenetic protein-4 (BMP-4), vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF; Peprotech, Rocky Hill, NJ); Scramble and iNOS siRNA (Integrated DNA Technologies, Coralville, IA).
Cell culture and iEC generation
BJ human neonatal foreskin fibroblast cells (BJ fibroblasts) (Stemgent), WT and iNOS−/− murine embryonic fibroblasts (MEF; Cell Biologics Inc, Chicago IL) were cultured and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum and 1% penicillin/streptomycin (5% CO2, 37°C). We transdifferentiated human and mouse fibroblasts to endothelial cells (iEC) using our previously established protocol 7. Briefly, cells were treated with PIC (30 ng/mL) daily for 7 days, and the medium was gradually transitioned to a 10% knockout serum replacement. Cells were treated with endothelial cell growth medium 2 (EGM-2; Lonza) containing 20 ng/mL BMP4, 50 ng/mL VEGF, and 20 ng/mL bFGF for another 7 days. For maintenance of transdifferentiation, the cells were cultured for another 2 weeks in the presence of bFGF, VEGF, and 0.1 mmol/L 8-bromoadenosine-3′:5′-cyclic monophosphate sodium salt (8-Br-cAMP), with the medium changed every 2 days. In some experiments, inhibitors of iNOS or NFκB were present throughout the transdifferentiation protocol; they were re-added when the transdifferentiation medium was refreshed.
RNA extraction and Real-Time PCR
mRNAs were extracted from cultured cells and reverse transcribed into cDNA and then specific primers for targeted genes were used for PCR amplification. For real-time PCR, a SYBR-green PCR system or Taqman system was used and ran on QuantStudio real-time PCR system from Life Technologies. Gene expression data using the ΔΔCt method was normalized to the expression of the 18S gene and plotted as relative fold changes.
Western blot and immunoprecipitation
Western blot analysis was performed as previously described 6. Immunoprecipitation was performed using the Pierce Classic Magnetic IP/Co-IP Kit (Thermo Scientific, Waltham, MA) following manufacturer’s manual.
Chromatin immunoprecipitation (ChIP) –qPCR
ChIP was performed using a previously established method 6, 7. Recovery of genomic DNA was calculated as the ratio of copy numbers in the immunoprecipitate to the input control. Primers of the CD144 promoter were custom designed and synthesized from IDT, as follows: primer sequence (5′ to 3′), (F): CAGCCTCTTGGTTCTTCTGG; (R): GGAGTCAAGTGACCCAGCTC.
Fluorescent assay for nitric oxide (NO)
BJ fibroblasts were stained with DAF-FM DA to access intracellular NO. Cultured cells exposed to different experimental conditions were incubated with DAF-FM DA 10 μM at 37 °C for 30 min. Fluorescence was quantified by measuring fluorescence intensity at 495 nm excitation and 515 nm emission by a fluorescence plate reader (Tecan M1000 PRO).
Immunofluorescent staining
After BJ fibroblasts were cultured in 8 well chamber slides and exposed to experimental conditions, cells were fixed, permeabilized, blocked with 5% normal donkey serum, and stained for ring finger protein 1A (RING1A) and iNOS. The nuclei were stained with DAPI for 10 min.
Analysis of S-nitrosylation by mass spectrometry (MS)
Protein from both control and PIC treated BJ fibroblasts was immunoprecipated using RING1A antibody, resolved on 4–12% gradient SDS/PAGE gel and stained with Coomassie Blue. After destaining, the RING1A band was cut and sent to the mass spectrometry-proteomics core facility (Baylor College of Medicine) for MS analysis. The gel band was further destained and double digested with trypsin and GluC (NEB) in situ. Peptides in the gel were extracted, quantified and 200 ng of each sample was analyzed using the Eksigent nanoLC system and then the ABCIEX TripleTOF 5600 mass spectrometer. The Ekisgent nanoLC and the ABCIEX TripleTOF 5600 mass spectrometer were controlled by Analyst software version 1.6 (ABCIEX Inc.). The MS spectra (m/z 100–2000) were acquired with internal mass calibration. Raw data were converted to MGF format and then imported into Peaks software v6.0 for further peak generation and database search with the SPIDER function. The search sequence was E3 ubiquitin-protein ligase RING1A of human (NP_002922.2 GI:51479192).
Construction of RING1A overexpression plasmid and site directed mutagenesis
The human RING1A cDNA fragment was amplified with forward primer (5′-CAGGAATTCGAATGACGACGCCGGCGAATG-3′) and reverse primer (5′-GAGGTCGACGTCACTTTGGATCCTTGGTGGGAGC-3′) from pCMV6-XL4-RING1A (OriGene, # SC118280) and ligated into EcoRI/SalI digested pCMV-Tag2C to generate pCMV-Tag2C-RING1A. We further created pCMV-Tag2C-RING1A-C398A using a PCR-based method 11, 12. Briefly, PCR reaction was performed using primer pair: C398A Mutation-Forward (5′-CGGCCACTGGAGCTGGCCTATGCTCCCACCAAGGATCCAAAG-3′) and C398A Mutation-Reverse (5′-TTGGTGGGAGCATAGGCCAGCTCCAGTGGCCGGGACAC-3′). The PCR products were treated with DpnI and transformed into E. coli Top10 competent cells by heat shock. Single clones were sequenced to examine the C398A mutation.
Flow cytometry
Cells were trypsinized and filtered through a 40 μm nylon mesh to generate single cell suspensions. Cells were stained with antibody for cell surface marker detection. Data were analyzed by flowjo software.
Data analysis
Results are expressed as the mean±SEM. Statistical comparisons between 2 groups were performed via Student t test. One-way ANOVA was used to compare the means of multiple groups. P<0.05 was considered statistically significant.
RESULTS
iNOS is induced during transdifferentiation
Our lab has previously established a non-viral methodology to transdifferentiate fibroblasts to iECs7. We activate innate immunity with a toll-like receptor 3 (TLR3) agonist PIC to induce epigenetic plasticity. In this state of epigenetic plasticity, fibroblasts are exposed to a media containing endothelial cytokines and small molecules that favor endothelial phenotype. The iECs that we obtain using this protocol are highly similar to human dermal microvascular endothelial cells as assessed by immunohistochemical markers, acetylated LDL uptake, the generation of capillary-like networks in matrigel, the generation of NO and angiogenic cytokines, and by RNAseq analysis 7.
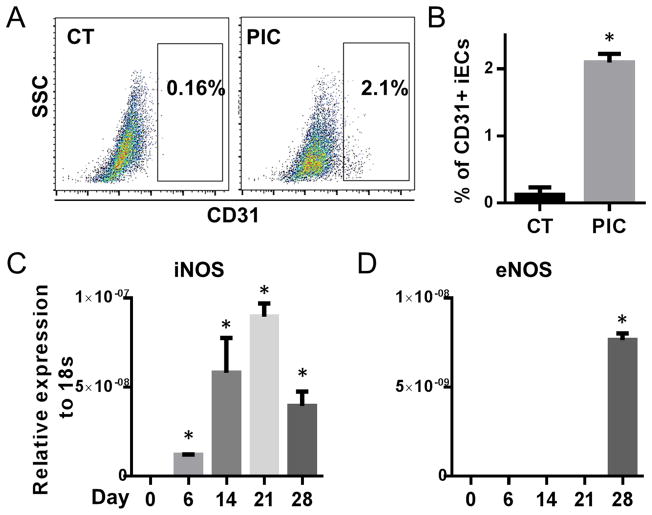
We examined NOS gene expression at day 0, 6, 14, 21, 28; by day 28 of the transdifferentiation protocol, CD31+ iECs can be detected, as quantified by FACS analysis (Fig. 1A&B). We observed that the expression of iNOS increased throughout transdifferentiation, peaking at day 21 (Fig. 1C). By contrast, the expression of eNOS was not detectable until day 28 when there was evidence of transdifferentiation to a mature endothelial cell phenotype (Fig. 1D). Throughout the whole process nNOS was not detectable (data not shown).
Figure 1. Transdifferentiation is associated with innate immune activation and iNOS expression.
BJ fibroblasts at passage 8 were treated with the 28 day transdifferentiation protocol to generate iECs. (A) Representative FACS data of CD31+ iECs at day 28. Cells were treated with the transdifferentiation protocol in the presence or absence of PIC. (B) Quantification of percentage of CD31+ cells by FACS analysis. Relative gene expression levels of (C) iNOS and (D) eNOS during transdifferentiation at day 0, 6, 14, 21 and 28 by RT-PCR. *P<0.05, vs gene expression at day 0. *P<0.05, vs vehicle treated-CT. Data are shown as the means ± SEM and are representative of 3 independent experiments.
NO is generated during transdifferentiation
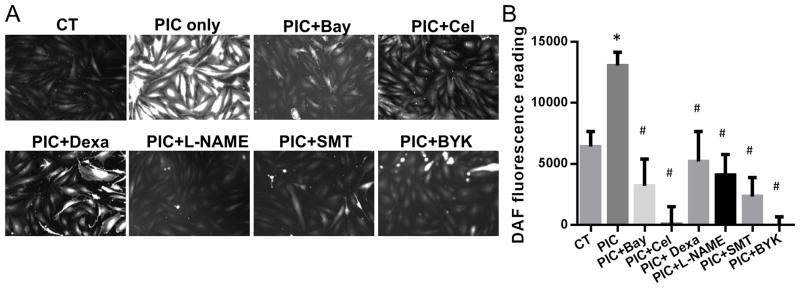
To determine if the upregulation of iNOS expression increased the generation of NO, we used DAF-FM DA staining. DAF-FM DA is essentially nonfluorescent until it reacts with NO to form a fluorescent benzotriazole 13. On day 3 of the transdifferentiation protocol, we observed a significant increase in DAF-FM DA fluorescence in both image analysis (Fig. 2A) and fluorescence readings (Fig. 2B) in the group treated with PIC compared with the vehicle-treated control. An inhibitor of all NOS isoforms L-NAME, as well as the more specific iNOS inhibitors (S)-Methylisothiourea sulfate (SMT) and BYK 191023 dihydrochloride (BYK) each reduced NO generation. In addition, the NFκB inhibitors Bay117082, celastrol or dexamethasone each blocked iNOS upregulation and NO synthesis induced by PIC (Figs. 2A&B, 3D).
Figure 2. NO generation during transdifferentiation is blocked by inhibitors of iNOS and NFκB.
Fiibroblasts were treated with the transdifferentiation protocol with or without PIC for three days. In addition, fibroblasts were treated with vehicle (control treatment, i.e. CT), or with NOS inhibitors L-NAME (100 μM), SMT (5 μM) or BYK (100 nM); or the NFκB inhibitors Bay117082 (Bay; 3 μM), celastrol (Cel; 2.5 μM) or dexamethasone (Dexa; 100 nM). (A) Representative images of DAF-FM DA staining after day 3. (B) Fluorescence intensity reading at 515 nm of DAF-FM DA of different groups by fluorescent plate reader. *P<0.05, vs vehicle treated-CT. #P<0.05, vs PIC treatment group. Data are shown as the means ± SEM and are representative of 3 independent experiments.
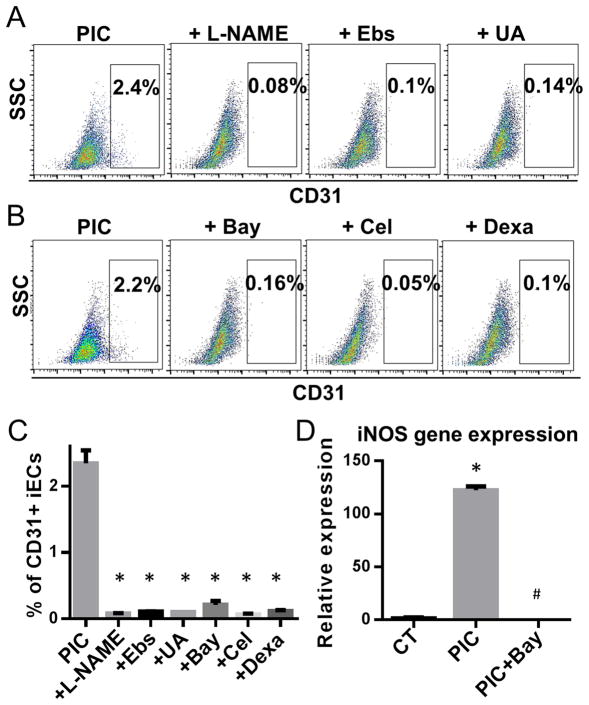
Figure 3. NOS and NFκB inhibitors abrogate transdifferentiation.
Human fibroblasts were treated with the transdifferentiation factors and PIC for three days, in the presence or absence of antagonists to the nitric oxide synthase (NOS) pathway or to NFκB activation. We used the NOS inhibitor L-NAME (100 μM), or the NO scavengers Ebselen (Ebs; 10 μM) or uric acid (UA; 100 μM); or the NFκB inhibitors Bay117082 (Bay; 3 μM), celastrol (cel; 2.5 μM) or dexamethasone (dexa; 100 nM). (A) Representative FACS data of CD31+ cells obtained with the transdifferentiation protocol, in the presence of L-NAME, Ebselen or uric acid. (B) Representative FACS data of CD31+ iEC population with Bay117082, celastrol or dexamethasone. (C) Quantification of percentage of CD31+ cells of (A) and (B). *P<0.05, vs PIC group. (D) Relative fold change in gene expression levels of iNOS at day three during transdifferentiation. Cells were treated with the transdifferentiation cocktail in the presence or absence of PIC or with PIC plus Bay117082. *P<0.05, vs vehicle treated-CT. #P<0.05, vs PIC treatment. Data are shown as the means ± SEM and are representative of 3 independent experiments.
Transdifferentiation requires iNOS induction and NO generation
To determine if generation of NO is required for transdifferentiation, we performed the transdifferentiation studies in the presence of the NOS inhibitor L-NAME or in the presence of NO scavengers uric acid or ebselen. Each of these agents substantially reduced transdifferentiation, as assessed at day 28 by FACS for CD31+ iECs (Fig. 3A&C). This indicates that NO generation is required for efficient transdifferentiation.
We hypothesized that by inhibiting NFκB activity, we would suppress the induction of iNOS and thereby reduce the efficiency of transdifferentiation. Indeed an inhibitor of NFκB activity, Bay117082, decreased iNOS gene expression (Fig. 3D) and reduced the yield of iECs (Fig. 3B&C). Two other known antagonists of NFκB-mediated iNOS induction, celastrol and dexamethasone, also reduced the yield of iECs by 90% (Fig. 3B&C). These studies confirm that NFκB-mediated induction of iNOS is required for effective transdifferentiation.
Genetic deficiency of iNOS abolishes transdifferentiation
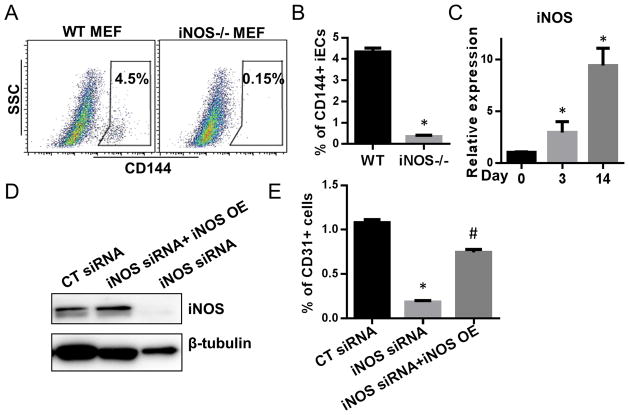
To further test the role of iNOS in transdifferentiation, we assessed the transdifferentiation of WT and iNOS −/− murine embryonic fibroblasts (MEFs) to iECs. We confirmed the increasing expression of iNOS during transdifferentiation in WT MEF at day 0, 3 and 14 (Fig. 4C), whereas iNOS gene expression was not detected in iNOS−/− MEF. We observed that transdifferentiation was nearly abolished in the iNOS−/− MEF. To confirm this work in human cells, we knocked down iNOS in BJ fibroblasts by siRNA (Fig. 4D). This knockdown of iNOS substantially reduced the yield of iECs (Fig. 4E). The effect of siRNA knockdown was reversed by iNOS overexpression using a pcDNA3.1-his-iNOS plasmid 14 (Fig. 4E). These data suggest that iNOS is required for transdifferentiation.
Figure 4. Transdifferentiation is abrogated by iNOS deficiency.
(A) Representative FACS data of CD144+ iEC after transdifferentiation of WT and iNOS−/− MEFs. (B) Quantification of CD144+ cells. *P<0.05, iNOS−/− MEFs vs WT MEFs. (C) Relative fold change in gene expression levels of iNOS during transdifferentiation of WT or iNOS−/− MEFs at day 0, 3 and 14 by RT-PCR. (D) Western blot detection of iNOS expression in BJ fibroblasts transfected with scramble-CT siRNA (10nM); with iNOS siRNA(10nM) plus pcDNA3.1-his-iNOS plasmid 14; or with 10 nM of iNOS siRNA only. (E) Quantification of CD31+ cells derived from BJ fibroblasts treated with scramble-CT siRNA, with iNOS siRNA, or with iNOS siRNA plus pcDNA3.1-his-iNOS plasmid. Data are shown as the means ± SEM and are representative of 3 independent experiments.
RING1A is S-nitrosylated during transdifferentiation
Our data indicated that the NFκB-iNOS-NO pathway is required for transdifferentiation. We were interested to know if this pathway exerted its effect on transdifferentiation by post-translational modification of epigenetic modifiers, so as to enhance epigenetic plasticity. Because NO species may react with cysteine to form S-nitrosylated (SNO) proteins, and peroxynitrite can react with L-tyrosine to form 3-nitro-L-tyrosine (3-NT), we quantified SNO and 3-NT containing proteins as an index of iNOS activity. As shown in western analysis, cell lysate protein SNO (Fig. 5A) and 3-NT levels are significantly increased as early as day 3 after the induction of transdifferentiation (Online Figure I).
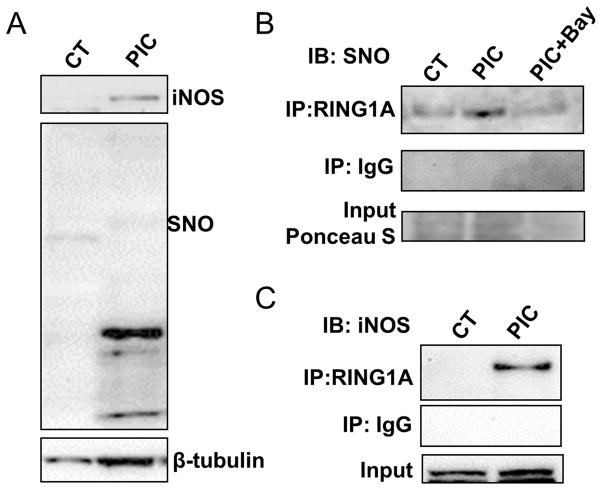
Figure 5. iNOS binds and S-nitrosylates Ring1A.
BJ fibroblasts were treated with vehicle; PIC; or with PIC and the NFκB inhibitor Bay117082 for 3 days and proteins were collected for western blot or immunoprecipitation application. (A) Western blot detection of iNOS and SNO protein expression. (B) Proteins from cell lysate were immunoprecipitated Online with SNO or control IgG antibody and subject to western blot detection of RING1A. (C) Proteins from cell lysate were immunoprecipitated with RING1A or control IgG antibody and subject to western blot detection of SNO protein. (D) Proteins from cell lysate were immunoprecipitated with RING1A or control IgG antibody and subject to western blot detection of iNOS protein. Data are shown as the means ± SEM and are representative of 3 independent experiments.
It has been previously reported that HDAC2, RING1A, CBX1 can be S-nitrosylated 15–18. Accordingly, we turned our attention to detecting S-nitrosylation of epigenetic modifiers focusing first on HDAC2, RING1A and CBX1. We observed that RING1A was SNO modified at day 3 during transdifferentiation (Fig. 5B), but not HDAC2 or CBX1 (Online Figure II). We confirmed the RING1A S-nitrosylation by immunoprecipitation of RING1A, followed by probing with SNO antibody. We found that in PIC treated BJ fibroblasts RING1A was S-nitrosylated (Fig. 5C). Consistent with our hypothesis, NFκB inhibitor Bay117082 could block this S-nitrosylation (Fig. 5C), suggesting that NFκB-iNOS pathway is upstream of RING1A S-nitrosylation.
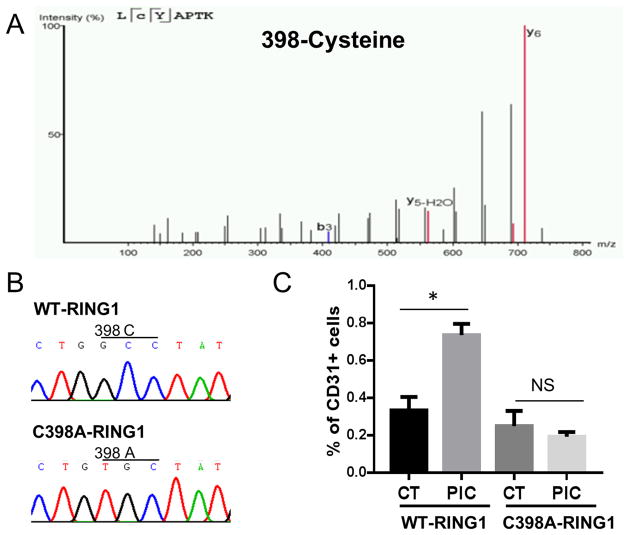
To identify the specific cysteine that is S-nitrosylated, we performed MS analysis of the RING1A immunoprecipitated from cell lysate of fibroblasts treated with vehicle or PIC. The MS studies revealed Cysteine at 398 position is heavily S-nitrosylated (Fig. 6A) by PIC stimulation. In addition, nitroso-cysteine modifications could also be detected in 279 and 332 position cysteines, although the modification level is low (Online Figure III).
Figure 6. MS analysis reveals Cys398 S-nitrosylation of RING1A. Overexpression of C398A-RING1A mutant blocks transdifferentiation.
(A) Representative MS/MS fragmentation spectrum for the Cys398-containing peptide. MS spectra of the cysteine-containing peptides identified after MS analysis. Peptide sequence is shown at the top left of each spectrum, with the annotation of the identified matched amino terminus-containing ions (b ions) in black and the carboxyl terminus-containing ions (y ions) in red. The spectrum confirms the identity of the peptides LCYAPTK and the labeled C as S-nitrosylated cysteine. (B) Sequence confirmation of pCMV-Tag2C-RING1A and pCMV-Tag2C-RING1A-C398A plasmid. (C) BJ fibroblasts were transfected with either pCMV-Tag2C-RING1A-C398A (WT-RING1A) or pCMV-Tag2C-RING1A-C398A (C398A-RING1A) plasmid and then subject to transdifferentiation protocol. At day 3, proteins were collected and immunoprecipated with Flag or control IgG antibody. Products were examined by western blot detection of SNO protein. (D) BJ fibroblasts were transfected with either pCMV-Tag2C-RING1A-C398A (WT-RING1A) or pCMV-Tag2C-RING1A-C398A (C398A-RING1A) plasmid and then subject to transdifferentiation protocol with or without PIC. At day 28, CD31+ iECs were quantified by FACS analysis. *P<0.05. Data are shown as the means ± SEM and are representative of 3 independent experiments.
To further examine whether S-nitrosylation of RING1A at 398 cysteine is critical for transdifferentiation, we performed site directed mutagenesis. We first constructed plasmids containing wildtype (pCMV-Tag2C-RING1A) or mutant RING1A (C398A mutation, pCMV-Tag2C-RING1A-C398A plasmid). The GCC codon encoding the 398 position cysteine was mutated into a TGC codon encoding alanine, which we confirmed by sequencing (Fig. 6B). Then both plasmids were transfected into BJ fibroblasts, which were then subjected to the transdifferentiation protocol with or without PIC. As shown in Fig. 6C, C398A-RING1A is minimally S-nitrosylated after PIC, suggesting that the 398 position cysteine is the major site of S-nitrosylation. Subsequently, we assessed the effect of overexpression of the mutant RING1A on transdifferentiation. Fibroblasts overexpressing C398A-RING1A were resistant to transdifferentiation (Fig. 6D), by comparison to fibroblasts overexpressing the wildtype form of RING1A. Altogether, these data suggest that S-nitrosylation of RING1A at 398 cysteine is critical for transdifferentiation.
iNOS translocates to the nucleus and binds RING1A
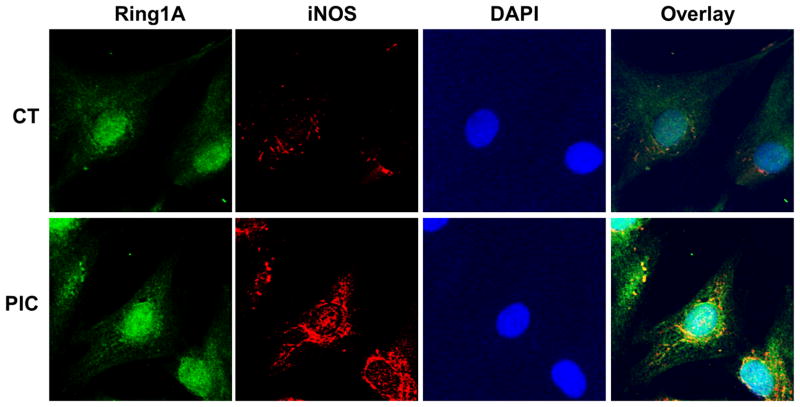
A recent study indicates that iNOS may bind to its protein target, providing some specificity of S-nitrosylation 14. To determine if iNOS physically associates with RING1A, we immunoprecipitated RING1A and probed for iNOS (Fig. 5D). These experiments indicated that iNOS does bind to RING1A. Further, we examined the cellular localization of iNOS and RING1A. Under basal conditions, iNOS was primarily observed in the cytoplasm. After PIC treatment, we observed iNOS translocated to the perinuclear and nuclear region. Co-localization of iNOS and RING1A in the nucleus (Fig. 7) can also be detected. A physical association of iNOS with the epigenetic modifier RING1A could provide for specificity of the RING1A-SNO modification.
Figure 7. iNOS colocalizes with RING1A during transdifferentiation.
BJ fibroblasts treated with or without PIC for three days were fixed and stained with anti-RING1A antibody (green), anti-iNOS antibody (red) and DAPI. Images were taken under the confocal microscope to analyze the colocalization of RING1A and iNOS.
RING1A nitrosylation reduces its chromatin binding as well as suppressive chromatin marks in endothelial promoters
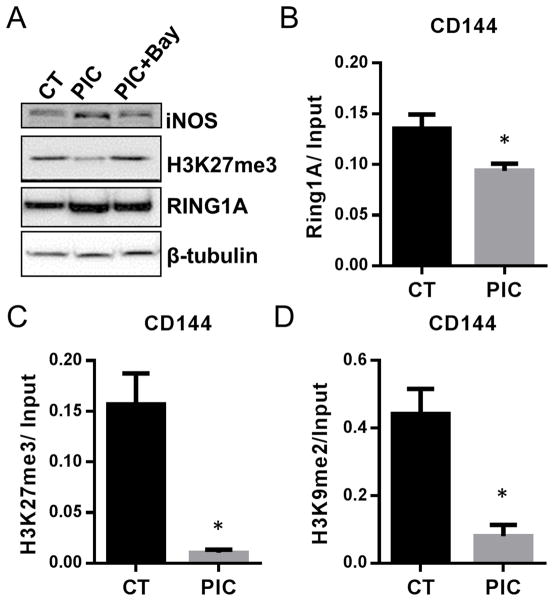
RING1A is a critical component of polycomb repressive complex (PRC) 1 and is required for stabilizing repressive epigenetic marks such as H3K27trimethylation (H3K27me3) 19, 20. To examine whether the SNO modification of RING1A has any impact on cellular H3K27me3 level, we performed western blot and found that cellular H3K27me3 levels were decreased in PIC treated cells. This effect of PIC was abrogated by using an NFκB inhibitor Bay117082 to block iNOS induction (Fig. 8A). This result suggested that the RING1A-SNO modification may destabilize the PRC complex to reduce H3K27 trimethylation.
Figure 8. Innate immune activation decreases RING1A binding to chromatin and reduces repressive histone markers.
BJ fibroblasts treated with or without PIC for three days during the transdifferentiation protocol. (A) Western blot examination of iNOS, H3K27me3, RING1A and SNO protein expression. (B) RING1A, (C) H3K27me3 and (D) H3K9me2 binding to the CD144 promoter were analyzed by ChIP-PCR. *P<0.05, vs vehicle treated-CT. Data are shown as the means ± SEM and are representative of 3 independent experiments.
To confirm that SNO modification of RING1A impairs its binding to chromatin, and thereby reduces H3K27me3, we used ChIP assay. Specifically, we examined the promoter region of CD144, an endothelial-specific gene which is fully repressed in fibroblasts. After PIC treatment for 3 days, RING1A binding to CD144 promoter is decreased (Fig. 8B) together with decreased levels of H3K27me3 (Fig. 8C). In addition, H3K9me2 is also decreased in the CD144 promoter (Fig. 8D). These studies are consistent with a functional effect of S-nitrosylation to reduce RING1A association with the chromatin and thereby to decrease the suppressing histone marks at promoters of endothelial genes during transdifferentiation.
DISCUSSION
A novel mechanism underpinning transdifferentiation
The new and salient findings of this paper are that 1) During transdifferentiation of fibroblasts to endothelial cells, the expression of iNOS is induced, and NO is generated; 2) The generation of NO is required for efficient transdifferentiation; 3) NO facilitates transdifferentiation in part by blocking repressive epigenetic markings; 4) This effect is achieved at least in part by iNOS binding to and nitrosylating RING1A, and 5) The S-nitrosylation of RING1A causes it to dissociate from the chromatin, impairing the ability of the polycomb repressive complex to maintain H3K27 trimethylation. Most importantly, we have identified a critical role for NO in transdifferentiation. Our results indicate that iNOS serves as an S-nitrosylase that directly binds to an epigenetic modifier to regulate chromatin remodeling during transdifferentiation. The temporal and spatial characteristics of the S-nitrosylation may provide the specificity of the epigenetic effect. We have previously shown that innate immunity is required for efficient nuclear reprogramming 6. This is the first demonstration that NO generated during innate immune signaling regulates epigenetic plasticity.
These observations significantly extend our previous work which revealed that innate immune activation is required for cellular metamorphoses. Specifically we showed previously that TLR3 stimulation facilitates nuclear reprogramming of human fibroblasts to iPSCs 6 or their transdifferentiation into endothelial cells 7. We found that TLR3 stimulation by PIC increased the expression of HAT family members and reduced the expression of HDAC family members and Dot1L. These changes would be expected to increase epigenetic plasticity 21, 22 an expectation that was confirmed by ChIP-qPCR studies of the relevant promoter regions 6, 7.
iNOS is an effector of innate immunity and epigenetic plasticity
Innate immune activation of NFκB induces iNOS expression 23, 24. Subsequently, iNOS generates NO which can activate guanylate cyclase to produce the second messenger cGMP. In addition, NO may S-nitrosylate cysteine residues of proteins to modify their function25. For example, S-nitrosylation of caspase 3 regulates apoptosis 26. Others have shown that S-nitrosylation of the epigenetic modifier HDAC2 in embryonic cortical neurons regulates dendritic growth and branching 15. In addition, iNOS has been reported to translocate into the nucleus both in malignant 27 and normal cells 28, 29 but the biological implications of this translocation are obscure. Some epigenetic modifiers have been shown to be capable of S-nitrosylation in screening assays 16–18, including RING1A and CBX1. However, it is not known whether these proteins are post-translationally modified in cells and whether the modifications modulate their function.
Thus we hypothesized that iNOS-derived NO is a major effector of innate immunity in transdifferentiation. We found that iNOS is indeed upregulated during transdifferentiation (Fig. 1) in association with increases in intracellular NO (Fig. 2), and with increased generation of SNO- (Fig. 5A) and 3-NT modifications of cellular proteins (Online Figure I). Notably, pharmacological antagonists that reduced intracellular NO levels consistently diminished transdifferentiation efficiency (Fig. 3A). Furthermore, inhibitors of NFκB blocked the upregulation of iNOS, reduced NO levels, and also impaired transdifferentiation efficiency (Fig. 3B&D). Together these studies indicated that iNOS-generated NO is critical for transdifferentiation.
To further confirm iNOS is required for effective transdifferentiation, we derived MEFs from iNOS−/− mice 30. These mice have a knock out of the iNOS calmodulin binding domain encoded by exon 12–13 and are not capable of generating NO after LPS challenge. Accordingly, we observed that iNOS−/− MEFs are resistant to transdifferentiation (Fig. 4A). We then observed similar results in human fibroblasts by transient knockdown of iNOS. Overexpression of iNOS in the knockdown cells restores the transdifferentiation capacity (Fig. 4E). Taken together, the genetic and pharmacological studies suggest that iNOS-generated NO is required for transdifferentiation.
Nitrosylation of RING1A is required for transdifferentiation
Next, we screened several epigenetic modifiers that have been reported to be S-nitrosylated. Of these, only RING1A is heavily S-nitrosylated at the 398 position cysteine during innate immune activation as assessed by MS (Fig. 6A) and Co-IP studies (Fig. 5). The S-nitrosylation of a protein can be achieved through several different mechanisms 10, 25, 31, 32. Recently it has been shown that protein S-nitrosylation can be achieved in a target-selective manner at a sequence motif recognition site by an inducible heterotrimeric S-nitrosylase complex 14. This protein complex consists of iNOS, S100A8, and S100A9 and recognizes a conserved I/L-X-C-X2-D/E motif. Of note, although RING1A lacks this motif, our Co-IP data indicates that iNOS associates with and S-nitrosylates RING1A (Fig. 5D). Immunostaining also confirms the colocalization of iNOS and RING1A in the nucleus (Fig. 7). This physical association is likely to provide some specificity of the S-nitrosylation. Indeed, we were not able to rescue the transdifferentiation process in iNOS−/− MEFs using the NO donor SIN1 (data not shown). These experimental results indicate a critical role of the physical association of iNOS with RING1A for transdifferentiation.
RING1A is a major component of the PRC1 33, which acts as a transcriptional repressor 34. A sequential recruitment of PRC2-PRC1 has been described 35, 36 in that PRC1 recognizes the H3K27me3 marker that is imposed by PRC2 37–40. Maintenance of H3K27 trimethylation requires PRC1 induced H2A monoubiquitination 19, 20. Knockout of RING1A reduces H3K27me3 marks34. In this regard, we observed that PIC reduced binding of RING1A to, and H3K27 trimethylation of, the endothelial specific gene promoter CD144 (Fig. 8B&C). These observations suggest the when RING1A is S-nitrosylated, it dissociates from the chromatin, thereby releasing the PRC complex and destabilizing repressive marks such as H3K27me3.
Physiological and pathobiological relevance of our findings
iNOS is a potent cellular defense mechanism, generating reactive nitrogen species that also have cytotoxic effects. We now reveal another role for iNOS in innate immunity, as a facilitator of epigenetic plasticity and phenotypic fluidity. This too may be an important mechanism for cellular defense, as phenotypic fluidity in the face of pathogens or damage may provide a survival advantage.
In some instances, the epigenetic plasticity associated with innate immune activation may have a physiological role. For example, in the setting of experimental myocardial infarction, a substantial number of cardiac fibroblasts may transdifferentiate to endothelial cells to enhance the angiogenic response to ischemic injury 41. Epigenetic plasticity induced by innate immunity might also participate in pathological transdifferentiation. For example, the chronic inflammation associated with gastroesophageal reflux disease is accompanied by a pre-malignant transdifferentiation of the stratified squamous esophageal epithelium to a columnar epithelium reminiscent of the small intestine42.
Indeed, it has been long recognized by clinicians that cancers may arise in the setting of chronic inflammation or infection 43, 44. Furthermore, iNOS is often expressed in pre-malignant states and in cancer 45–48. An understanding of the role by which chronic inflammation leads to epigenetic plasticity may provide a therapeutic avenue in the prevention of cancer. Strategies for direct reprogramming of somatic cells may be enhanced by insights into the mechanisms by which innate immune signaling drives epigenetic plasticity.
Conclusion
We have identified a novel mechanism by which innate immune signaling facilitates transdifferentiation. The activation of iNOS and the generation of NO are associated with iNOS binding to, and S-nitrosylating RING1A. This modification causes a dissociation of RING1A from the chromatin. This effect is associated with a reduction in H3K27 trimethylation, a repressive chromatin marking. These effects would be predicted to increase the probability of an open configuration of the chromatin and thereby facilitate transdifferentiation. These insights have relevance to cancer biology in that pathological transformation frequently occurs in the setting of chronic inflammation. Our work also has relevance to cardiovascular regeneration, given the importance of physiological transdifferentiation in the response to injury.
Supplementary Material
Novelty and Significance.
What Is Known?
Transdifferentiation describes the process by which a somatic cell undergoes a change in phenotype.
Transdifferentiation may contribute to disease, such as with endothelial-to-mesenchymal transition, which participates in cardiac fibrosis.
Transdifferentiation may contribute to regeneration, such as with mesenchymal-to-endothelial transition (EndoMT), which contributes to angiogenesis in ischemic myocardium
What New Information Does This Article Contribute?
Transdifferentiation of fibroblasts to endothelial cells requires expression of inducible nitric oxide synthase (iNOS).
RING1A, a component of the polycomb repressive complex, is bound by iNOS and nitrosylated.
The nitrosylation of RING1A reduces repressive epigenetic markings in lineage specific gene promoters, facilitating EndoMT
Transdifferentiation of one cell type to another plays a role in disease, as well as in development and regeneration. The process of transdifferentiation from one cell lineage to another requires epigenetic changes that unlock those genes that are needed for expression of the new phenotype (and which suppress those genes that were responsible for the previous phenotype). An understanding of the epigenetic changes that govern these changes in gene expression (and thus cell phenotype) will lead to new therapeutic avenues. We have shown that activation of innate immunity causes global changes in the expression of epigenetic modifiers that can enhance epigenetic plasticity. In the current paper, we show that iNOS-generated NO, which is one of the major effectors of innate immunity, also plays a critical role in mediating the epigenetic plasticity that is required for the transdifferentiation of human fibroblasts to endothelial cells. The nitrosylation of RING1A, a component of the polycomb repressive complex, causes the dissociation of this complex from the chromatin, thereby reducing repressive epigenetic markings, such as H3K27 trimethylation. These insights may lead to new approaches to enhance therapeutic transdifferentiation, e.g. mesenchymal-to-endothelial transition to enhance vascularity in ischemic syndromes.
Acknowledgments
We thank Dr. Paul L. Fox for kindly providing us the pcDNA3.1-his-iNOS plasmid. We thank Dr. Hon-Chiu Eastwood Leung for his expertise in helping with the mass spectrometry analysis.
SOURCES OF FUNDING
This work supported in part by grants to Dr. Cooke from the National Institutes of Health (U01 HL100397) and the Cancer Prevention and Research Institute (RP150611). SM is supported by a postdoctoral fellowship from the American Heart Association.
Nonstandard Abbreviations and Acronyms
- EC
endothelial cells
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- iNOS
Inducible nitric oxide synthase
- PIC
polyinosinic: polycytidylic acid
- ChIP
chromatin immunoprecipitation
- NO
nitric oxide
- RING1A
ring finger protein 1A
- MS
mass spectrometry
- TLR3
toll-like receptor 3
- MEF
murine embryonic fibroblast
- PRC
polycomb repressive complex
Footnotes
DISCLOSURES
Dr. Cooke is an inventor on patents owned by Stanford University related to the manipulation of innate immune signaling for nuclear reprogramming so as to induce pluripotency, transdifferentiation, or other therapeutic cellular modifications.
References
- 1.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai T-n, Baban D, Ragoussis J, Huang Y, Han J-DJ, Zeng L, Hu Y, Xu Q. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proceedings of the National Academy of Sciences. 2012;109:13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. 2013;33:1366–1375. doi: 10.1161/ATVBAHA.112.301167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayed N, Wong WT, Ospino F, Meng S, Lee J, Jha A, Dexheimer P, Aronow BJ, Cooke JP. Transdifferentiation of human fibroblasts to endothelial cells: Role of innate immunity. Circulation. 2015;131:300–309. doi: 10.1161/CIRCULATIONAHA.113.007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Predonzani A, Cali B, Agnellini AH, Molon B. Spotlights on immunological effects of reactive nitrogen species: When inflammation says nitric oxide. World journal of experimental medicine. 2015;5:64–76. doi: 10.5493/wjem.v5.i2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nature reviews Microbiology. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 10.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein s-nitrosylation: Purview and parameters. Nature reviews Molecular cell biology. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Gu Q, Lin L, Li S, Zhong S, Li Q, Cui Z. Nucleoporin 62-like protein activates canonical wnt signaling through facilitating the nuclear import of beta-catenin in zebrafish. Molecular and cellular biology. 2015;35:1110–1124. doi: 10.1128/MCB.01181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC biotechnology. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stroissnigg H, Trancikova A, Descovich L, Fuhrmann J, Kutschera W, Kostan J, Meixner A, Nothias F, Propst F. S-nitrosylation of microtubule-associated protein 1b mediates nitric-oxide-induced axon retraction. Nature cell biology. 2007;9:1035–1045. doi: 10.1038/ncb1625. [DOI] [PubMed] [Google Scholar]
- 14.Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL, Fox PL. Target-selective protein s-nitrosylation by sequence motif recognition. Cell. 2014;159:623–634. doi: 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 16.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of s-nitrosylation and denitrosylation by resin-assisted capture. Nature biotechnology. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YI, Giovinazzo D, Kang HC, Lee Y, Jeong JS, Doulias PT, Xie Z, Hu J, Ghasemi M, Ischiropoulos H, Qian J, Zhu H, Blackshaw S, Dawson VL, Dawson TM. Protein microarray characterization of the s-nitrosoproteome. Molecular & cellular proteomics: MCP. 2014;13:63–72. doi: 10.1074/mcp.M113.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seth D, Stamler JS. The sno-proteome: Causation and classifications. Current opinion in chemical biology. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pengelly AR, Kalb R, Finkl K, Muller J. Transcriptional repression by prc1 in the absence of h2a monoubiquitylation. Genes & development. 2015 doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. Histone h2a monoubiquitination promotes histone h3 methylation in polycomb repression. Nature structural & molecular biology. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 21.Chen T, Dent SY. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nature reviews Genetics. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of klf4. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie QW, Kashiwabara Y, Nathan C. Role of transcription factor nf-kappa b/rel in induction of nitric oxide synthase. The Journal of biological chemistry. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 24.Lawrence T. The nuclear factor nf-kappab pathway in inflammation. Cold Spring Harbor perspectives in biology. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous s-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein s-nitrosylation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall HE, Hess DT, Stamler JS. S-nitrosylation: Physiological regulation of nf-κb. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8841–8842. doi: 10.1073/pnas.0403034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TW, Chen GG, Xu H, Yip JH, Chak EC, Mok TS, Yim AP. Differential expression of inducible nitric oxide synthase and peroxisome proliferator-activated receptor gamma in non-small cell lung carcinoma. European journal of cancer. 2003;39:1296–1301. doi: 10.1016/s0959-8049(02)00733-5. [DOI] [PubMed] [Google Scholar]
- 28.Jones RJ, Jourd’heuil D, Salerno JC, Smith SM, Singer HA. Inos regulation by calcium/calmodulin-dependent protein kinase ii in vascular smooth muscle. American journal of physiology Heart and circulatory physiology. 2007;292:H2634–2642. doi: 10.1152/ajpheart.01247.2006. [DOI] [PubMed] [Google Scholar]
- 29.Giordano A, Tonello C, Bulbarelli A, Cozzi V, Cinti S, Carruba MO, Nisoli E. Evidence for a functional nitric oxide synthase system in brown adipocyte nucleus. FEBS letters. 2002;514:135–140. doi: 10.1016/s0014-5793(02)02245-7. [DOI] [PubMed] [Google Scholar]
- 30.Laubach VE, Shesely EG, Smithies O, Sherman PA. Mice lacking inducible nitric oxide synthase are not resistant to lipopolysaccharide-induced death. Proceedings of the National Academy of Sciences. 1995;92:10688–10692. doi: 10.1073/pnas.92.23.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell DA, Marletta MA. Thioredoxin catalyzes the s-nitrosation of the caspase-3 active site cysteine. Nature chemical biology. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 33.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by prc1, a polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 34.Satijn DP, Gunster MJ, van der Vlag J, Hamer KM, Schul W, Alkema MJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. Ring1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Molecular and cellular biology. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Molecular cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. Variant prc1 complex-dependent h2a ubiquitylation drives prc2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone h3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 38.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of zeste/esc complexes have a histone h3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 39.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 41.Ubil E, Duan J, Pillai IC, Rosa-Garrido M, Wu Y, Bargiacchi F, Lu Y, Stanbouly S, Huang J, Rojas M, Vondriska TM, Stefani E, Deb A. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514:585–590. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stairs DB, Nakagawa H, Klein-Szanto A, Mitchell SD, Silberg DG, Tobias JW, Lynch JP, Rustgi AK. Cdx1 and c-myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward barrett’s esophagus. PloS one. 2008;3:e3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Molecular cancer research: MCR. 2006;4:221–233. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- 44.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cianchi F, Cortesini C, Fantappie O, Messerini L, Schiavone N, Vannacci A, Nistri S, Sardi I, Baroni G, Marzocca C, Perna F, Mazzanti R, Bechi P, Masini E. Inducible nitric oxide synthase expression in human colorectal cancer: Correlation with tumor angiogenesis. The American journal of pathology. 2003;162:793–801. doi: 10.1016/S0002-9440(10)63876-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechner M, Lirk P, Rieder J. Inducible nitric oxide synthase (inos) in tumor biology: The two sides of the same coin. Seminars in cancer biology. 2005;15:277–289. doi: 10.1016/j.semcancer.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Janakiram NB, Rao CV. Inos-selective inhibitors for cancer prevention: Promise and progress. Future medicinal chemistry. 2012;4:2193–2204. doi: 10.4155/fmc.12.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W, Liu LZ, Loizidou M, Ahmed M, Charles IG. The role of nitric oxide in cancer. Cell research. 2002;12:311–320. doi: 10.1038/sj.cr.7290133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.