Abstract
There is great interest in drugs that are capable of modulating multiple aging pathways, thereby delaying the onset and progression of aging. Effective strategies for drug development include the repurposing of existing drugs already approved by the FDA for human therapy. FDA approved drugs have known mechanisms of action and have been thoroughly screened for safety. Although there has been extensive scientific activity in repurposing drugs for disease therapy, there has been little testing of these drugs for their effects on aging. The pool of FDA approved drugs therefore represents a large reservoir of drug candidates with substantial potential for anti-aging therapy. In this paper we employ FINDSITEcomb, a powerful ligand homology modeling program, to identify binding partners for proteins produced by temperature sensing genes that have been implicated in aging. This list of drugs with potential to modulate aging rates was then tested experimentally for lifespan and healthspan extension using a small invertebrate model. Three protein targets of the rotifer Brachionus manjavacas corresponding to products of the transient receptor potential gene 7, ribosomal protein S6 polypeptide 2 gene, or forkhead box C gene, were screened against a compound library consisting of DrugBank drugs including 1347 FDA approved, non-nutraceutical molecules. Twenty nine drugs ranked in the top 1 % for binding to each target were subsequently included in our experimental analysis. Continuous exposure of rotifers to 1 µM naproxen significantly extended rotifer mean lifespan by 14 %. We used three endpoints to estimate rotifer health: swimming speed (mobility proxy), reproduction (overall vitality), and mitochondria activity (cellular senescence proxy). The natural decline in swimming speed with aging was more gradual when rotifers were exposed to three drugs, so that on day 6, mean swimming speed of females was 1.19 mm/s for naproxen (P = 0.038), 1.20 for fludarabine (P = 0.040), 1.35 for hydralazine (P = 0.038), as compared to 0.88 mm/s in the control. The average reproduction of control females in the second half of their reproductive lifespan was 1.08 per day. In contrast, females treated with 1 µM naproxen produced 1.4 offspring per day (P = 0.027) and females treated with 10 µM fludarabine or 1 µM hydralazine produced 1.72 (P = <0.001) and 1.66 (P = 0.001) offspring per day, respectively. Mitochondrial activity naturally declines with rotifer aging, but B. manjavacas treated with 1 µM hydralazine or 10 µM fludarabine retained 49 % (P = 0.038) and 89 % (P = 0.002) greater mitochondria activity, respectively, than untreated controls. Our results demonstrate that coupling computation to experimentation can quickly identify new drug candidates with anti-aging potential. Screening drugs for anti-aging effects using a rotifer bioassay is a powerful first step in identifying compounds worthy of follow-up in vertebrate models. Even if lifespan extension is not observed, certain drugs could improve healthspan, slowing age-dependent losses in mobility and vitality.
Keywords: Aging, Re-purposing drugs, Rotifera, Lifespan, Healthspan, TRP genes
Introduction
There is great interest in finding drugs capable of extending human lifespan and healthspan (Armanios et al. 2015). Compounds are sought that are capable of modulating multiple aging pathways, thereby preventing a broad-spectrum of age-related diseases. Conserved aging pathways have been identified using experimental animal models (de Cabo et al. 2014), and some compounds capable of delaying the onset and progression of aging have been identified; these include spermidine, rapamycin, metformin and resveratrol.
One of the most effective strategies for drug development is the repurposing of existing drugs that have been approved by the FDA for human therapy (Ashburn and Thor 2004; Pantziarka et al. 2014). FDA approved drugs have known mechanisms of action and have been thoroughly screened for safety. Moreover, many drugs have much broader ranges of action than their licenses suggest. FDA approved drugs are likely to have low risks, so that even small benefits might provide an acceptable risk/benefit ratio. Although there has been extensive commercial activity in repurposing drugs for disease therapies (Naylor and Schonfeld 2015), there has been little testing of these drugs for their effects on aging. The pool of FDA approved drugs therefore represents a large reservoir of drug candidates with substantial potential for anti-aging therapy. Repurposing existing drugs could have a rapid impact on human aging if drugs can be shown to have efficacy in slowing the progression of age-related diseases. A strategy for rapidly finding such drugs is to screen the suite of existing drugs computationally and then to test the predictions experimentally using a short-lived invertebrate model. Promising results can then be followed up in vertebrate and mammalian experimental models, then confirmed in human clinical trials.
Rotifers are experimentally tractable animal models for studying the biology of aging and representatives of the supra-phylum Lophotrochozoa (Snell 2014). The brachionid rotifers that we study are about 0.4 mm long aquatic herbivores, capable of both asexual and sexual reproduction. Their lifespan in the laboratory at 22 °C is about 2 weeks, enabling rapid screening of many treatments using cohort life tables (Snell et al. 2014). For example, libraries of natural products from marine algae have been screened for bioactivity using this rotifer model (Snare et al. 2013). Rotifers, therefore, are useful animal models for initial drug screening for life and healthspan extension before moving to vertebrate models.
Several rotifer genes and pathways capable of extending lifespan have been identified (Oo et al. 2010; Ozaki et al. 2010; Snell et al. 2012, 2014b; Gribble and Welch 2012; Snell and Johnston 2014; Johnston and Snell 2016). Prominent among them are nutrient sensing and stress resistance pathways. A less appreciated pathway with substantial effects on aging is the temperature-sensing pathway. Like many animals, rotifer lifespan is extended by exposure to moderately lower temperatures as a result of combined thermodynamic effects on metabolic reactions and changes in gene expression (Johnston and Snell 2016). These authors used a combination of life table experiments, stressor challenge experiments, and RNAi knockdown treatments to identify putative temperature sensing genes likely involved in regulating aging rates. Four of 12 temperature-sensing genes were identified as especially promising drug targets for life and healthspan extension. Among these, we have explored three rotifer temperature sensing genes and computationally investigated drug binding partners with the potential to modulate their expression.
Many bioinformatics and computational approaches for drug-protein interaction discovery have been developed (Cavasotto and Orry 2007; Cheng et al. 2012; Gottlieb et al. 2011; Laarhoven and Marchioro 2013; Lee and Zhang 2011; von Eichborn et al. 2011; Yamanishi et al. 2008; Yamanishi et al. 2010). Some methods use protein sequences and a priori knowledge of binding drugs of a protein of interest (Laarhoven and Marchioro 2013; Yamanishi et al. 2010), whereas docking-based approaches require high-resolution protein structures that in most cases are not available (Cavasotto and Orry 2007). The recently developed FINDSITEcomb, a ligand homology modeling approach that has been extensively experimentally validated (Srinivasan et al. 2014; Zhou and Skolnick 2013), has several advantages over other state-of-the-art methods for predicting drug–protein interactions: First, it does not require known drug interactions for a protein target as is required by many sequence-based methods; second, it does not require high resolution protein structures required by docking-based methods; third, it is much more efficient than docking-based methods; and last, and most importantly, it has better accuracy for ranking drug–target interactions than other methods (Zhou and Skolnick 2013). FINDSITEcomb has been applied in the creation of the DR.PRODIS database (Zhou et al. 2015), which contains comprehensive drug–protein interactions predicted for the entire Human proteome.
The hypothesis of this paper is that FINDSITEcomb can identify drug candidates capable of life or healthspan extension, and that these can be rapidly tested experimentally using rotifers as an animal model.
Materials and methods
FINDSITEcomb algorithm for computational screening of FDA approved drugs
FINDSITEcomb (Zhou and Skolnick 2013) is a computational method that is capable of screening millions of compounds on a desktop computer within a few hours. The basic assumption of FINDSITEcomb is that similar pockets bind to similar ligands regardless of evolutionary relationship. However, evolutionarily related proteins have a high chance of being similar structurally. For a given protein of unknown ligands and unknown structure, FINDSITEcomb firstly builds a structure model of the protein target using state-of-the-art threading method (Skolnick et al. 2004; Zhou and Zhou 2005) and one of the best protein structure modeling approaches, TASSER (Zhang and Skolnick 2004). The modeled structure is subsequently employed in searching against a library of pockets with experimentally determined ligands [PDB holo structures (Bernstein et al. 1977)] in the FINDITEfilt component (Zhou and Skolnick 2013)—an updated version of FINDSITE approach (Brylinski and Skolnick 2008); and is then searched against a library of modeled structures of proteins having experimentally determined binders but no experimentally determined structure of the ligand–protein complex structures [DrugBank (Wishart et al. 2006) & ChEMBL (Gaulton et al. 2012)] in the FINDSITEX (Zhou and Skolnick 2012) component. The ligands of the top 100 ranked pockets from PDB, of top ranked proteins from DrugBank & ChEMBL (called “template ligands”) are used as “seeds” to search against a compound library by a similarity-based approach. The ligands in the compound library are independently ranked by the three components (FINDSITEfilt using the PDB, FINDSITEX using DrugBank & FINDSITEX using ChEMBL) according to their similarity to the respective template ligands. The combined ranking gives a final prediction. In practice, the top 1 % of ranked ligands of the compound library are considered for further experimental test.
In this work, three protein targets of Brachionus manjavacas corresponding to products of the transient receptor potential gene 7 (GARS01012197.1, TRP7), ribosomal protein S6 polypeptide 2 gene (GARS01003002.1, S6P), or forkhead box C gene (GARS01006072.1, FhBC), were screened against a compound library consisting of DrugBank drugs including 1347 FDA approved, non-nutraceutical molecules. Together with the ZINC8 (Irwin and Shoichet 2005) background, a total of 74,378 molecules are screened by FINDSITEcomb. FDA approved drugs ranked within the top 1 % (i.e. higher than 740th) for each target are subsequently considered for further experimental validation. Candidate drugs are listed in Table 1.
Table 1.
Drugs predicted to bind to proteins produced by three temperature sensing Brachionus manjavacas genes, TRP7, S6P, and FhBC
| Gene | Drug | Ranking | Mechanism | Names | Source |
|---|---|---|---|---|---|
| TRP7 | Tadalafil | 12 | PDE-5 inhibitor | Cialis, Adcirca | Selleck Chemicals |
| Sildenafil | 14 | cGMP PDE-5 inhibitor | Viagra | Selleck Chemicals | |
| Vardenafil | 15 | PDE-5/PDE-1 inhibitor | Levitra | Selleck Vhemicals | |
| Enprofylline | 16 | Xanthine derivative, A1/A2 adenosine receptor antagonist | LGM Pharma | ||
| Enoximone | 20 | PDE-3 inhibitor | Perfan | Sigma-Aldrich | |
| Amrinone | 44 | PDE-3 inhibitor | Inocor | Sigma-Aldrich | |
| Hydralazine | 46 | Smooth muscle relaxant- K-channel activator | Apresoline | Selleck Chemicals | |
| Lenalidomide | 47 | TNF-a secretion inhibitor | Revlimid | Selleck Chemicals | |
| Dyphylline | 56 | Xanthine derivative, adenosine receptor antagonist, PDE inhibitor | Dilor, Lufyllin | Selleck Chemicals | |
| Pentoxifylline | 68 | Xanthine derivative, adenosine receptor antagonist, PDE inhibitor | Trental | Selleck Chemicals | |
| S6P | Lactulose | 68 | Synthetic disaccharide- promote gut bacterial growth | Selleck Chemicals | |
| Fludarabine | 185 | STAT1 activation inhibitor, DNA synthesis inhibitor | Fludara | Selleck Chemicals | |
| Naproxen | 189 | COX-1/COX-1 inhibitor | Aleve, Naprosyn | Selleck Chemicals | |
| Calcium gluceptate | 236 | Calcium supplement | Selleck Chemicals | ||
| Fusidic acid | 351 | Bacteriostatic antibiotic–protein synthesis inhibitor | Sigma-Aldrich | ||
| Adenosine | 361 | Purine nucleoside- energy transfer, signal transduction | Selleck Chemicals | ||
| Vidarabine | 362 | Inhibits viral DNA synthesis | Vira-A | Selleck Chemicals | |
| Ibuprofen | 381 | COX-1/COX-1 Inhibitor | Advil, Dolgesic | Selleck Chemicals | |
| Mupirocin | 408 | isoleucyl tRNA synthetase inhibitor | Bactroban, Centany | Selleck Chemicals | |
| γ-Hydroxybutyric acid | 444 | CNS depressant- GHB agonist, weak GABA antagonist | Xyrem | Controlled Subst | |
| FhBC | Lamotrigine | 13 | 5-HT inhibitor, Na-channel blocker | Lamictal | Selleck Chemicals |
| Mexiletine | 21 | Na-channel inhibitor | Mexitil | Selleck Chemicals | |
| l-Carnitine | 29 | Fatty acid transport, lipid breakdown | Selleck Chemicals | ||
| Leucovorin | 85 | Rescues low levels of folic acid | Selleck Chemicals | ||
| Fludarabine | 119 | STAT1 activation inhibitor, DNA synthesis inhibitor | Fludara | Selleck Chemicals | |
| Methotrexate | 178 | Folic acid metabolism inhibitor | Trexall | Selleck Chemicals | |
| Adenosine | 218 | Purine nucleoside- energy transfer, signal transduction | Selleck Chemicals | ||
| Vidarabine | 219 | Inhibits viral DNA synthesis | Vira-A | Selleck Chemicals | |
| Nafarelin | 293 | GRH analogue, LH-RH agonist, decreases LH and FSH | Synarel | Sigma-Aldrich | |
| Colistin | 320 | Polymyxin antibiotic, breaks down bacterial membranes | Xylistin, Koolistin | Selleck Chemicals |
Rotifer culture
All experiments were performed using the rotifer species B. manjavacas. This strain was originally collected from the Azov Sea in Russia, and has been continuously cultivated in the Snell laboratory since 1983, with resting eggs being periodically collected, dried, and stored. Before each experiment, B. majavacas resting eggs were hatched in 25 mL, 15 ppt artificial sea water (ASW, Instant Ocean) under constant fluorescent illumination (2000 lux) at 25 °C. Under these conditions, hatching begins after 18–20 h, resulting in a uniform cohort of neonates. Hatchlings were fed the green alga Tetraselmis suecica cultured in modified F medium (Guillard 1983) in a 560 mL chemostat with 25 % daily medium replacement, at 25 °C and constant fluorescent illumination of 2000 lux. All cultures were kept in percival I-41VL incubators to maintain stable environmental conditions (Table 2).
Table 2.
Effect of TRP agonists and antagonists on B. manjavacas reproduction and lifespan
| Compound | Target | Source | Conc. (μM) | Reproduction | Lifespan |
|---|---|---|---|---|---|
| Capsaicin | TRPV1 agonist, hot taste of chili peppers | Sigma-Aldrich | 1 | NS | NS |
| 10 | NS | NS | |||
| Resiniferatoxin | TRPV1 agonist, analog of capsaicin | Sigma-Aldrich | 0.5 | NS | ↓40 % |
| 1 | NS | ↓56 % | |||
| Icilin | TRPM8 Super-agonist, similar effects to menthol | Sigma-Aldrich | 1 | ↑16 % | NS |
| 5 | NS | NS | |||
| 10 | NS | - | |||
| WS-12 | Selective TRPM8 agonist | Sigma-Aldrich | 1 | ↑6 % | NS |
| 5 | NS | NS | |||
| Allyl isothiocyanate | TRPA1 and TRPV1 agonist, pungent mustard taste | Sigma-Aldrich | 1–22 °C | NS | NS |
| 1–16 °C | – | ↓12 % | |||
| HC030031 | Selective TRPA1 blocker, lC50 = 5 μM | Sigma-Aldrich | 5–22 °C | NS | NS |
| 5–16 °C | – | ↓16 % | |||
| SB705498 | Antagonizes heat activation of hTRPV1 | Selleck | 0.5 | NS | NS |
| 1 | NS | NS | |||
| AMG-517 | Selective TRPV1 antagonist | Selleck | 0.5 | NS | NS |
| 1 | NS | NS |
Experimental design and treatments
Full cohort life table experiments were conducted with 120 newly hatched female B. manjavacas per treatment. Animals were kept in 24-well plates, with each well containing five females in 1 mL of medium. The medium contained 6 × 105 T. suecica cells/mL in 15 ppt ASW, any drug treatments, and 20 µM 5-fluoro-2-deoxyuridine (FDU), added to prevent the hatching of asexual eggs (Snell et al. 2014b). In experiments where drugs were dissolved in DMSO, the control medium also contained DMSO at the same concentration. Plates were incubated at 22 °C in the dark. Animals were checked daily and mortality was recorded until all animals were dead. All animals were transferred to new plates with fresh medium on day 8 to replenish their food supply, FDU, and drug treatment.
Reproductive life table experiments were performed in a similar fashion, with a few modifications. Single females were kept in each well of 24-well plates in 1 mL of medium, for a total of 24 females per treatment. The medium contained 2 × 105 T. suecica cells/mL in 15 ppt ASW and drug treatment. No FDU was added to allow for normal egg hatching. Offspring were produced parthenogenetically and were counted and removed daily. The original maternal females were transferred to new plates with fresh medium on day 6. For reproductive screens exploring the effects of TRP effector drugs, offspring were counted and removed for only the first 3 days. The total number of offspring over 3 days was used to calculate the intrinsic population growth rate (r) for each treatment.
Survival screens
Survival screens were conducted with a cohort of 84 B. manjavacas hatchlings per treatment and 164 hatchlings in the control. Animals were kept in 24-well plates with each well containing 7 females in 1 mL of medium. The medium contained 6 × 105 T. suecica cells/mL in 15 ppt ASW, any drug treatments, and 20 µM FDU. Plates were incubated at 28 °C in the dark. On days 2, 4, and 6, 5 μL additional FDU (1 mg/mL) was added to each well to prevent the hatching of eggs. On day 6, 100 μL 6.6 × 106 cells/mL T. suecica was added to each well to replenish food supply. On day 8 or 10, the number of living animals was counted and survival was recorded as average percent surviving in each well.
Estimation of rotifer swimming speed
To analyze swimming speed at different ages, rotifer hatchlings were cultured in 6-well plates with approximately 40 females per well in 5 mL medium. The medium contained 6 × 105 T. suecica cells/mL in 15 ppt ASW, any drug treatments, and 20 µM FDU. Plates were incubated at 22 °C in the dark. Animals were transferred to new plates with fresh medium on day 8. On days 0, 2, 4, 6, 8, 10, and 12, 15 females from each treatment were chosen randomly and transferred to 5 mL 15 ppt ASW to rinse away algae cells. These animals were then transferred to painted microscope slides, with five females in each painted well in 12 μL ASW. Swimming behavior in each well was recorded for 30 s at 10× magnification using a PixeLink camera attached to a stereomicroscope. The Tracker Video Analysis and Modeling Tool program (http://physlets.org/tracker/) was used to track the movement of each individual animal and calculate velocity in mm/s. Swimming speed was estimated for 6–10 individuals from each treatment at each time point.
Estimation of mitochondrial activity
MitoTracker® Red (Invitrogen), which has an excitation of 581 nm and an emission of 644 nm (Snell et al. 2014b), was used to measure mitochondrial activity. Female rotifers were cultured in 24-well plates with 6 × 105 T. suecica cells/mL in 15 ppt ASW, appropriate drug treatments, and 20 µM FDU for 4 days. Plates were incubated at 22 °C in the dark. On day 4, animals were rinsed in 15 ppt ASW for 2 h to clear their guts and eliminate auto-fluorescent algae. After clearing, the animals were incubated in 5 μM MitoTracker® Red in the dark for 30 min. The stain was then rinsed away with ASW. The rotifers were anesthetized with 1 mL club soda, fixed with 20 μL 20 % formalin, and rinsed with ASW. Images were taken at 200× magnification with an Alexa 568 nm excitation filter using a Zeiss Imager Z1 microscope with a 5 ms exposure. The average pixel intensity of each animal was measured using ImageJ. The entire animal was selected and measured, and the pixel intensity of the background was subtracted. Twenty animals were measured for each treatment.
Statistics
Survival curves from full life tables were evaluated using the JMP Pro 11 (SAS Institute) reliability and survival analysis with a Wilcoxon’s test to compare control and treatments. Eight day survival screens were compared by ANOVA with Dunnett’s test to compare treatments to control. A similar analysis was used for swimming speed, MitoTracker, and reproduction.
Results
The algorithm FINDSITEcomb (Zhou and Skolnick 2013) was used to generate a list of FDA approved drugs that are predicted to bind to proteins produced by the S6Ka, TRP7, and FhBC rotifer putative temperature sensing genes. Only the top 1 % of the strongest binders were considered. In practice this translates to ten drugs for each gene (Table 1). We selected four drugs to test for each gene, based on availability, price, and mode of action. Drugs like fludarabine that appeared on the list of two genes were favored. We first tested the effects of selected drugs from this list on rotifer asexual reproduction using a 3 day dose–response screen (data not shown). This allowed us to estimate the toxicity of each drug and the likely therapeutic dose to produce lifespan extension. These results led us to conclude that all drugs should be tested at concentrations of 1 and 10 µM.
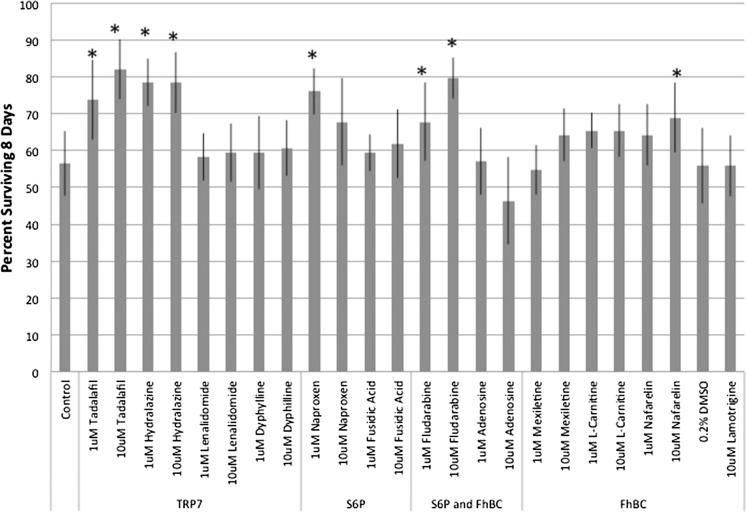
As rotifers are aquatic animals, all exposures were with drugs dissolved in the medium. The effects of 12 drugs on the survival of B. manjavacas are shown in Fig. 1. Survival after 8 days of continuous drug exposure was compared to either the control (dilution medium) or a solvent control (0.2 % DMSO), if the drug required DMSO to dissolve in the test medium. Survival was similar in both the control and solvent control. Exposure to two drugs putatively binding to the TRP7 protein, tadalafil and hydralazine at 1 and 10 µM, significantly increased rotifer survival 31–43 % over the control. Similarly, 1 µM naproxen putatively bound to the S6P protein, significantly increasing survival 35 %. Fludarabine putatively binds to both the S6P and FhBC proteins and significantly increased rotifer survival by 21 and 43 % at 1 and 10 µM, respectively. Nafarelin at 10 µM putatively bound to the FhBC protein and significantly increased rotifer survival by 22 %.
Fig. 2.
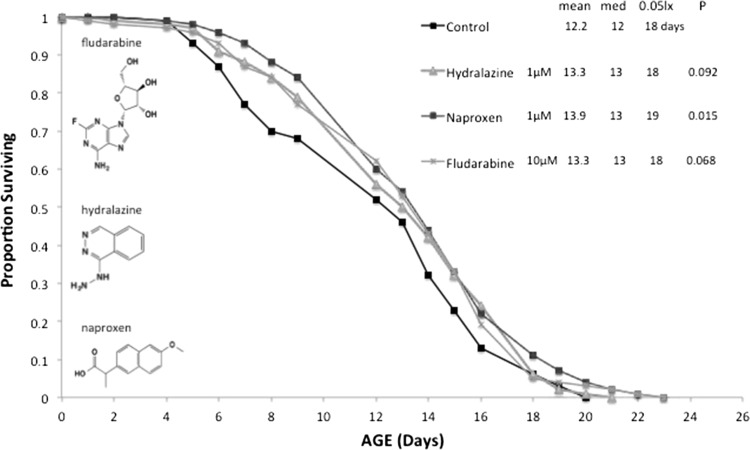
B. manjavacas survival in treatments of continuous 1 uM hydralazine, 1 µM naproxen or 10 µM fludarabine exposure. Proportion surviving refers to the age-specific survival from an initial cohort of 120 rotifers. Mean, median and age in days when 5% of the cohort remains alive is shown in the figure legend. P refers to the probability that the survival curve differs significantly from the control by Wilcoxon’s test. (Color figure online)
Fig. 3.
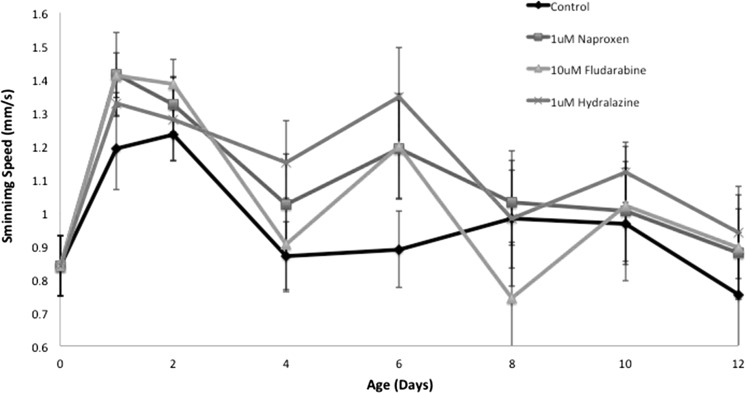
Effect of continuous exposure to 1 µM naproxen, 10 µM fludarabine, or 1 µM hydralazine on delaying the age-induced decline of rotifer swimming speed. (Color figure online)
Fig. 4.
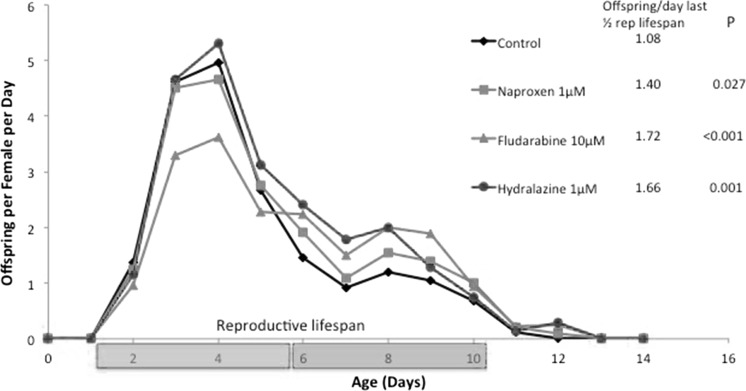
Effect of drugs on age-specific asexual reproduction of B. manjavacas females. Reproductive lifespan is divided into the first half (gray bar) and second half (yellow bar). Offspring produced per day of the second half of the reproductive lifespan is listed in the figure legend. P refers to the probability that offspring production is significantly higher than control by ANOVA and Dunnet’s test. (Color figure online)
Fig. 5.
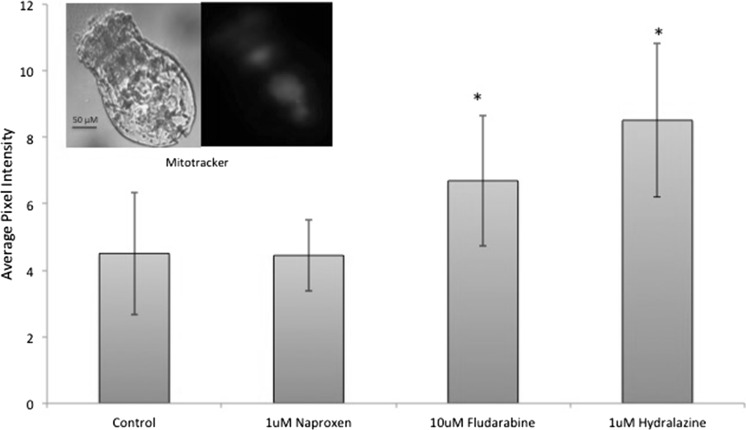
Fludarabine and hydralazine enhance mitochondrial activity at age 4 days. Y-axis is average pixel intensity of fluorescence at 620 nm. Asterisks indicate the probability by ANOVA and Dunnet’s test that the treatments have higher fluorescence than the control. (Color figure online)
Fig. 6.
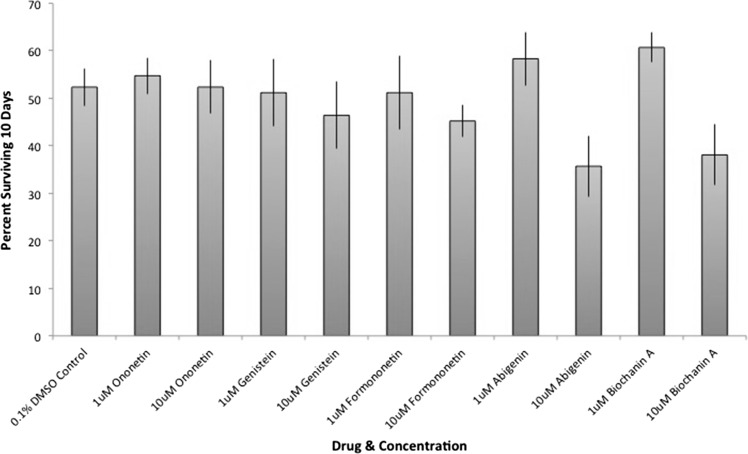
10-day survival screen for 5 ononetin analogs. (Color figure online)
Fig. 1.
Percent rotifers surviving after 8 days exposure to drugs putatively binding to proteins of 3 rotifer temperature-sensing genes. Asterisks indicate treatments with significantly higher survival than controls (P < 0.05). (Color figure online)
The results from the 8 day survival screen were followed up in a full cohort life table experiment (Fig. 2). Cohorts of 120 females were continuously exposed to 1 µM hydralazine, 1 µM naproxen, or 10 µM fludarabine, and compared to survival in a no drug control. The naproxen treatment significantly extended rotifer mean lifespan by 14 %, whereas the lifespans of hydralazine and fludarabine exposed rotifers were not significantly different from controls. One explanation for differences in the results of the 8 day survival screen and the life table experiment, is that the former was conducted at 28 °C, whereas the latter was conducted at 22 °C.
In addition to life extension, we also are interested in drugs capable of extending rotifer healthspan. We have developed three endpoints that estimate rotifer health: swimming speed (mobility proxy), reproduction (overall vitality), and mitochondria activity (cellular senescence proxy). B. manjavacas females swim continuously throughout their life, initially at an average of 0.84 mm/s as juveniles, increasing to 1.23 mm/s at age 2 days, followed by a decline back to 0.86 mm by age 4 days (Fig. 3). Swimming speed remains steady at about 0.9 mm/s through age 10 days, and then slowly declines until senescent females stop swimming, fall to the bottom and die at age about 14 days. When treated with naproxen, fludarabine or hydralazine, maximum swimming speed peaked at age 1.5 days at 1.38, 1.38 and 1.28 mm/s, respectively. The decline in swimming speed was more gradual in the drug treatments, so that on day 6, mean swimming speed of females was 1.19 mm/s for naproxen, 1.20 for fludarabine, 1.35 for hydralazine, as compared to 0.88 mm/s in the control. These differences are significant by an F-test, with P values of 0.038–0.040. These data suggest that treatment with these three drugs slows the decline in rotifer swimming speed in older age classes.
Reproduction is an endpoint that often correlates with female health and vitality. In toxicology tests, rotifer reproduction typically is suppressed by toxicant stress at lower concentrations than effects on survival are observed (Snell and Janssen 1995). After a brief juvenile period of 1 day, rotifer females begin asexual reproduction and continue until age 12 days, so that their reproductive lifespan is about 80 % of their entire lifespan (Fig. 4). Reproduction peaks on day 4 at about five offspring/female/day and then gradually declines until reproduction ceases on day 12. Their reproductive lifespan can be divided into two halves, days 1–5 and days 6–10. We compared the effects of the drugs naproxen, fludarabine and hydralazine on reproduction in the second half of the reproductive lifespan. The rationale is that if these drugs improve the health of females in older age classes, we might observe higher reproductive rates. The average number of offspring produced by control females in the second half of their reproductive lifespan was 1.08 per day. In contrast, females treated with 1 µM naproxen produced 1.4 offspring per day. Likewise, females treated with 10 µM fludarabine or 1 µM hydralazine produced 1.72 and 1.66 offspring per day, respectively. All three of these reproductive rates are significantly higher than control with P values of 0.027, <0.001, and 0.001, respectively.
The third endpoint that we used to estimate rotifer healthspan is mitochondria activity. Using the fluorochrome Mitotracker as a proxy for overall mitochondrial activity, we quantified the decline in fluorescence in older age classes. Preliminary experiments indicated that declines in mitotacker fluorescence can be seen at age 4 days, about 1/3 of the rotifer lifespan. B. manjavacas treated with 1 µM hydralazine or 10 µM fludarabine retained 49 % (P = 0.038) and 89 % (P = 0.002) greater mitochondria activity, respectively, than untreated controls (Fig. 5).
Discussion
The significance of these results is that they provide a proof of concept for coupling computation to experimentation to quickly identify new drug candidates with anti-aging potential. Exploring the pool of FDA approved drugs significantly shortens drug development cycles because the safety these compounds in humans is already established (Ashburn and Thor 2004). Drug repurposing also reduces the cost and associated risks typical of traditional drug discovery (Pantziarka et al. 2014). Most drugs bind to multiple targets (Zhou et al. 2015), opening possibilities that they have as yet undiscovered effects beyond their licensed targets. A sensible place to identify new targets for novel anti-aging therapies is in the physiological processes that underlie aging. Even if lifespan extension is not observed, certain drugs could improve healthspan, slowing age-dependent losses in mobility and vitality. Screening drugs for anti-aging effects using a rotifer bioassay is a powerful first step in identifying compounds worthy of follow-up in vertebrate models (Fig. 6).
The feasibility of structure-based drug repurposing has been demonstrated by Taylor et al. (2014). They identified druggable targets for the platelet collagen receptor, GPVI, which promotes pathological thrombus formation through inappropriate platelet aggregation. If small molecules could be found that modulate platelet formation with minimal side effects, the impact of cardiovascular disease may be mitigated. To find drugs that bind to GPVI, they used a computer model that docked a FDA-approved drug library into the GPVI collagen-binding site. They found that the drugs losartan and cinanserin inhibited GPVI-mediated platelet activation selectively, competitively, and dose-dependently. Even though losartan is licensed as an angiotensin II receptor antagonist drug used to treat high blood pressure, it also binds to GPVI receptor. Similarly, cinanserin is a 5-HT2A and 5-HT2C receptor antagonist, used to treat atypical pneumonia, but also binds to GPVI. The efficacy of these two drugs as antiplatelet agents needs further testing, but their potential for this new therapeutic use was rapidly identified by computer modeling followed by experimental testing on isolated human platelets.
Since many aging pathways have yet to be identified, it is sensible to screen a wide range of drugs for anti-aging effects. A variety of drug screens have been conducted using Caenorhabditis elegans (Collins et al. 2006), with varying degrees of success. Evason et al. (2005) screened 19 FDA-approved drugs and identified ethosuximide, an anticonvulsant in humans, which extended mean lifespan in wild-type C. elegans by 17 %. Ethosuximide and trimethadione exposure at 4 mg/mL also delayed functional declines in body movement and pharyngeal pumping. Kumar et al. (2016) screened 15 FDA-approved drugs for their effects on the lifespan of C. elegans and found that exposure to 2.4 mm of the antihypertensive drug captopril extended lifespan 23 %. In both of these studies, drugs with mechanisms of action thought to impact aging pathways were chosen for screening. Since many aging pathways remain unknown, this approach is unavoidably biased toward pathways already characterized. Kwok et al. (2006) described a method to conduct a screen of 14,100 small molecules for bioactivity and found that 308 produced morphologically recognizable phenotypes in C. elegans. Although this was an unbiased high-throughput screen, none of these phenotypes could be related to aging processes. The strength of using FINDSITEcomb to prioritize drugs for screening for anti-aging effects as we did in this study is that this algorithm provides a ranked list of small molecules to target proteins. If one screens the top 50 molecules, roughly 21 % will have binding affinities in the micromolar range or better (Srinivasan et al. 2014). Choosing drugs to screen based on their binding properties is more likely to produce novel hits than screening drugs based on their putative modes of action in known aging pathways.
Work by Johnston and Snell (2016) implicated three novel rotifer genes in the determination of lifespan and healthspan. These observations motivated us to screen a library of FDA drugs for compounds that are predicted to bind to proteins produced by these genes. Our intent was to determine whether some of these drugs could be repurposed for anti-aging therapy. Predicted binding partners were experimentally assessed for anti-aging efficacy to identify new drug targets. For example, the transient receptor potential cation channel gene family (TRP) is a non-selective calcium permeant cation channel involved in osmotic, mechano- and thermosensitivity (Virens et al. 2004). It regulates intracellular Ca2+ levels, maintenance of functional intercellular barriers, expression of chemokines and cytokines related to proinflammatory pathways in adipocytes, and production of IL-8 in mice (Kottgen et al. 2008). Although TRPs play several vital roles in regulating cell metabolism, they have not been implicated in aging. Likewise, ribosomal protein S6 kinase alpha-1 (S6 K) is a family of protein kinases involved with signal transduction and the regulation of several cellular pathways (Frödin and Gammeltoft 1999). In mice their putative function is to mediate the activation of several mitogenic and stress-induced transcription factors. These in turn mediate cell proliferation, survival, and differentiation by modulating mTOR signaling and repressing pro-apoptotic function of BAD and DAPK1 proteins. mTOR is a well known aging pathway, but the role of S6 K in its regulation is poorly understood. Proteins coded by forkhead box C genes are transcriptional activators that regulate a variety of developmental pathways in humans, including organ growth, artery morphogenesis and blood vessel remodeling (van der Horst and Burgering 2007). These proteins also affect insulin-signaling pathways, but again their connection to aging is not well understood. The significance of this work is that these three genes have been identified as potential targets for anti-aging drug therapy and several candidate drugs have been experimentally identified that are capable of extending lifespan or healthspan in a rotifer animal model.
The FINDSITEcomb algorithm identified several binding partners for the TRP, S6 K and FhBC proteins and focused our attention on particular drugs. Naproxen is a nonsteroidal anti-inflammatory drug (NSAID) for relieving pain, fever, swelling, and stiffness. It is a nonselective COX inhibitor, reversibly inhibiting both the COX-1 and COX-2 enzymes (Hinz et al. 2008). Naproxen also may have anti-viral activity against influenza by blocking the RNA-binding groove of viral nucleoprotein, thus preventing the formation of ribonucleoprotein complexes (Lejal et al. 2013). Although we do not know whether these same mechanisms of action apply to rotifers, continuous treatment with 1 µM naproxen extended rotifer lifespan by 14 %. Fludarabine is a chemotherapy drug used to treat hematological malignancies like chronic lymphocytic leukemia (Rai et al. 2000). It is a purine analog, that inhibits DNA synthesis by interfering with ribonucleotide reductase and DNA polymerase. Exposure of rotifers to 10 µM fludarabine significantly improved reproduction and mitochondrial function in older age classes, these two endpoints serving as overall vitality and cellular senescence proxies, respectively. Hydralazine is an antihypertensive drug that relaxes smooth muscle, causing vasodilation in arteries and arterioles (Candelaria et al. 2010). This promotes a decrease in peripheral resistance and lowers blood pressure. It also has therapeutic value as a DNA methyltransferase inhibitor. Treatment of rotifers with 1 µM hydralazine slowed the loss of mobility (swimming speed) with aging. There has been no prior indication that any of these three drugs might be useful for anti-aging therapy. Our work identified these three drugs as potentially useful for anti-aging therapy from thousands of FDA approved compounds. Follow up studies of the efficacy of these drugs in slowing aging in vertebrate models is clearly warranted. But taken as a whole, these studies demonstrate the utility of FINDSITEcomb as a prioritization tool to identify repurposed FDA approved drugs (as well as new chemical entities) that might show promising anti-aging properties. When combined with a screen in a model animal such as rotifers promising leads can be rapidly identified which can then be subject to subsequent testing in vertebrate animal models.
Acknowledgments
We are grateful for the support of the National Institute of Aging, Grant R01 AG037960-02. We also express our appreciation for the technical assistance provided by Patricia Chang and Amelia Matthews. The computational aspects of this work were partially supported by NIH Grant R35GM118039 and a gift from the Mann family.
References
- Armanios M, de Cabo R, Mannick J, Partridge L, van Deursen J, Villeda S. Translational strategies in aging and age-related disease. Nat Med. 2015;21:1395–1399. doi: 10.1038/nm.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, et al. The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. FINDSITE: a threading-based method for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci USA. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelaria M, Herrera A, Labardini J, González-Fierro A, Trejo-Becerril C, Taja-Chayeb L, Pérez-Cárdenas E, Cruz-Hernández E, Arias-Bofill D, Vidal S, Cervera E, Dueñas-Gonzalez A. Hydralazine and magnesium valproate as epigenetic treatment for myelodysplastic syndrome. Preliminary results of a phase-II trial. Ann Hematol. 2010;90(4):379–387. doi: 10.1007/s00277-010-1090-2. [DOI] [PubMed] [Google Scholar]
- Cavasotto C, Orry A. Ligand docking and structure-based virtual screening in drug discovery. Curr Top Med Chem. 2007;7:1006–1014. doi: 10.2174/156802607780906753. [DOI] [PubMed] [Google Scholar]
- Cheng F, et al. Predicting of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8(5):e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Evason K, Kornfeld K. Pharmacology of delayed aging and extended lifespan of Caenorhabditis elegans. Exp Gerontol. 2006;41:1032–1039. doi: 10.1016/j.exger.2006.06.038. [DOI] [PubMed] [Google Scholar]
- de Cabo R, Carmona-Gutierrez D, Bernier M, Hall MN, Madeo F. The search for antiaging interventions: from elixirs to fasting regimens. Cell. 2014;157:1515–1526. doi: 10.1016/j.cell.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evason K, Huang C, Yamben I, Covey DF, Kornfeld K. Anticonvulsant medications extend worm life-span. Science. 2005;307:258–262. doi: 10.1126/science.1105299. [DOI] [PubMed] [Google Scholar]
- Frödin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase in signal transduction. Mol Cell Endocrinol. 1999;151(1-2):65–77. doi: 10.1016/S0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucl Acid Res. 2012;40(D1):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb A, et al. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:414–496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble KE, Welch DBM. Lifespan extension by caloric restriction is determined by type and level of food reduction and by reproductive mode in Brachionus manjavacas (Rotifera) J Gerontol Ser A. 2012;68(4):349–358. doi: 10.1093/gerona/gls170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Berg CJJr., editor. Culture of marine invertebrates. Stroudsburg, PA: Hutchinson Ross; 1983. [Google Scholar]
- Hinz B, Cheremina O, Besz D, Zlotnick S, Brune K. Impact of naproxen sodium at over-the-counter doses on cyclooxygenase isoforms in human volunteers. Int J Clin Pharmacol Ther. 2008;46(4):180–186. doi: 10.5414/CPP46180. [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RK, Snell TW. Moderately lower temperatures greatly extend the lifespan of Brachionus manjavacas (Rotifera): thermodynamics or gene regulation? Exp Gerontol. 2016;78:12–22. doi: 10.1016/j.exger.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottgen M, Buchholz B, Garcia-Gonzalez MA, Kotsis F, Fu X, Doerken M, Boehlke C, Steffl D, Tauber R, Wegierski T, Nitschke R, Suzuki M, Kramer-Zucker A, Germino GG, Watnick T, Prenen J, Nilius B, Kuehn EW, Walz G. TRPP2 and TRPV4 form a polymodal sensory channel complex. J Cell Biol. 2008;182:437–447. doi: 10.1083/jcb.200805124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dietrich N, Kornfeld K. Angiotensin converting enzyme (ACE) inhibitor extends Caenorhabditis elegans lifespan. PLoS Genet. 2016;12(2):e1005866. doi: 10.1371/journal.pgen.1005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok TCY, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- Laarhoven TV, Marchioro E. Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS One. 2013;8(6):e66952. doi: 10.1371/journal.pone.0066952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Zhang Y. BSP-SLIM: a blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins. 2011;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejal N, Tarus B, Bouguyon E, Chenavas S, Bertho N, Delmas B, Ruigrok RW, Di Primo C, Slama-Schwok A. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother. 2013;57(5):2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S, Schonfeld JM. Therapeutic drug repurposing, repositioning and rescue. Part I—overview. Drug Discov World Winter. 2015;2015:54–62. [Google Scholar]
- Oo AKS, Kaneko G, Hirayama M, Kinoshita S, Watabe S. Identification of genes differentially expressed by calorie restriction in the rotifer (Brachionus plicatilis) J Comp Physiol B. 2010;180:105–116. doi: 10.1007/s00360-009-0389-6. [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Kaneko G, Kanagawa Y, Watabe S. Calorie restriction in the rotifer Brachionus plicatilis enhances hypoxia tolerance in association with the increased mRNA levels of glycolytic enzymes. Hydrobiologia. 2010;649:267–277. doi: 10.1007/s10750-010-0269-9. [DOI] [Google Scholar]
- Pantziarka P, Bouche G, Meheus L, Sukhatme V, Sukhatme VP, Vikas P. The repurposing drugs in oncology (ReDO) project. eCancer. 2014;8:442. doi: 10.3332/ecancer.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai KR, Peterson BL, Appelbaum FR, Kolitz J, Elias L, Shepherd L, Hines J, Threatte GA, Larson RA, Cheson BD, Schiffer CA. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N Engl J Med. 2000;343:1750–1757. doi: 10.1056/NEJM200012143432402. [DOI] [PubMed] [Google Scholar]
- Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR_3 threading algorithm. Proteins. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- Snare DJ, Fields AM, Snell TW, Kubanek J. Lifespan extension of rotifers by treatment with red algal extracts. Exp Gerontol. 2013;48:1420–1427. doi: 10.1016/j.exger.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW. Rotifers as models for the biology of aging. Int Rev Hydrobiol. 2014;99:84–95. doi: 10.1002/iroh.201301707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW, Janssen CR. Rotifers in ecotoxicology: a review. Hydrobiologia. 1995;313(314):231–247. doi: 10.1007/BF00025956. [DOI] [Google Scholar]
- Snell TW, Johnston RK. Glycerol extends the lifespan of Brachionus manjavacas (Rotifera) and protects against stressors. Exp Gerontol. 2014;57:47–56. doi: 10.1016/j.exger.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW, Fields AM, Johnston RK. Antioxidants can extend lifespan of Brachionus manjavacas (Rotifera), but only in a few combinations. Biogerontology. 2012;7:261–275. doi: 10.1007/s10522-012-9371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW, Johnston RK, Gribble KE, Mark Welch DB. Rotifers as experimental tools for investigating aging. Invertebr Reprod Dev. 2014 doi: 10.1080/07924259.2014.925516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell TW, Johnston RK, Rabeneck B, Zipperer C, Teat S. Joint Inhibition of TOR and JNK pathways interacts to extend the lifespan of Brachionus manjavacas (Rotifera) Exp Gerontol. 2014;52:55–69. doi: 10.1016/j.exger.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, et al. Experimental validation of FINDSITEcomb virtual ligand screening results for eight proteins yields novel nanomolar and picomolar binders. J Cheminform. 2014;6(1):16–29. doi: 10.1186/1758-2946-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L, Vasudevan SR, Jones CI, Gibbins JM, Churchill GC, Campbell RD, Coxson CH. Discovery of novel GPVI receptor antagonists by structure-based repurposing. PLoS One. 2014;9(6):e101209. doi: 10.1371/journal.pone.0101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8(6):440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- von Eichborn J, et al. PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res. 2011;39:1060–1066. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriens J, Watanabe H, Janssens A, Droogmans G, Voets T, Nilius B. Cell swelling, heat, and chemical agonists use distinct pathways for the activation of the cation channel TRPV4. Proc Natl Acad Sci USA. 2004;101:396–401. doi: 10.1073/pnas.0303329101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucl Acid Res. 2006;34:668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y, et al. Prediction of drug–target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24:i232–i240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y, et al. Drug–target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics. 2010;26(12):i246–i254. doi: 10.1093/bioinformatics/btq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on genomic scale. Proc Natl Acad Sci USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. FINDSITEX: a structure-based, small molecule virtual screening approach with application to all identified human GPCRs. Mol Pharm. 2012;9(6):1775–1784. doi: 10.1021/mp3000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. FINDSITEcomb: a threading/structure-based, proteomic-scale virtual ligand screening approach. J Chem Inf Model. 2013;53(1):230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. SPARKS 2 and SP3 servers in CASP 6. Proteins. 2005;7:152–156. doi: 10.1002/prot.20732. [DOI] [PubMed] [Google Scholar]
- Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug–protein interactions and side effects for the human proteome. Sci Rep. 2015;5:11090. doi: 10.1038/srep11090. [DOI] [PMC free article] [PubMed] [Google Scholar]