Abstract
Hyaluronan (HA) plays an essential role in cartilage where it functions to retain aggrecan. Previous studies have suggested that aggrecan is anchored indirectly to the plasma membrane of chondrocytes via its binding to cell-associated HA. However, reagents used to test these observations such as hyaluronidase and HA oligosaccharides are short term and may have side activities that complicate interpretation. Using the CRISPR/Cas9 gene editing approach, a model system was developed by generating HA-deficient chondrocyte cell lines. HA synthase-2 (Has2)-specific single guide RNA was introduced into two different variant lines of rat chondrosarcoma chondrocytes; knockout clones were isolated and characterized. Two other members of the HA synthase gene family were expressed at very low relative copy number but showed no compensatory response in the Has2 knockouts. Wild type chondrocytes of both variants exhibited large pericellular matrices or coats extending from the plasma membrane. Addition of purified aggrecan monomer expanded the size of these coats as the proteoglycan became retained within the pericellular matrix. Has2 knockout chondrocytes lost all capacity to assemble a particle-excluding pericellular matrix and more importantly, no matrices formed around the knockout cells following the addition of purified aggrecan. When grown as pellet cultures so as to generate a bioengineered neocartilage tissue, the Has2 knockout chondrocytes assumed a tightly-compacted morphology as compared to the wild type cells. When knockout chondrocytes were transduced with Adeno-ZsGreen1-mycHas2, the cell-associated pericellular matrices were restored including the capacity to bind and incorporate additional exogenous aggrecan into the matrix. These results suggest that HA is essential for aggrecan retention and maintaining cell separation during tissue formation.
Keywords: hyaluronan, hyaluronan synthase-2, aggrecan, chondrocytes
1. Introduction
One event associated with osteoarthritis (OA)1 is the loss of aggrecan proteoglycan from the extracellular matrix (ECM) of articular cartilage [1-3]. In addition to aggrecan, a significant loss of hyaluronan (HA) is also observed in human and animal models of OA [4-8]. For example, cultured explants of human articular cartilage treated with IL-1α displayed a loss of HA coincident with a loss of aggrecan within the same layers of the tissue [8]. Primary chondrocytes treated with IL-1β or IL-1α as models of OA, adopt an enhanced catabolic metabolism that generates a loss of the prominent HA / aggrecan-rich glycocalyx or pericellular coat [8, 9]. As the loss of ECM components is associated with the enhanced catabolism of OA chondrocytes, we hypothesize that the re-establishment of a HA / aggrecan-rich pericellular matrix will provide positive signals and promote a steady-state metabolism. We further propose that HA is the limiting factor necessary for this pericellular matrix repair to be successful. To test these hypotheses we have developed a variety of approaches to selectively enhance the accumulation of cell-associated HA as well as aggrecan. What is lacking in our studies, and the field in general, is a suitable, stable, HA-deficient chondrocyte cell line—a model chondrocyte that is also amenable to plasmid transfection or viral transduction. Experiments using primary murine, bovine or human OA chondrocytes, made HA-free by treatment with a hyaluronidase are always a race against time (6-12 hours) before endogenous biosynthesis re-establishes pericellular HA levels.
We and others have used the rat chondrosarcoma (RCS) cell line as a chondrocyte model system [9-14]. RCS cells are a continuous long-term cultured line derived from the Swarm rat chondrosarcoma tumor [15, 16]. Several different RCS cell lines are in use by investigators [9, 12-14] and each differs in various aspects of the chondrocyte phenotype. To our knowledge, these cells lines were all derived (grown out) from tumors propagated from the Swarm rat chondrosarcoma but by different investigators at different times. The cells of the RCS cell line used often in our laboratory (labeled for this study as RCS-o) are more elongated than round and synthesize less aggrecan. However, these cells display HA / aggrecan-dependent pericellular matrices that are retained at the cell surface via CD44; retention of the HA / aggrecan-rich matrix can be blocked with the use of anti-CD44 antibodies, HA oligosaccharides [11] or overexpression of the CD44 intracellular domain [9]. These cells also exhibit the capacity for CD44-mediated endocytosis of HA [10, 12]. Both of these properties are similar to what we observe using primary cultures of bovine articular chondrocytes [11, 17-19]. The clustered regularly interspersed short palindromic repeat (CRISPR) technology using RNA-guided Cas9 endonuclease is a powerful gene editing tool. In 2014, Yang et al. [13] developed a Cas9 stable transfectant clone of a different RCS cell line and used these cells to generate a CRISPR/Cas9 mediated knockout of aggrecan. We obtained the wild type parental RCS cells (labeled in this study as RCS-Cas9). These cells are more round in morphology, exhibit high aggrecan biosynthesis, prominent HA / aggrecan pericellular matrices but express less CD44. Both RCS cell lines display relevant phenotypic aspects that we observe in primary cultures of articular chondrocytes and thus serve as useful models.
HA in eukaryotic cells is synthesized by one or more of three HA synthases, HAS1, HAS2 and HAS3 [20, 21]. HA is synthesized directly at the inner surface of the plasma membrane and extruded through the plasma membrane into the pericellular space. HA can remain anchored to the plasma membrane by continued interaction with the HA synthase and/or via binding to cell surface receptors such as CD44 [22, 23]. We have used displacement of pericellular coats or 3H-labeled HA by HA oligosaccharides or unlabeled high molecular mass HA, to differentiate displaceable HA (receptor-bound) from non-displaceable HA (assumed to be HAS-bound) [11, 22, 24, 25]. For example, during the initial biosynthesis of HA by chick embryo chondrocytes, all membrane-associated 3H-HA was non-displaceable for the first 4 hours, switching to predominately displaceable at times thereafter [24]. Although all three HA synthases have the capacity to synthesize HA, we demonstrated that primary human chondrocytes utilize the HAS2 synthase as the predominant HA synthesizing isoform [26]. Has2 knockout in mice results in embryonic lethality due to disruption of cardiac development [27]. Conditional inactivation of Has2 of early limb bud mesenchyme by introduction of the Prx1-Cre transgene results in skeletal deformities and severely shorten limbs due to abnormal and disorganized growth plates and a decrease in aggrecan deposition into the ECM [28]. Has1 and Has3 did not appear to compensate for the HA deficiency in the conditional inactivation mice although this was not determined directly.
In this study we have developed a single guide RNA (sgRNA) to target a Cas9 dependent cleavage within exon 2 of the rat Has2 gene. We have successfully generated Has2 mutations in two different RCS cell lines, RCS-o and RCS-Cas9—mutations that blocked the synthesis of HA in the resultant cloned cells. Has2 knockout cells lost the ability to assemble a HA / aggrecan-rich pericellular matrix and lost the capacity to retain exogenously added, purified aggrecan. Other questions addressed were the effect of HA loss on cell-cell spacing during neocartilage formation, changes in aggrecan synthesis and retention, and the potential for compensation by the Has1 and Has3 synthases.
2. Results
2.1. Selection and screening for Has2 knockout clones
Following transfection of RCS-o and RCS-Cas9 cells with the PX458 plasmid containing a 20 nt sgRNA sequence targeting Has2, Cas9 and GFP, the GFP+ cells were selected and propagated following dilution cloning by the FACS cell sorter (Fig. 1, panels A and B, top left). Following growth of the individual clones, cells were analyzed for cell surface-associated HA by staining with a HA binding protein (HABP) and fluorescent microscopy. The micrographs in Figure 1A, 1B, show the presence of HA by Alexa-Fluor-488 fluorescent detection of HABP on wild type RCS-o and RCS-Cas9 cells. Shown at higher magnification is the HABP staining for the RCS-o WT cells counterstained with DAPI (panel A) and the RCS-Cas9 cells that co-express mCherry (Panel B). Some of the GFP+ cells generated clones still exhibited HABP staining of cell surface HA (RCS-o Has2 KO clones 1 and 3) and likely represent unsuccessful knockouts. However, ∼80% of the GFP+ cells no longer exhibited HABP staining of cell surface HA and several were selected for further analysis. The conditioned media of RCS-o Has2 KO clones 4 and 7 as well as RCS-Cas9 Has2 KO clones 3 and 7 showed a nearly non-detectable level of HA by ELISA (Fig. 1C; shown as percent of control RCS media HA). However, RCS-o Has2 KO clones 1 and 3 exhibited both cell surface HABP staining and the culture media contained HA, albeit at reduced levels compared to RCS-o WT cells. The conditioned media was next analyzed for proteoglycan content using the DMMB assay to measure sulfated glycosaminoglycan (sGAG). In Fig. 1D, more sGAG accumulated in the medium of monolayer cultures of the RCS-Cas9 Has2 KO clones as compared to RCS-Cas9 WT cells. In another experiment, when sGAG recovered from the cell layer was added to the conditioned medium fraction (Fig. 1E) the total sGAG produced by Has2 KO clones and the RCS WT cells was equivalent. This demonstrates that the WT RCS-Cas9 cells retain a substantial proportion of proteoglycan to the cell surface whereas the Has2 KO clones release a substantial proportion of proteoglycan directly into the medium.
Fig. 1. Selection and screening of transfected RCS cells for Has2 knockout clones.
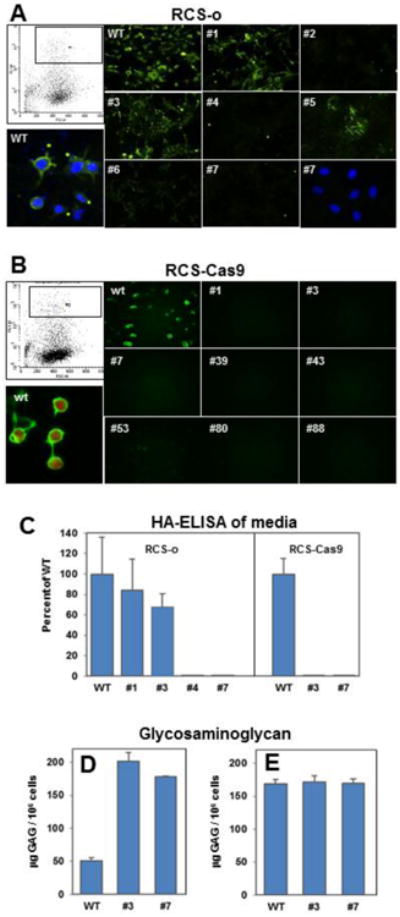
Panel A: Representative flow cytometric cell sorting of RCS-o transfected chondrocytes to select GFP+ cell is shown in upper left. WT RCS-o and GFP+ cloned RCS cells were stained with b-HABP to detect cell surface associated HA; individual clone numbers are indicated. WT RCS-o cells with DAPI counterstain and positive HABP staining for cell surface HA is shown in lower left. Clone #2, #4 and #7 are negative for HABP staining; lower right panel shows DAPI staining of the same field of Clone #7 cells. Panel B: Representative flow cytometric cell sorting of RCS-Cas9 transfected chondrocytes to select GFP+ cell is shown in upper left. WT RCS-Cas9 and GFP+ cloned RCS cells were stained with b-HABP to detect cell surface associated HA; individual clone numbers are indicated. WT RCS-Cas9 cells show positive HABP staining for cell surface HA and mCherry fluorescence is shown in lower left. Panels A and B show representative images from 3 independent experiments. Panel C. Media from the cultures of WT and Has2 KO clones was analyzed by HA ELISA. Values represent the percent of the mean value for WT RCS-o or WT RCS-Cas9 detected in the media of the numbered clones from two independent experiments. Panel D: Release of sGAG by RCS-Cas9 WT and Has2 KO clones into the conditioned media was determined by the DMMB assay. Values represent the mean μg GAG per 106 cells from two independent experiments. Panel E: Total sGAG in the cultures is presented as the sum of sGAG in the conditioned media + obtained from the cell layer of RCS-Cas9 WT and Has2 KO clones by trypsin treatment. Values represent the mean μg GAG per 106 cells from two independent experiments.
Figure 2 shows the results of Pas1 restriction digest analysis (panels B and D) of PCR products (panels A and C) of WT and selected Has2 KO clones generated from RCS-o (panels A and B) and RCS-Cas9 (panels C and D) cells. The WT cells have the Pas1 digestion site, and the two bands resulting from cleavage are seen at the 171 bp and 283 bp expected sizes. The Pas1 site should be destroyed by the Cas9 cleavage so only the one 454 bp product is seen in the Has2 KO clones (panels B + D). Loss of Pas1 susceptibility indicates that these clones contained indel mutations at the target site. Loss of Pas1 susceptibility correlated with the loss of HABP staining shown in Figure 1. The Has2 sgRNA target site of RCS-Cas9 clone #7, one that showed complete loss of Pas1 susceptibility, was sequenced. DNA sequencing and alignment of the targeted rat Has2 sequence in RCS-Cas9 clone #7 showed one allele with a single base deletion and the other allele with 2 bases deleted, both disrupting the open reading frame (Fig. 2E).
Fig. 2. Detection and determination of Has2 gene mutation.
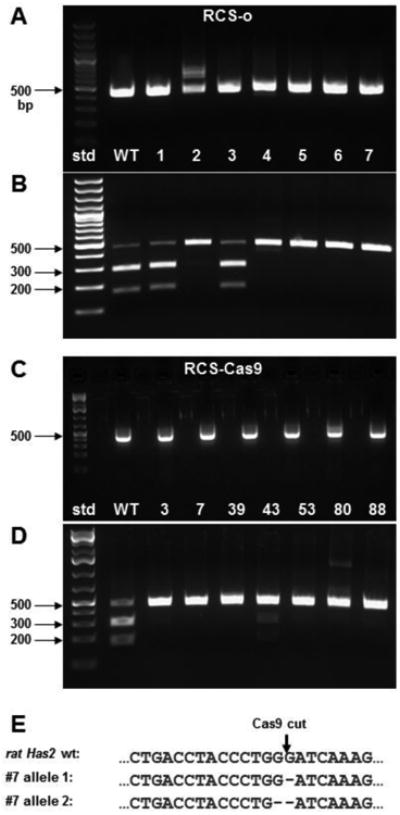
A 454 bp region within exon2 of rat Has2 that contains the Cas9 mutation site was PCR amplified from genomic DNA of each cell clone as a template. The amplified product was gel-purified and then digested with Pas1. The final products were analyzed by agarose gel electrophoresis. DNA standards (std) are labeled. Panel A. Products amplified from RCS-o WT cells and clones 1 thru 7. Panel B. Pas1 digestion products of the amplified 454 bp products shown in panel A. Panel C. Products amplified from RCS-Cas9 WT cells and clones 3, 7, 39, 43, 53, 80 and 88. Panel D. Pas1 digestion products of the amplified 454 bp products shown in panel C. Panel E. Alignment of the targeted rat Has2 genomic sequences from RCS-Cas9 clone #7, allele 1 and allele 2 with the WT RCS-Cas9 cells. Deleted bases are indicated.
2.2. Cell biological characterization of WT and Has2 knockout clones
RCS-o WT chondrocytes, even after single-cell cloning, exhibit a variety of cell shapes from polygonal to elongated-ovoid to round (Fig. 3A). This pattern was reflected in the staining of RCS-o actin cytoskeleton with TRITC-phalloidin (Fig. 3B, E, F). Focal adhesions were noted in both polygonal and elongated ovoid cells. The RCS-Cas9 WT chondrocytes were more uniform with a mixture of polygonal and round cells (Fig. 3C). Like the RCS-o, RCS-Cas9 cells exhibited a light, more cortical cytoskeleton typical of chondrocytes (Fig. 3D, G, H). The occurrence of stress fibers was only rarely detected in the two WT cultures. In both the RCS-o and RCS-Cas9 Has2 KO clones, the cells were more spread and often contained stress fibers as well as enhanced focal adhesions (Fig. 3I - L). These features were more obvious for the RCS-Cas9 Has2 KO clone #7 as the WT cells were more uniformly round.
Fig. 3. Initial characterization of WT and Has2 knockout clones.
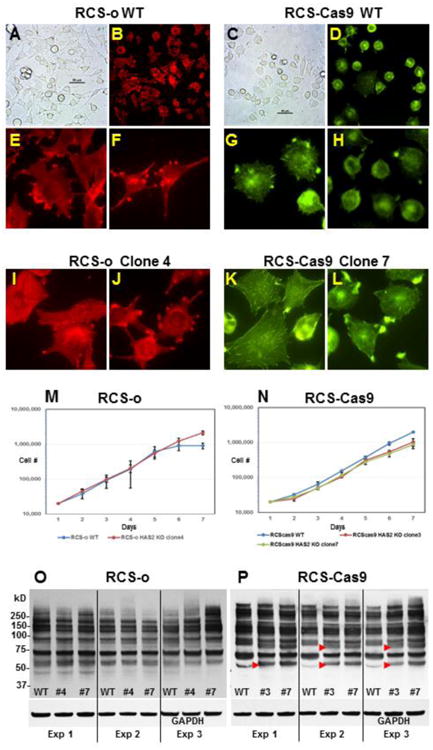
Panels A and C: Phase contrast microscopy of RCS-o WT and RCS-Cas9 WT cells. Panels B, E, F: TRITC-phalloidin staining of RCS-o WT cells. Panels D, G, H: Fluorescein phalloidin staining of RCS-Cas9 WT cells. Panels B and D represent lower power view of phalloidin-stained cells. Panels I and J: TRITC-phalloidin staining of RCS-o Clone #4 cells that lack cell surface HA. Panels K and L: Fluorescein phalloidin staining of RCS-Cas9 clone #7 cells with mutated Has2 alleles. Panel M: Growth curves for RCS-o WT and RCS-o Has2 KO clone #4 cells plotted on a log scale showing the mean value from 2 independent experiments. Panel N: Growth curves for RCS-Cas9 WT, and RCS-Cas9 Has2 KO clone #3 and clone #7 plotted on a log scale showing the mean value from 3 independent experiments. Panels O and P: Representative western blots using the RL2 antibody to detect GlcNAcylated proteins expressed in WT and Has2 KO clones of RCS-o or RCS-Cas9 cells as labeled, from three independent experiments. Red arrowheads indicate bands with changes in intensity.
An analysis of cell proliferation growth curves revealed that the RCS-o cells (both WT and Has2 KO, Fig. 3M) had a slightly faster growth rate than the RCS-Cas9 clones (Fig. 3N). Nonetheless, little difference was observed between WT and Has2 KO clones in either the RCS-o line or the RCS-Cas9 cells. Next, a western blot analysis was performed to determine whether cellular proteins in the Has2 KO clones were altered due to enhanced transfer of N-acetylglucosamine (GlcNAcylation) as compared to WT. This experiment was designed to test whether the elimination of HA biosynthesis would result in increased GlcNAcylation due to changes in cytosolic UDP-GlcNAc and UDP-GlcUA; the former of which is a precursor for GlcNAcylation transferases [29]. As shown in Figure 3O (RCS-o) and Figure 3P (RCS-Cas9), using an antibody that can detect GlcNAcylated proteins, only subtle changes were observed between WT and Has2 KO clones of each line, each replicated in three independent experiments. However, some distinct changes were noted (red arrow heads) especially between lysates of RCS-Cas9 WT and Has2 KO clones. Both Has2 KO clone #3 and #7 consistently exhibited the same additional bands or enhanced bands as compare to WT. These results emphasize that while major changes in GlcNAcylation did not occur upon loss of Has2, minor, perhaps more selective changes did occur including the possibility of altered proteins hidden beneath other protein bands.
Real-time qRT-PCR was used to determine changes in gene expression relevant to extracellular matrix production in chondrocytes. RCS-Cas9 Has2 KO clones #3 and #7 exhibited little change in aggrecan (Fig. 4A) or type II collagen (Fig. 4B) mRNA expression and a modest increase in type X collagen (Fig. 4F) as compared to their respective WT cells. RCS-o Has2 KO clone #4 showed a 1.5-fold increase in aggrecan, a 2.4-fold increase in collagen II and an 8-fold increase in collagen X mRNA. These increases were not reflected to the same extent in the RCS-o Has2 KO clone #7. Each of the Has2 KO clones from both RCS lines displayed small changes in Has2 mRNA expression (Fig. 4E). Two additional genes relevant to this model are Has1 and Has3, the two other genes with the potential capacity to compensate for deficiencies in Has2-mediated production of HA. As shown in Figure 4C and 4D, Has1 or Has3 mRNA levels differed little comparatively to WT especially for the RCS-o Has2 KO clones. The RCS-Cas9 Has2 KO clones showed a 1.7-fold increase in Has1 and Has3 (for clone #7) as compared to WT cells.
Fig. 4. Effect of Has2 deficiency on the expression of Acan, Col2a and Col10a as well as Has1, Has2 and Has3 mRNA.
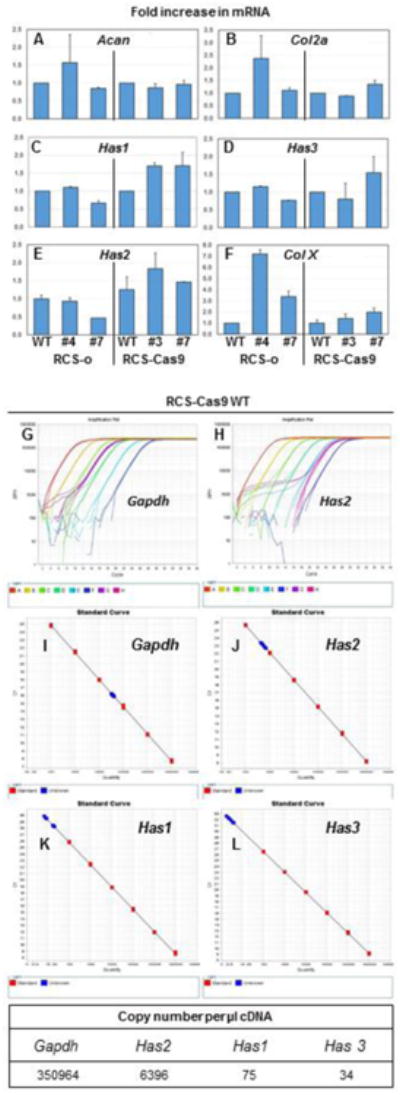
Panels A - F: Fold change in mRNA is shown for RCS-o Has2 KO clone #4 and clone #7 as well as RCS-Cas9 Has2 KO clone #3 and clone #7 versus the WT cells. Panel A: fold change in Acan mRNA. Panel B: fold change in Col2a mRNA. Panel C: fold change in Has1 mRNA. Panel D: fold change in Has3 mRNA. Panel E: fold change in Has2 mRNA. Panel F: fold change in Col10a (Col X) mRNA. Bars depict the average and range of two independent experiments (each analyzed in duplicate). Panel G, H: Representative amplification plots for Gapdh and Has2 as labeled. Panels I, J, K, L: Standard curves used to determine mRNA copy numbers (red squares), with samples shown in blue on the standard curves for Gapdh, Has2, Has1 and Has3, as labeled. Absolute copy numbers for RCS-Cas9 WT cells were performed twice. Bottom panel shows a representative example of absolute copy numbers of Gapdh, Has2, Has1 and Has3 per μl cDNA for RCS-Cas9 WT cells.
Our previous work in human chondrocytes suggested that HAS1 and HAS3 mRNA are present at very low copy numbers [26]. If this were the same in RCS chondrocytes, the ΔΔCt comparison used for comparison of Has2 KO clones to WT might be misleading. We thus performed absolute copy number quantitation by qRT-PCR using small, purified, specific DNA templates to generate standard curves. An example reaction is shown in Figure 4G-H. Copy numbers for Gapdh, Has2, Has1 and Has3 derived from RCS-Cas9 WT chondrocytes (blue squares) were determined by comparison to their respective standard curves (red squares, Fig. 4I - L). The data demonstrate that Has2 is present in RCS chondrocytes at approximately 2 orders of magnitude higher copy number than Has1 and Has3; Gapdh is more than 3 orders of magnitude higher. Together with the lack of staining for cell surface-associated HA by HABP and the negligible levels of HA in the media of the Has2 KO clones there is no evidence that Has1 or Has3 provided any major compensatory contribution of HA biosynthesis to the Has2 KO chondrocytes.
2.3. Effect of Has2 deficiency on pericellular matrix assembly
Our previous studies demonstrated that RCS-o chondrocytes synthesize and retain a HA/aggrecan-rich pericellular matrix similar to what we detect on primary cultures of bovine and human articular chondrocytes [9, 11]. Recently, we observed that purified bovine aggrecan monomer could become incorporated into an existing RCS (or bovine chondrocyte) pericellular matrix and substantially expand the size of the cell coat [12]. This observation provided a useful assay to visualize the retention of aggrecan to live chondrocytes in real time. Both RCS-o WT (Fig. 5A) and RCS-Cas9 WT (Fig. 5G) cells exhibit pericellular matrices that exclude small particles used for detection. In both RCS-o and RCS-Cas9 WT cells, the addition of exogenous aggrecan resulted in a dramatic increase in the size of this excluded zone pericellular matrix (Fig. 5D, J) as the aggrecan became incorporated within the existing matrix. In contrast, the Has2 KO clones from both backgrounds failed to exhibit a particle-excluding pericellular matrix (Fig. 5B, H, I), even though the expression of Acan and sGAG content were not apparently altered (Fig. 1E, 4A). Moreover, the addition of exogenous aggrecan to the Has2 KO RCS clones also did not result in pericellular matrix formation (Fig. 5E, K, L). The bovine aggrecan monomer is added at a saturating concentration and thus functions to normalize and compensate for any differences that may occur in the biosynthesis of endogenous aggrecan. These results demonstrate that HA, synthesized via HAS2, is required for the assembly and retention of aggrecan within a chondrocyte pericellular matrix.
Fig. 5. Effect of Has2 deficiency on pericellular matrix assembly.
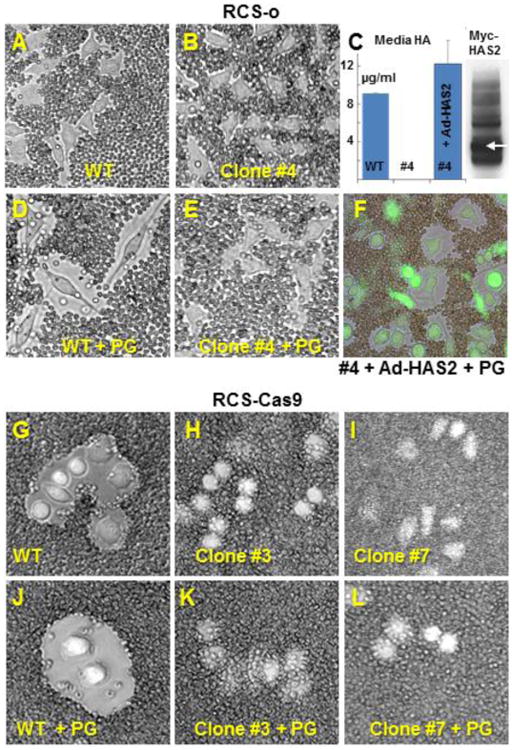
All panels (except panel C) show the application of the particle exclusion assay to RCS chondrocytes to reveal the pericellular matrix. Panel A: RCS-o WT chondrocytes endogenous coats. Panel D: RCS-o WT cells incubated with 2.0 mg/ml purified aggrecan proteoglycan (+PG) show enhanced pericellular matrix. Panel B. RCS-o Has2 KO clone #4 cells without (Panel B) or with incubation with exogenous aggrecan (Panel E +PG) exhibit no coats. Panel C: RCS-o Has2 KO clone #4 cells were transduced with Adeno-ZsGreen1-mycHas2. The bar graph shows HA in the media of the cultures of RCS-o WT cells, no detectable HA in RCS-o Has2 KO clone #4 and, significant production of HA by the RCS-o Has2 KO clone #4 cells following Ad-Has2 transduction. The western blot shows the expression of the myc epitope in the HAS2 fusion protein monomer (white arrow) and multimers detected in lysates of the transduced cells. Panel F: Addition of exogenous aggrecan (+PG) to the RCS-o Has2 KO clone #4 cells transduced with Ad-Has2 results in coat formation. Panel G: RCS-Cas9 WT chondrocytes endogenous coats. Panel J: RCS-Cas9 WT cells plus exogenous aggrecan (+PG) show enhanced pericellular matrix. Panels H, I: RCS-Cas9 Has2 KO clone #3 and clone #7 cells show no coats. Panel K, L: RCS-Cas9 Has2 KO clone #3 and clone #7 cells plus exogenous aggrecan (+PG) show no change in coat size. Images shown are representative of 3 independent experiments.
To further validate this conclusion, the expression of Has2 in the RCS-o Has2 KO clone #4 was rescued by transduction of these cells with an Adeno-ZsGreen1-mycHas2 construct (Ad-Has2). HA-ELISA of samples from the media of these cultures demonstrated that Ad-Has2 transduction of RCS-o Has2 KO clone #4 chondrocytes resulted in HA levels similar to the production of HA by RCS-o WT cells; no detectable HA was synthesized by clone #4 chondrocyte (Fig. 5C). The western blot of lysates from Ad-Has2 transduced clone #4 cells shows the expression of the ∼64 kD mycHAS2 fusion protein monomer (white arrow) as well as multimers required for HA biosynthesis. Following the addition of exogenous aggrecan to the RCS-o Has2 KO clone #4 cells transduced with Ad-Has2 (Zsgreen1 positive cells, Fig. 5, panel F), large pericellular matrix exclusion zones were recovered. This further supports the requirement for HA production by HAS2 in the assembly and retention of aggrecan within the chondrocyte pericellular matrix.
2.4. The effect of Has2 deficiency on cartilage neogenesis
The effect of altered HA/aggrecan matrix assembly and retention was determined using two in vitro 3-D culture models often used for generating bioengineered cartilage [30-33]. First, RCS-Cas9 WT and RCS-Cas9 Has2 KO clones #3 and #7 were cultured in alginate beads for 7 days (Fig. 6). In these beads, the alginate provides a scaffold to separate individual cells or cell clusters and newly-synthesized endogenous extracellular matrix pushes out into the scaffold with time. Toluidine Blue stains both the scaffold (alginate polysaccharide) as well as the chondrocytes (darker blue cells) in sections of these bead cultures (Fig. 6). In bead cultures of the RCS-Cas9 WT cells, distinct, clear areas surrounding the chondrocytes were consistently observed within the alginate scaffold matrix. However, few cleared zones were observed surrounding either the RCS-Cas9 Has2 KO clone #3 or clone #7 chondrocytes. In these cultures, the alginate scaffold remains in close proximity to the KO cells. To determine whether HA-associated extracellular matrix was present in the cleared zone surrounding the WT cells, the alginate bead sections were stained for HA (using a biotinylated HABP probe) and proteoglycan (using Safranin O / fast green). Visible in examples of increasing power (Fig. 6), HABP staining revealed the presence of HA in the WT RCS-Cas9 alginate bead cultures, streaming out from the cell surface into the surrounding pericellular matrix. As expected, no HABP staining was detected in the RCS-Cas9 Has2 KO (clones #3 and #7) alginate bead cultures. Safranin O-positive staining, indicative of the presence of aggrecan, was observed only in the WT RCS-Cas9 alginate bead cultures. Interestingly, the pattern for aggrecan staining by Safranin O matched the localization of HA. One conclusion from these results is that the retention of HA/aggrecan complexes to the plasma membrane of the chondrocytes facilitates the expansion of the pericellular spaces within the scaffold. Moreover, the lack of Safranin O staining in the Has2 KO clones suggests that aggrecan aggregation with HA is necessary to retain the proteoglycan within the alginate bead scaffold.
Fig. 6. Effect of Has2 deficiency on cartilage neogenesis in alginate bead cultures.
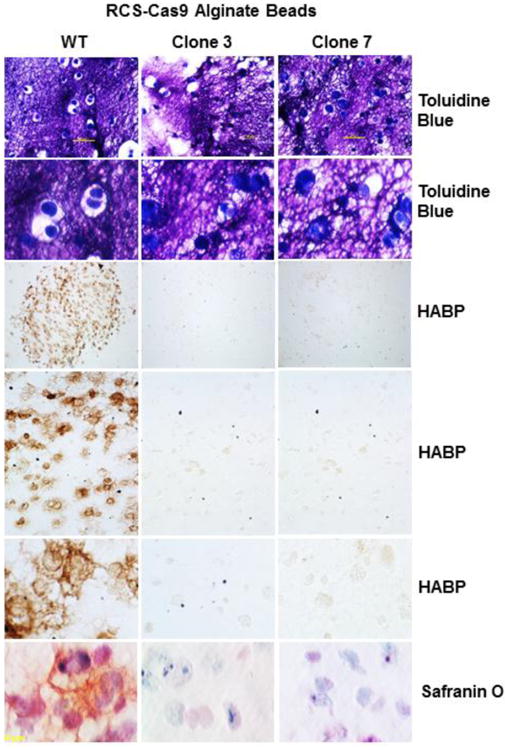
Matrix assembly and retention in 3-D cultures were examined using RCS-Cas9 WT cells and RCS-Cas9 Has2 KO clones 3 and 7 as labeled. Upper six Panels: Cells were cultured in alginate beads for 7 days, and paraffin sections of the beads were stained with Toluidine Blue. Lower magnification images (10× objective lens) are shown in the upper row; higher magnification views (40× objective) in second row. Sections were stained with biotin-HABP revealing HA in the matrix as a brown stain in rows three, four and five, shown at low (4× objective), medium (20× objective) and higher (40× objective) magnification. Safranin O/fast green staining of alginate bead sections are shown in the bottom row (40× objective).
In a second 3-D model the RCS-Cas9 WT cells and RCS-Cas9 Has2 KO clones #3 and #7 were grown as pellet cultures. No exogenous scaffold is used in these cultures and thus all tissue organization and cell spacing is dependent on the endogenous production of matrix macromolecules. As revealed in Toluidine Blue stained paraffin sections at low power, these cells formed intact, rounded pellets within two days of culture (Fig. 7). Pellets cartilages formed using primary bovine articular chondrocytes were included for comparison in the left side column. The pellet cartilages formed by the Has2 KO clones were generally slightly smaller in size than the cartilage-like structure generated by RCS-Cas9 WT cells and exhibited an apparent deficit in Toluidine Blue staining on day 2 (Fig. 7). At higher magnification, the deficit in Toluidine Blue staining of the Has2 KO clones could be seen to be due in part to a condensation or aggregation of the cells, resulting in more intervening white spaces. This condensation of the Has2 KO clones was more evident in 11-day pellets shown in the lower panels of Fig. 7. The RCS-Cas9 WT cells, like the bovine chondrocytes, were tightly packed but more evenly spaced in both 2-day and 11-day cultures. In the 2-day pellets, Toluidine Blue staining of WT cells revealed dark staining of the cell bodies but also wispy blue matrix between cells. This was more prominent in the bovine chondrocytes and substantially less in the Has2 KO clones. Additionally, both the RCS-Cas9 WT and bovine chondrocytes exhibited strong staining for HA and positive staining for proteoglycan production as detected by Safranin O reagents. In comparison, RCS-Cas9 Has2 KO clones #3 and #7 grown in 2-day pellet cultures exhibited only background staining for HA and proteoglycan (only background fast green counterstain detected).
Fig. 7. Effect of Has2 deficiency on cartilage neogenesis in scaffold-free pellet cultures.
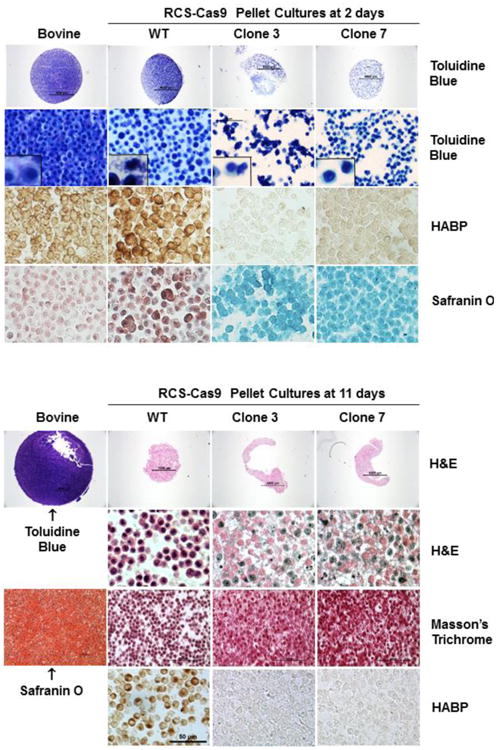
RCS-Cas9 WT cells, RCS-Cas9 Has2 KO clones 3 and 7 were grown in pellet cultures for 2 days (top group of 16 panels) or 11 days (lower group of panels). Paraffin sections were stained with Toluidine Blue, HABP, Safranin O/fast green, H&E or Masson's Trichrome as labeled. Sections from pellet cultures formed with bovine articular chondrocytes are shown in the left column. In the top group of panels (2 days), the top row shows mid-pellet sections stained with Toluidine Blue at low magnification (4× objective). At higher magnification (20× objective) in the second row, the packing density of the bovine chondrocyte cartilage pellet is seen with the early deposition of matrix. In row two, the RCS-Cas9 WT pellet cultures show cell-to-cell spacing and some pale matrix; high magnification inset shows three cells. The RCS-Cas9 Has2 KO clone 3 and 7 pellet cultures show more variable cell-to-cell spacing and the initial formation of compressed cell clusters. Row three shows representative HABP staining of HA in pellets made from all four cell types (40× objective). The bottom row shows Safranin O/fast green staining of pellets made from all four cell types (40× objective). In the lower group of panels (11 days), the top row shows mid-pellet sections at low magnification (4× objective), the bovine cartilage stained with Toluidine Blue A and the RCS-Cas9 pellets stained with H&E. At higher magnification (40× objective) the RCS-Cas9 WT pellet shows cell-cell spacing but the cells within the RCS-Cas9 Has2 KO pellets are compressed. Staining with Masson's Trichrome also revealed the cell compression in the RCS-Cas9 Has2 KO pellets (20× objective). Staining the bovine cartilage pellet with Safranin O shows the deposition of ECM by 11 days (20× objective). After 11 days, HA was detected in the RCS-Cas9 WT pellets but no HABP staining was detected in the RCS-Cas9 Has2 KO pellets (bottom row; 40× objective).
By 11 days, the bovine cartilage pellets had increased in overall size as compared to 2 day cultures and stained intensively for proteoglycans as visualized by both Toluidine Blue and Safranin O. No such expansion was observed for the any of the RCS-Cas9 cultures. The 11-day WT pellets were smaller than at 2-days but remained intact, retained their high cellularity and clear, recognizable spacing between the cells. Although cells within the WT pellets continued to exhibit positive staining for HA, they no longer stained well for proteoglycan. In contrast, in the cartilage pellets formed by both of the Has2 KO clones, spacing between neighboring cells was substantially reduced. Masson's Trichrome staining was used to better delineate these observations and, viewed at lower magnification, the pellet neocartilage generated by the two Has2 KO clones appeared highly compressed with very little space between cells. Additionally, the Has2 KO clones were negative for HA using the HABP staining protocol. The more unexpected results from these pellet cultures was the consistent failure of both Has2 KO clones to maintain the same uniform cell-to-cell spacing like that of the RCS-Cas9 WT pellets—an event that occurs early on in the 3-D cultures. This could be due to the lack of HA, a reduced cell retention of aggrecan or, some as-yet-characterized change in cell metabolism that occurs downstream as a result of Has2 deletion.
3. Discussion
The data from this study demonstrated that HA biosynthesis, by way of HAS2, is required for the retention of aggrecan. This was observed using Has2 knockout chondrocytes developed from two different RCS cell lines. The Has2 KO clones failed to establish an expanded pericellular coat following the addition of purified aggrecan monomer to the (as compared to the wild type control cells) and failed to elicit staining by the application of a HABP probe which is essentially a biotinylated aggrecan G1 domain [34]. These data were further validated by the rescue of aggrecan retention by of one of the Has2 KO cells following transduction with Ad-Has2.
The lack of HA and aggrecan matrix assembly did not dramatically affect the proliferative capacity of these Has2 KO RCS cells and there was no apparent change in migratory capacity (data not shown) although this is not a major feature of chondrocytes. Has2 knockouts did elicit some cell shape changes including differences in the organization of the actin cytoskeleton but the behavioral ramifications of these changes have not, as yet, been elucidated. In a recent study by Evanko et al. (2015) the HA matrix of myofibroblasts was associated with CD44 and the cortical actin network to affect fibroblast adhesion rather than through focal adhesions [35]. Enhanced stress fibers were observed when pericellular HA in these myofibroblasts was reduced.
The CRISPR-Cas9 gene editing system was highly efficient with ∼80% of the GFP-positive RCS clones giving rise to successful Has2 KO cells. The high frequency level of mutations in both the RCS cell lines expressing Cas9 and the RCS-o cells reflects the efficiency of either approach. The ability of chondrosarcoma cells to be sorted and cloned enabled the isolation of cells lines for comprehensive analysis and further experimentation. The very low transfection efficiency and cloning potential of primary bovine and human articular chondrocytes has impeded the molecular analyses of these cells. Development and study of these permanent cell lines with targeted Has2 mutation will provide definitive evidence of the function of HA in chondrocyte metabolism and matrix assembly. Our findings indicate an increase in type X collagen in the RCS Has2 KO cells, especially those derived in the RCS-o line. The expression of type X collagen, while usually associated with chondrocyte hypertrophy [36], is also observed in OA cartilage, especially in areas of fibrillation and chondrocyte cloning [37]. The continuing differentiation chondrocytes to hypertrophy could lead to joint deformity as well as articular cartilage degeneration [38]. It is noteworthy that the RCS-o cells are morphologically more elongated than the RCS-Cas9 cells—a morphology similar of de-differentiated primary chondrocytes or some OA chondrocytes. It will be of interest to determine whether the enhanced expression of type X collagen by RCS-o Has2 KO cells is part of a critical linkage associated with this phenotype
Although two other HA synthase genes are expressed in these chondrocytes, no major compensation by Has1 or Has3 was observed following the inhibition of HA production through HAS2. This might suggest that cells cannot detect or respond to deficiencies in HA or, that Has1 or Has3, as minor Has gene isoforms, are not responsive in chondrocytes. In past studies when bovine or human chondrocytes were stimulated with osteogenic protein-1 (BMP7) or IL-1α there was a pronounced increase in HAS2 but no change in HAS3 mRNA [8, 39]. The expression of HAS1 was below the detection limits available at the time. Human chondrocytes made HA-deficient via hyaluronidase or by HA oligosaccharide treatment also expressed an increase in HAS2 but no change in HAS3 mRNA [40]. This would suggest that HAS3 (and likely HAS1) are overall less responsive to changes in the metabolism or HA levels in chondrocytes.
The knockout of Has2 in mice results in embryonic lethality due to failed heart septation [27]. Conditional inactivation of Has2 in the early limb bud mesenchyme by introduction of the Prx1-Cre transgene results in skeletal deformities and severely shorten limbs due to abnormal and disorganized growth plate and a decrease in aggrecan deposition into the ECM [28]. HAS1 and HAS3 did not appear to compensate for the HA deficiency in the conditional inactivation mice although this was not determined directly. The structural organization of the ECM in the Has2-deficient growth plates suggests the HAS2 production of HA is necessary for the normal progression of chondrocyte maturation and more importantly, expansion of the lacunar space during hypertrophy [41]. The growth plates in the conditional Has2 knockout mouse exhibited increased cell density and a decrease in the amount of ECM separating the chondrocytes. Reduced retention of aggrecan in the growth plates of these Has2-deficient mice was observed. In this study, Has2 KO chondrocytes in alginate beads failed to develop spaces within the alginate scaffold and in scaffold-free pellet cultures, spaces between individual Has2 KO cells also appeared to be compressed. The straightforward conclusion that can be drawn from both sets of data is that, (1) cell-associated aggrecan is important for cell-cell spacing (i.e., not just aggrecan in the more distant ECM) and; (2) HA is critical to retaining aggrecan within the cell-associated environment for this spacing event. However, as we have seen in this study, intracellular changes in metabolism, such as those due to altered GlcNAc transfer to select proteins, may also be induced due to inactivation of Has2.
We anticipate that these newly-developed RCS Has2 KO chondrocytes will be useful to address questions related to why it is necessary for aggrecan and HA to be retained at the chondrocyte cell surface. If the principal role of aggrecan is as a shock-absorber within the more distance interterritorial ECM, why is there a need for cell-bound HA/aggrecan-rich pericellular matrix? In the current study we have identified one possibility; that is, a role in maintaining cell-cell spacing or spacing within a lacunar environment. We recently demonstrated another function of aggrecan binding to cell-associated HA; the binding of aggrecan regulates the HA endocytosis by CD44 [12]. HA endocytosis becomes proportionately enhanced as aggrecan undergoes progressive degradation during its extracellular turnover. However, in this endocytosis study there were challenges with the cell model. To investigate HA endocytosis, aggregates of FITC-HA and aggrecan monomer were prepared in the test tube and then added to hyaluronidase-treated RCS-o chondrocytes. However, as incubation times proceeded, renewed biosynthesis of endogenous HA and aggrecan would compete with the carefully size-defined exogenous FITC-HA / aggrecan complexes and begin to affect the endocytosis rates. These Has2 KO RCS cells should provide a better model for studying HA endocytosis especially when we can rescue HAS2 expression by way of adenovirus transduction. Interestingly, there appears to be a “saturation” of the cell surface HA when using Ad-ZsGreen1-mycHas2. The subsequently formed pericellular coats with the addition of aggrecan are large but not as extensive as we observed on some primary chondrocytes [22, 39]. Although not as yet validated, it appears that much of the transgene-derived HAS2 synthesized HA is released into the media. The Adeno-Tet-mycHas2 we have developed (Ishizuka et al., submitted) should be able to better define this.
One critical function for cell-surface retained HA and aggrecan is to function as means for chondrocytes to detect or “sense” changes in the composition of the ECM, especially changes associated with disease states such as OA [42]. We have hypothesized that during OA, aggrecan is proteolytically degraded and lost, resulting in enhanced HA endocytosis [12, 17], resulting in affected chondrocytes deficient in their HA/aggrecan-rich pericellular matrix. Also, the procatabolic response to ECM fragments may be down-regulated by pericellular HA [43-46]. The Has2 KO RCS chondrocytes developed in this study, now permanently HA-deficient, will help to address the mechanisms used by chondrocytes to sense and respond to such a deficiency. We did not observe a compensatory response to replenish lost HA by the Has2 KO chondrocytes. However, elimination of Has2 could potentially alter the cytoplasmic levels of glucosamine, UDP-GlcUA and UDP-GlcNAc. Using the RL2 antibody that detects GlcNAcylated proteins we could demonstrate only minor observable changes in the profile of GlcNAcylated cellular proteins between WT and Has2 KO RCS cells. Nonetheless, such subtle changes, especially to a particular regulatory protein altered by GlcNAc transfer, could have major effects on cellular metabolism and candidates including Akt, NF-κB and YY1 [47] are worth investigating in future studies using this Has2-negative model system. Another potential marker that will be examined in the future is the cytoplasmic enzyme, UDP-glucose dehydrogenase (UGDH) [48]. UGDH is the essential enzyme to convert UDP-glucose into UDP-glucuronic acid (UDP-GlcUA). HA is the glycosaminoglycan that is more sensitive to UDP-GlcUA cytosolic concentration and, therefore cells regulate HA synthesis, in part, by controlling the UDP-GlcUA availability [29, 49]. For example, transient transfection of chick embryo chondrocytes with UGDH resulted in increased size of the pericellular coats and more HA released into the culture media [50]. In addition, increases in tissue HA and increases in UGDH are seen during embryonic limb development [51]. Such a local rise in tissue HA may lead to a switch from intercellular cohesion to cell dissociation and increased cell-to-cell spacing [52, 53]. Since UGDH seems to be tandemly regulated with HAS2 in embryonic chick chondrocytes, future studies will examine changes in UGDH as a surrogate to HAS2 expression.
How the loss of HA affects other signaling events remains to be determined. Given that there are effects on the actin cytoskeleton in Has2 KOs derived from both RCS lines, it is likely that we will observe changes in cell signaling. Pericellular HA may be involved in co-activity with one or more signaling networks that regulate chondrocyte maturation [28, 42, 54, 55]. Our future studies will look to determine whether there are changes in CD44 and other potential HA receptors. We expect that some events will be dependent on HA and the assembly of the HA-aggrecan pericellular matrix such as cell-shape or spacing dependent signaling whereas other will require the interaction of HA with receptors such as CD44. Nonetheless, it should be remembered that the two lines of RCS cells used in this study are not primary chondrocytes. These lines were derived from the Swarm rat chondrosarcoma tumor and, as with any cell line, may exhibit metabolic and cell signaling properties more consistent with transformed cells but also, other features that serve as a good model system of chondrocytes.
4. Experimental procedures
4.1. Cell culture
The RCS-o cell line was a gift of Dr. James H Kimura (formerly of Rush University Medical Center) and represented an early clone of cells that eventually became known as long-term culture RCS [16]. The RCS-Cas9 cells, that co-express Cas9 and mCherry genes [13], were a generous gift of Dr. Gary Gibson (Henry Ford Hospital). Both RCS line chondrocytes were cultured as high density monolayers (2.0 × 106 cells/ cm2) in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS; Hyclone) and 1% L-glutamine and penicillin-streptomycin, and incubated at 37° C in a 5% CO2 environment. The RCS cells were passaged at confluence using 0.25% trypsin/2.21 mM EDTA (Corning CellGrow).
4.2. Generation of Has2 knockouts in RCS cells
The pSpCas9(BB)-2A-GFP (PX458; Plasmid #48138), containing Cas9 and GFP marker, was a gift from Feng Zhang and purchased from Addgene [56]. A 20 nucleotide sgRNA sequence targeting exon 2 of rat Has2 was designed using the website http://crispr.mit.edu/. This sgRNA sequence contains a Pas1 enzyme restriction site; beneficial for future screening of potential knockout clones. The complementary sgRNA oligonucleotides (forward): 5′-CACCGCCATCACGACTTTGATCCCA-3′ and (reverse): 5′-AAACTGGGATCAAAGTCGTGATGGC-3′, (synthesized by Integrated DNA Technology) were annealed and cloned into the PX458 plasmid, and DNA sequences verified. Both RCS-o and RCS-Cas9 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfected cells were identified by the expression of GFP observed under fluorescence microscopy.
For flow cytometry studies, 2 days after sgRNA transfection, RCS chondrocytes were released from monolayers with trypsin/EDTA followed by inactivation in DMEM containing 10% FBS. Live cells were applied and sorted using a FACScan™ cytometer (Becton Dickinson) with CELL Quest software. For 10,000 cells, log fluorescence channel versus cells per channel was plotted to identify the GFP-positive cells. These were sorted as single cells into individual wells of 96 well plates using the FACScan cytometer; the plates were monitored daily and allowed to expand in the wells for about 10 days and eventually passaged to T75 flasks.
4.3. Restriction digestion and DNA sequencing of knockout site
RCS-o and RCS-Cas9 WT and Has2 knockout cells were plated at 70% confluency in 12 well plates in DMEM + 10% FBS and cultured for 48 h. Cells were released from monolayers with trypsin/EDTA followed by inactivation in DMEM containing 10% FBS and pelleted at 1500rpm for 5 min. The genomic DNA of each cell clone was extracted using QuickExtract DNA extraction solution (Epicentre, QE0905T) according to the manufacturer's instructions. Primers were used to PCR amplify a 454 bp region within exon2 of rat Has2 that contains the Cas9 mutation site. The primers used were (forward): 5′-TTTGGAGTGTCTCTCCTCCT-3′ and (reverse): 5′-GCGAACTTTCTTTATGGGACTC-3′. PCR was performed using AccuPrime Pfx DNA polymerase (ThermoFisher Scientific) with the following cycling conditions: initial denaturation of 95°C for 2 min, followed by 30 cycles of 15 seconds denaturation at 95°C, 30 seconds annealing at 55°C, and 40 seconds extension at 68°C. The PCR amplified product was run on an agarose gel and then gel-purified using the QIAquick Gel Extraction Kit (Qiagen); it was digested with Pas1 restriction enzyme (ThermoFisher Scientific) for 2 h to yield 171 bp and 283 bp cleavage products to screen for Has2 mutant clones. To identify the mutation in the RCS-Cas9 clone #7, the same PCR product was also amplified but with a different DNA polymerase, TaKaRa LA Taq® DNA polymerase (Clontech), and this PCR product cloned into the pCR4-TOPO vector (Invitrogen) according to the manufacturer's instructions. 16 colonies were picked and sent for DNA sequencing in order to identify the specific Has2 gene mutation in the RCS-Cas9 clone #7.
4.4. Cell biology of Has2 knockout RCS cells
Visualization of cell surface HA
The Has2 KO clones were next screened for the loss of cell surface associated HA. Cells were plated at 20, 000 cells/well in 4-well chamber slides in DMEM + 10% FBS and cultured for 48 h. The cells were fixed in 4% paraformaldehyde for 1 hr at 4°C, rinsed in PBS and then incubated with 4 ug/ml biotinylated hyaluronic acid binding protein (b-HABP from bovine nasal cartilage; Calbiochem, # 385911-50UG) in PBS with 1% BSA at 4°C overnight. Then the cells were rinsed and incubated with Alexa-Fluor-488-Streptavidin (green fluorescence; Jackson ImmunoResearch Labs, 016540-084) for 1 hr at RT in PBS with 1% BSA, rinsed and mounted using a media containing DAPI nuclear stain (FluoroGel II; Electron Microscopy Sciences). Cells were visualized using a Nikon Eclipse E600 microscope equipped with Y-Fl Epi-fluorescence, a 60× 1.4 n.a. oil-immersion objective and FITC (green) and DAPI (blue) filters. Images were captured digitally in real time using a Retiga 2000R digital camera and processed using NIS Elements BR imaging software (Nikon).
For HABP staining of sections from alginate beads or pellet cultures, after deparaffinization and hydration, sections were rinsed with enhanced Tris at pH 8.0 followed by PBS rinses. The sections were next incubated with 4 ug/ml b-HABP (Calbiochem) in PBS with 1% BSA at 4°C overnight. Sections were rinsed with PBS and the biotin detected using the Streptavidin-peroxidase and the DAB peroxidase substrate (Vectastain Elite ABC kit; Vector Laboratories) according to the manufacturer's protocol.
HA ELISA
Hyaluronan in the media of cell cultures was analyzed using the DuoSet HA ELISA kit for hyaluronan (DY3614-05; R&D Systems, Inc, Minneapolis, MN) following the manufacturer's instructions. Results are plotted for RCS-o Has2 KO clones 1, 3, 4 and 7 as percent of RCS-o WT, and RCS-Cas9 Has2 KO clones 3 and 7 as a percent of RCS-Cas9 WT media HA.
Determination of sulfated glycosaminoglycans
RCS-Cas9 cells were plated at 1 million cells/ well in a 6-well dish in DMEM + 10% FBS. 48 h later, the medium samples were collected and equal volumes were analyzed. In other experiments, the medium samples were collected, and the cell layer was treated with 0.25% trypsin with EDTA (Corning) for 30 min. The trypsinate was collected, centrifuged at 430 × g for 5 min, and cell pellet discarded. Each sample was analyzed in duplicate by adding 20 μl to a 96 well plate, followed by 180 μl of dimethylmethylene blue (DMMB) reagent [57, 58]. After 5 min of incubation, absorbance was determined at 525 nm with Multiskan Ascent microplate reader. Shark chondroitin sulfate (Sigma) dissolved from 0 to 2 μg in culture medium was used to produce a standard curve to calculate the sulfated glycosaminoglycans (sGAG) content.
Growth curves
RCS-Cas9 WT and Has2 knockout cells were plated at 20,000 cells/well in a 6 well plate. For each cell clone, six replica wells were plated. Cells in one well were counted in duplicate each day using the Countess™ automated cell counter (Invitrogen) and cell number recorded over the course of 6 days.
Phalloidin staining
RCS-o and RCS-Cas9 WT and Has2 KO clones were plated in a 4-well chamber slide, cultured overnight, fixed for 1 h at 4°C in 4% paraformaldehyde, rinsed with PBS and then permeabilized for 10 min at room temperature with 0.2% Triton-X. For RCS-o cells, samples were then mounted with a medium containing TRITC-phalloidin (Vector Laboratories). For RCS-Cas9, cells were incubated in 100 mM glycine in PBS for 10 min at room temperature followed by incubation with Fluorescein Phalloidin (ThermoFisher Scientific) in PBS with 1% BSA at room temperature for 30 min. Cells were rinsed and mounted using a media containing DAPI nuclear stain (FluoroGel II; Electron Microscopy Sciences). Cells were visualized using a Nikon Eclipse E600 microscope equipped with Y-Fl Epi-fluorescence (Melville, NY), a 1.4 n.a. oil-immersion objective and a Texas Red or FITC (green) filter. Images were captured digitally in real time using a Retiga 2000R digital camera and processed using NIS Elements BR imaging software (Nikon).
Western blotting
Total protein was extracted using Cell-Lysis-Buffer (Cell Signaling) containing protease inhibitor cocktail (Thermo Scientific). Equivalent protein concentrations were loaded into 4-12% NuPAGE® Novex® Tris-acetate gradient mini gels. Following electrophoresis, proteins were transferred to a nitrocellulose membrane using a Criterion blotter apparatus (BioRad), and the nitrocellulose membrane was then blocked in 5% non-fat dry milk in Tris buffered saline containing 0.1% Tween 20 (TBS-T). Both anti-myc antibody (Cell Signaling, clone #9B11) and mouse anti-O-linked N-acetylglucosamine antibody (clone RL2, lot PB197682, ThermoFisher) were detected with HRP-conjugated donkey anti-mouse antibody (SA1-100, ThermoFisher) followed by chemiluminescence detection (Novex ECL, Invitrogen).
4.5. Real time reverse transcription-polymerase chain reaction (RT-PCR)
RCS-o and RCS-Cas9 WT and knockout cells were plated at 70% confluency in 12 well plates in DMEM + 10% FBS and cultured for 48 h. Total RNA was isolated according to the manufacturer's instructions for the use of Trizol® reagent. Total RNA was reverse transcribed to cDNA using the iScript cDNA Synthesis Kit (BioRad). Quantitative PCR was performed using Sso-Advanced SYBR-Green Supermix (BioRad) and amplified on a StepOnePlus Real-Time PCR System (Applied Biosystems) to obtain cycle threshold (Ct) values for target and internal reference cDNA levels. The rat-specific primer sequences are follows: GAPDH, (forward) 5′-GATACTGAGAGCAAGAGAGAGG-3′ and (reverse) 5′-TCTGGGATGGAATTGTGAGG-3′; HAS1, (forward) 5′-CCCGCTGGTCCAAATCTTAT- 3′ and (reverse) 5′-GAAAGGGAAGAGGCCAGAAA -3′; HAS2, (forward) 5′-GGCTGTGTCCAGTGCATAA-3′ and (reverse) 5′-CCTGTTGGTAAGGTGCCTATC-3′; HAS3, (forward) 5′-GGCAACTCAGTGGACTACATC-3′ and (reverse) 5′-TCCTCCAACACCTCCTACTT-3′; ACAN, (forward) 5′-GGTTCGAGTGAACAGCATCTAC-3′ and (reverse) 5′-GGAGCGAAGGTTCTGGATTT-3′; COL2A1, (forward) 5′-CATCGAGTACCGATCACAGAAG-3′ and (reverse) 5′-GCCCTATGTCCACACCAAAT-3′; COL10A1 (forward) 5′-AGCAAAGGCTACTTGGATCAG-3′ and (reverse) 5′-TGAGAAGGACGAGTGGACATA-3′. Real time RT-PCR efficiency (E) was calculated as E=10[-1/slope] [59]. The fold increase in copy numbers of mRNA was calculated as a relative ratio of target gene to GAPDH (ΔΔCt), following the mathematical model introduced by Pfaffl [60] as described previously [45, 61].
For quantitative RT-PCR with standard curves, a ∼100 bp PCR product of the respective genes was amplified. The PCR amplified product was run on an agarose gel and then gel purified using the QIAquick Gel Extraction Kit (Qiagen). DNA concentrations were determined by NanoDrop (Thermo Scientific), and these PCR products diluted to a series of dilutions from 107 to 102 in order to generate a standard curve. Quantitative PCR was performed using Sso-Advanced SYBR-Green Supermix (BioRad) and amplified on a StepOnePlus Real-Time PCR System (Applied Biosystems) to obtain cycle threshold (Ct) values for target and internal reference cDNA levels. The copy number of each gene was determined by absolute quantification method with standard curves using StepOne Software 2.2.2. (Applied Biosystems).
4.6. Generation of Adeno-ZsGreen1-mycHas2
The murine Has2 coding sequence (NM_008216) and NH2-terminal 6× myc tag in pCDNA3, kindly provided by Drs. Davide Vigetti and Alberto Passi [62], was PCR amplified using CloneAmp HiFi PCR Premix (Clontech). Primers were designed according to the manufacture's protocol as detailed in the Adeno-X Adenoviral System 3 User Manual (Clontech). The upstream primer included the ATG start site for the myc tag sequence: 5′-GTA ACT ATA ACG GTC GCT ATG GAG CAA AAG CTC ATT TCT-3′ and the downstream primer included the stop codon for murine Has2: 5′-ATT ACC TCT TTC TCC TCA TAC ATC AAG CAC CAT GTC ATA-3′. The PCR cycling conditions were: 30 cycles of 10 seconds denaturation at 98°C, 15 seconds annealing at 55°C, and 10 seconds extension at 72°C. The PCR amplified product (mycHas2) was purified using the NucleoSpin Gel and PCR Clean-Up Kit (Clontech). mycHas2 was then subcloned into the pAdenoX-CMV-ZsGreen1 linearized vector using the Adeno-X Adenoviral System 3 Kit (Clontech) to form Adeno-ZsGreen1-mycHas2. DNA sequences were verified for all PCR-amplified regions. Upon DNA sequencing, it was noted that Adeno-ZsGreen1-mycHas2 contained a 4× NH2-terminal myc tag sequence.
Adeno-ZsGreen1-mycHas2 was packaged and amplified in HEK293 cells (ATCC). Viral particles were purified using the Adeno-X Purification Kit (Clontech) and tittered using the Adeno-X Rapid Titer Kit (Clontech) to obtain a 4.89 × 109 IFU/ml Adeno-ZsGreen1-mycHas2 purified viral stock.
4.7. Isolation and purification of aggrecan from bovine articular cartilage
Purified bovine aggrecan was extracted from full thickness slices of 18-24 month bovine metacarpophalengeal joints and purified by CsCl density gradient centrifugation as described previously [12]. The D1 fraction obtained following centrifugation was dialyzed against distilled water for three days and then lyophilized until use. In most applications the purified aggrecan was dissolved at 2.0 mg/ml in serum-free DMEM.
4.8. Particle exclusion assay
RCS chondrocytes were cultured in DMEM plus 10% FBS for 48 h in 35-mm wells. For some culture wells, an additional overnight incubation in serum-free DMEM without or with 2 mg/ml aggrecan was carried out. Some cultures of RCS-o Has2 KO clone #4 chondrocytes were incubated with 10 IFU/ml Adeno-ZsGreen1-mycHas2 in serum-free medium for 24 h and then the medium was replaced with DMEM containing 10% FBS for 48 h. A final 24-hour incubation of these cultures was done in the presence or absence of 2 mg/ml aggrecan monomer. To visualize the pericellular matrix, the medium was replaced with a suspension of formalin-fixed erythrocytes in PBS/0.1% BSA [22]. Cells were photographed using a Nikon TE2000 inverted phase-contrast microscope and images were captured digitally in real time using a Retiga 2000R digital camera (QImaging). The presence of the cell-associated matrix is seen as the particle-excluded zone surrounding the chondrocytes.
4.9. Culture conditions for neocartilage formation
Alginate beads
To encapsulate RCS chondrocytes in alginate beads, cells were diluted in 1.2% alginate (Keltone LVCR; Kelco) in 0.15 M NaCl at 4 million cells/ml. This mixture was expressed through a 22-gauge needle dropwise into a 102 mM CaCl2 solution for instantaneous gelation and then cultured in DMEM with 10% FBS for 7 days [26, 32, 61]. The alginate beads were fixed in 4% paraformaldehyde in 0.1M cacodylate buffer containing 10 mM CaCl2 pH 7.4 for 4h, rinsed with 0.1 M cacodylate buffer containing 50 mM BaCl2 pH 7.4, dehydrated in a series of graded ethanols, cleared in xylene wash, and embedded in Paraplast Plus (Fisher). 10 μm sections were deparaffinized and stained with 0.5% Toluidine Blue O in 20% ethanol, HABP, or Safranin O/fast green.
Pellet cultures
To study neocartilage formation in vitro, RCS-Cas9 WT, Has2 KO clone 3 and Has2 KO clone 7 cells as well as primary bovine articular chondrocytes [19] were grown in pellet cultures [30, 33]. 2 million cells in DMEM containing 10% FBS were placed into 15 ml conical tubes and centrifuged at 200 × g into pellets. The media was changed every 2 days and the pellets were incubated for 2 days or 11 days before fixation and staining. The pellets were fixed with 4% paraformaldehyde in phosphate buffer pH 7.4, dehydrated in ethanols, cleared in xylene and embedded in Paraplast Plus (Fisher). 10 μm sections were deparaffinized, hydrated and stained with hematoxylin and eosin (H&E), Toluidine Blue, HABP, Safranin O or Masson's trichrome.
5. Conclusions
The CRISPR/Cas9 genome editing method was an efficient approach to generate HA-deficient chondrocyte cell lines. Using a sgRNA to target a Cas9 dependent cleavage within exon 2 of the rat Has2 gene, we generated Has2 mutations in several clones of RCS cells. The culture media of these Has2 knockout clones did not contain detectable HA and no cell surface associated HA was detected on these cells. The Has2 knockout RCS cells lost the ability to assemble a HA/aggrecan-rich pericellular matrix and lost the capacity to retain exogenously added aggrecan. These cellular properties were rescued following transduction of the Has2 knockout RCS cells with Adeno-ZsGreen1-mycHas2. The loss of HA production by these cells also resulted in a loss of cell-to-cell spacing during neocartilage formation. These data demonstrated that HA biosynthesis by way of HAS2 is required for the retention of aggrecan in the chondrocyte matrix. The isolation of these Has2 knockout chondrocyte cell lines provides a valuable tool to understand the function of HAS2 and HA produced by HAS2 in chondrocytes.
Highlights.
Hyaluronan synthase-2 (Has2) was targeted for mutation by CRISPR/Cas9 gene editing
Has2 knockout (KO) clones were generated in two rat chondrosarcoma (RCS) cell lines
No hyaluronan synthesis or pericellular matrix retention by the Has2 KO RCS clones
No compensation in expression of Has1 or Has3 was detected in Has2 KO RCS clones
Rescue by Ad-Has2 transduction restored aggrecan retention by Has2 KO RCS clones
Acknowledgments
The authors thank Michelle Cobb and Joani Zary Oswald for their technical assistance with this project. The authors thank Dr. Gary Gibson, Henry Ford Hospital (Detroit, MI) for graciously providing CRISPR/Cas9 rat chondrosarcoma cell line. Research reported in this publication was supported in part by the Brody School of Medicine Seed Grant Program (EBA) and by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R21-AR066581 (WK) and R01-AR039507 (CBK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Abbreviations: OA, osteoarthritis; ECM, extracellular matrix; HA, hyaluronan; HAS2, hyaluronan synthase-2; RCS, rat chondrosarcoma chondrocytes; sgRNA, single guide RNA; sGAG, sulfated glycosaminoglycan; DMMB, dimethylmethylene blue
Ethical statement: The authors have nothing to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rizkalla G, Reiner A, Bogoch E, Poole AR. Studies of the articular cartilage proteoglycan aggrecan in health and osteoarthritis. Evidence for molecular heterogeneity and extensive molecular changes in disease. J Clin Invest. 1992;90:2268–77. doi: 10.1172/JCI116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunziker EB, Lippuner K, Shintani N. How best to preserve and reveal the structural intricacies of cartilaginous tissue. Matrix Biol. 2014;39:33–43. doi: 10.1016/j.matbio.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Iozzo RV, Schaefer L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes MW, Bayliss MT, Muir H. Hyaluronic acid in human articular cartilage. Age-related changes in content and size. Biochem J. 1988;250:435–41. doi: 10.1042/bj2500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roughley P, Mort J. The role of aggrecan in normal and osteoarthritic cartilage. J Exp Orthop. 2015;1 doi: 10.1186/s40634-014-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales TI. Polypeptide regulators of matrix homeostasis in articular cartilage. In: Kuettner KE, Schleyerbach R, Peyron JG, Hascall VC, editors. Articular Cartilage and Osteoarthritis. New York: Raven Press; 1992. pp. 265–80. [Google Scholar]
- 7.Ng KC, Handley CJ, Preston BN, Robinson HC. The extracellular processing and catabolism of hyaluronan in cultured adult articular cartilage explants. Arch Biochem Biophys. 1992;298:70–9. doi: 10.1016/0003-9861(92)90095-e. [DOI] [PubMed] [Google Scholar]
- 8.Nishida Y, D'Souza AL, Thonar EJ, Knudson W. Stimulation of hyaluronan metabolism by interleukin-1alpha in human articular cartilage. Arthritis Rheum. 2000;43:1315–26. doi: 10.1002/1529-0131(200006)43:6<1315::AID-ANR14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 9.Mellor L, Knudson CB, Hida D, Askew EB, Knudson W. Intracellular domain fragment of CD44 alters CD44 function in chondrocytes. J Biol Chem. 2013;288:25838–50. doi: 10.1074/jbc.M113.494872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua Q, Knudson CB, Knudson W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci. 1993;106:365–75. doi: 10.1242/jcs.106.1.365. [DOI] [PubMed] [Google Scholar]
- 11.Knudson W, Aguiar DJ, Hua Q, Knudson CB. CD44-anchored hyaluronan-rich pericellular matrices: An ultrastructural and biochemical analysis. Exp Cell Res. 1996;228:216–28. doi: 10.1006/excr.1996.0320. [DOI] [PubMed] [Google Scholar]
- 12.Danielson BT, Knudson CB, Knudson W. Extracellular processing of the cartilage proteoglycan aggregate and its effect on CD44-mediated internalization of hyaluronan. J Biol Chem. 2015;290:9555–70. doi: 10.1074/jbc.M115.643171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang M, Zhang L, Stevens J, Gibson G. CRISPR/Cas9 mediated generation of stable chondrocyte cell lines with targeted gene knockouts; analysis of an aggrecan knockout cell line. Bone. 2014;69:118–25. doi: 10.1016/j.bone.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Yingst S, Bloxham K, Warner LR, Brown RJ, Cole J, Kenoyer L, et al. Characterization of collagenous matrix assembly in a chondrocyte model system. J Biomed Mater Res A. 2009;90:247–55. doi: 10.1002/jbm.a.32078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi HU, Meyer K, Swarm R. Mucopolysaccharide and protein--polysaccharide of a transplantable rat chondrosarcoma. Proc Natl Acad Sci U S A. 1971;68:877–9. doi: 10.1073/pnas.68.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kucharska AM, Kuettner KE, Kimura JH. Biochemical characterization of long-term culture of the Swarm rat chondrosarcoma chondrocytes in agarose. J Orthop Res. 1990;8:781–92. doi: 10.1002/jor.1100080602. [DOI] [PubMed] [Google Scholar]
- 17.Knudson W, Chow G, Knudson CB. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002;21:15–23. doi: 10.1016/s0945-053x(01)00186-x. [DOI] [PubMed] [Google Scholar]
- 18.Embry Flory JJ, Fosang AJ, Knudson W. The accumulation of intracellular ITEGE and DIPEN neoepitopes in bovine articular chondrocytes is mediated by CD44 internalization of hyaluronan. Arthritis Rheum. 2006;54:443–54. doi: 10.1002/art.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ariyoshi W, Knudson CB, Luo N, Fosang AJ, Knudson W. Internalization of aggrecan G1 domain neoepitope ITEGE in chondrocytes requires CD44. J Biol Chem. 2010;285:36216–24. doi: 10.1074/jbc.M110.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem. 1997;272:13997–4000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
- 21.Vigetti D, Viola M, Karousou E, De Luca G, Passi A. Metabolic control of hyaluronan synthases. Matrix Biol. 2014;35:8–13. doi: 10.1016/j.matbio.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Knudson CB. Hyaluronan receptor-directed assembly of chondrocyte pericellular matrix. J Cell Biol. 1993;120:825–34. doi: 10.1083/jcb.120.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knudson CB, Knudson W. Cartilage proteoglycans. Semin Cell Dev Biol. 2001;12:69–78. doi: 10.1006/scdb.2000.0243. [DOI] [PubMed] [Google Scholar]
- 24.Knudson CB. Hyaluronan in embryonic cell adhesion and matrix assembly. In: Laurent TC, editor. The Chemistry, Biology and Medical Applications of Hyaluronan and its Derivatives. London: Portland Press; 1998. pp. 161–8. [Google Scholar]
- 25.Knudson CB, Toole BP. Hyaluronate - cell interactions during differentiation of chick embryo limb mesoderm. Dev Biol. 1987;124:82–90. doi: 10.1016/0012-1606(87)90462-3. [DOI] [PubMed] [Google Scholar]
- 26.Nishida Y, Knudson CB, Nietfeld JJ, Margulis A, Knudson W. Antisense inhibition of hyaluronan synthase-2 in human articular chondrocytes inhibits proteoglycan retention and matrix assembly. J Biol Chem. 1999;274:21893–9. doi: 10.1074/jbc.274.31.21893. [DOI] [PubMed] [Google Scholar]
- 27.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–60. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, et al. Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development. 2009;136:2825–35. doi: 10.1242/dev.038505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hascall VC, Wang A, Tammi M, Oikari S, Tammi R, Passi A, et al. The dynamic metabolism of hyaluronan regulates the cytosolic concentration of UDP-GlcNAc. Matrix Biol. 2014;35:14–7. doi: 10.1016/j.matbio.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–72. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 31.Larson CM, Kelley SS, Blackwood AD, Banes AJ, Lee GM. Retention of the native chondrocyte pericellular matrix results in significantly improved matrix production. Matrix Biol. 2002;21:349–59. doi: 10.1016/s0945-053x(02)00026-4. [DOI] [PubMed] [Google Scholar]
- 32.Masuda K, Sah RL, Hejna MJ, Thonar EJ. A novel two-step method for the formation of tissue-engineered cartilage by mature bovine chondrocytes: the alginate-recovered-chondrocyte (ARC) method. J Orthop Res. 2003;21:139–48. doi: 10.1016/S0736-0266(02)00109-2. [DOI] [PubMed] [Google Scholar]
- 33.Ono Y, Sakai T, Hiraiwa H, Hamada T, Omachi T, Nakashima M, et al. Chondrogenic capacity and alterations in hyaluronan synthesis of cultured human osteoarthritic chondrocytes. Biochem Biophys Res Commun. 2013;435:733–9. doi: 10.1016/j.bbrc.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knudson CB, Toole BP. Fluorescent morphological probe for hyaluronate. J Cell Biol. 1985;100:1753–8. doi: 10.1083/jcb.100.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evanko SP, Potter-Perigo S, Petty LJ, Workman GA, Wight TN. Hyaluronan controls the deposition of fibronectin and collagen and modulates TGF-beta1 induction of lung myofibroblasts. Matrix Biol. 2015;42:74–92. doi: 10.1016/j.matbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LuValle P, Daniels K, Hay ED, Olsen BR. Type X collagen is transcriptionally activated and specifically localized during sternal cartilage maturation. Matrix. 1992;12:404–13. doi: 10.1016/s0934-8832(11)80037-5. [DOI] [PubMed] [Google Scholar]
- 37.Girkontaite I, Frischholz S, Lammi P, Wagner K, Swoboda B, Aigner T, et al. Immunolocalization of type X collagen in normal fetal and adult osteoarthritic cartilage with monoclonal antibodies. Matrix Biol. 1996;15:231–8. doi: 10.1016/s0945-053x(96)90114-6. [DOI] [PubMed] [Google Scholar]
- 38.Candela ME, Yasuhara R, Iwamoto M, Enomoto-Iwamoto M. Resident mesenchymal progenitors of articular cartilage. Matrix Biol. 2014;39:44–9. doi: 10.1016/j.matbio.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishida Y, Knudson CB, Eger W, Kuettner KE, Knudson W. Osteogenic protein-1 stimulates cell-associated matrix assembly by normal human articular chondrocytes. Arthritis Rheum. 2000;43:206–14. doi: 10.1002/1529-0131(200001)43:1<206::AID-ANR25>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Nishida N, Knudson CB, Knudson W. Extracellular matrix recovery by human articular chondrocytes after treatment with hyaluronan hexasaccharides or Streptomyces hyaluronidase. Modern Rheum. 2003;13:62–8. doi: 10.3109/s101650300009. [DOI] [PubMed] [Google Scholar]
- 41.Pavasant P, Shizari T, Underhill CB. Hyaluronan contributes to the enlargement of hypertrophic lacunae in the growth plate. J Cell Sci. 1996;109:327–34. doi: 10.1242/jcs.109.2.327. [DOI] [PubMed] [Google Scholar]
- 42.Wilusz RE, Sanchez-Adams J, Guilak F. The structure and function of the pericellular matrix of articular cartilage. Matrix Biol. 2014;39:25–32. doi: 10.1016/j.matbio.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno S, Im HJ, Knudson CB, Knudson W. Hyaluronan oligosaccharides induce matrix metalloproteinase 13 via transcriptional activation of NFkappaB and p38 MAP kinase in articular chondrocytes. J Biol Chem. 2006;281:17952–60. doi: 10.1074/jbc.M602750200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muto J, Yamasaki K, Taylor KR, Gallo RL. Engagement of CD44 by hyaluronan suppresses TLR4 signaling and the septic response to LPS. Mol Immunol. 2009;47:449–56. doi: 10.1016/j.molimm.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ariyoshi W, Takahashi N, Hida D, Knudson CB, Knudson W. Mechanisms involved in enhancement of the expression and function of aggrecanases by hyaluronan oligosaccharides. Arthritis Rheum. 2012;64:187–97. doi: 10.1002/art.33329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loeser RF. Integrins and chondrocyte-matrix interactions in articular cartilage. Matrix Biol. 2014;39:11–6. doi: 10.1016/j.matbio.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799:353–64. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarkin CE, Allen S, Wheeler-Jones CP, Bastow ER, Pitsillides AA. Reduced chondrogenic matrix accumulation by 4-methylumbelliferone reveals the potential for selective targeting of UDP-glucose dehydrogenase. Matrix Biol. 2011;30:163–8. doi: 10.1016/j.matbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigetti D, Ori M, Viola M, Genasetti A, Karousou E, Rizzi M, et al. Molecular cloning and characterization of UDP-glucose dehydrogenase from the amphibian Xenopus laevis and its involvement in hyaluronan synthesis. J Biol Chem. 2006;281:8254–63. doi: 10.1074/jbc.M508516200. [DOI] [PubMed] [Google Scholar]
- 50.Clarkin CE, Allen S, Kuiper NJ, Wheeler BT, Wheeler-Jones CP, Pitsillides AA. Regulation of UDP-glucose dehydrogenase is sufficient to modulate hyaluronan production and release, control sulfated GAG synthesis, and promote chondrogenesis. J Cell Physiol. 2011;226:749–61. doi: 10.1002/jcp.22393. [DOI] [PubMed] [Google Scholar]
- 51.Edwards JC, Wilkinson LS, Jones HM, Soothill P, Henderson KJ, Worrall JG, et al. The formation of human synovial joint cavities: a possible role for hyaluronan and CD44 in altered interzone cohesion. J Anat. 1994;185:355–67. [PMC free article] [PubMed] [Google Scholar]
- 52.Toole BP. Proteoglycans and hyaluronan in morphogenesis and differentiation. In: Hay ED, editor. Cell Biology of Extracellular Matrix. 2nd. New York: Plenum Press; 1991. pp. 305–39. [Google Scholar]
- 53.Maleski MP, Knudson CB. Hyaluronan meditated aggregation of limb bud mesoderm and mesenchymal condensation during chondrogenesis. Exp Cell Res. 1996;225:55–66. doi: 10.1006/excr.1996.0156. [DOI] [PubMed] [Google Scholar]
- 54.Peterson RS, Andhare RA, Rousche KT, Knudson W, Wang W, Grossfield JB, et al. CD44 modulates Smad1 activation in the BMP-7 signaling pathway. J Cell Biol. 2004;166:1081–91. doi: 10.1083/jcb.200402138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, et al. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133:4667–78. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 56.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samiric T, Parkinson J, Ilic MZ, Cook J, Feller JA, Handley CJ. Changes in the composition of the extracellular matrix in patellar tendinopathy. Matrix Biol. 2009;28:230–6. doi: 10.1016/j.matbio.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Ono Y, Ishizuka S, Knudson CB, Knudson W. Chondroprotective effecct of kartogenin on CD44-mediated functions in articular cartilage and chondrocytes. Cartilage. 2014;5:172–80. doi: 10.1177/1947603514528354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen TB, Uttenthal A, de Stricker K, Belak S, Storgaard T. Development of a novel quantitative real-time RT-PCR assay for the simultaneous detection of all serotypes of foot-and-mouth disease virus. Arch Virol. 2003;148:2005–21. doi: 10.1007/s00705-003-0145-2. [DOI] [PubMed] [Google Scholar]
- 60.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi N, Knudson CB, Thankamony S, Ariyoshi W, Mellor L, Im HJ, et al. Induction of CD44 cleavage in articular chondrocytes. Arthritis Rheum. 2010;62:1338–48. doi: 10.1002/art.27410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karousou E, Kamiryo M, Skandalis SS, Ruusala A, Asteriou T, Passi A, et al. The activity of hyaluronan synthase 2 is regulated by dimerization and ubiquitination. J Biol Chem. 2010;285:23647–54. doi: 10.1074/jbc.M110.127050. [DOI] [PMC free article] [PubMed] [Google Scholar]


