Abstract
Reduced representation bisulfite sequencing (RRBS) provides an efficient method for measuring DNA methylation at single base resolution in regions of high CpG density. This technique has been extensively tested on the HiSeq2500, which uses a 4-colour detection method, however it is unclear if the method will also work on the NextSeq500 platform, which employs a 2-colour detection system. We created an RRBS library and sequenced it on both the HiSeq2500 and NextSeq500, and found no significant difference in the base composition of reads derived from either machine. Moreover, the methylation calls made from the data of each instrument were highly concordant, with methylation patterns across the genome appearing as expected. Therefore, RRBS can be sequenced on the Nextseq500 with comparable quality to that of the HiSeq2500. All sequencing data are deposited in the GEO database under accession number GSE87097.
Keywords: HiSeq2500, NextSeq500, RRBS, Bisulfite sequencing
Specifications
| Organism/cell line/tissue | Mus musculus domesticus (C57BL/6 J) × Mus musculus castaneus F1 E9.5 whole embryo |
|---|---|
| Sex | Female |
| Sequencer or array type | HiSeq2500 and NextSeq500 |
| Data format | Raw (fastq) and calculated methylation calls |
| Experimental factors | N/A |
| Experimental features | Reduced Representation Bisulfite Sequencing (RRBS) of a single library made with the Ovation® RRBS Methyl-Seq System (NuGEN), sequenced on both the HiSeq2500 and NextSeq500. |
| Consent | All animal experiments were carried out in accordance with the Walter and Eliza Hall Institute of Medical Research (WEHI) Animal Ethics Committee guidelines under approval number AEC 2014.026. |
| Sample source location | Melbourne, Australia |
1. Direct link to deposited data
All sequencing data are deposited in GEO, accession number GSE87097.
2. Introduction
Addition of a methyl group to the 5th carbon of cytosine residues (5mC) is a frequent epigenetic modification that very strongly anti-correlates with transcription and is critical for mammalian development [1]. 5mC occurs predominantly in the CpG dinucleotide context and its abundance is tightly linked to the density of this motif. Compared to other dinucleotide combinations, CpGs are globally depleted throughout the genome, and instead occur in clusters known as CpG islands (CGIs). CGIs are typically located in the promoter regions of genes and, while CpG sparse genomic regions are highly methylated, CGIs are frequently hypomethylated [2]. High levels of CGI methylation at gene promoters correlates very strongly with repression of that gene, suggesting that CGIs are regulatory elements for repression by 5mC.
Treatment of DNA with bisulfite results in the deamination of unmethylated cytosine to uracil, leaving methylated cytosines unaffected [3], [4], and thereby providing a methyl-dependent DNA mutation that can be easily detected by next generation sequencing methods; known as bisulfite sequencing. In order to reduce the costs involved with bisulfite sequencing a restriction enzyme is often employed along with selection of the resulting small fragments of DNA, to enrich for fragments with high CpG content, thereby greatly reducing the percentage of the genome that needs to be sequenced to analyse CGI methylation. This technique is known as Reduced Representation Bisulfite Sequencing (RRBS) [5].
Illumina sequencing machines, such as the HiSeq2500, use a 4-colour system for detection of the four DNA bases, however the NextSeq500 uses a 2-colour system for detection of the bases (one colour for C, the other for T, both colours for A and no colour for G). Due to the loss of unmethylated cytosines, RRBS libraries have a highly skewed base composition and, while they have been effectively sequenced on the HiSeq2500 with 4-colour technology [6], there have been no reports of efficient RRBS using 2-colour technology. Here we sequence the same RRBS library on both the HiSeq2500 and NextSeq500 and compare the results, finding a high concordance between the data obtained from either instrument.
3. Experimental design, materials and methods
3.1. DNA extraction
Wild type Mus musculus domesticus C57BL/6J dams were mated to wild type Mus musculus castaneus sires. Pregnant females were sacrificed at E9.5 by CO2 asphyxiation and the embryos dissected from deciduae in PBS. Embryo samples were snap frozen in buffer RLT plus (Qiagen) and DNA was later extracted with the AllPrep DNA/RNA Mini Kit (Qiagen) using the manufacturer's instructions. DNA was treated with RNase A then purified through the DNA Clean and Concentrator column (Zymo).
3.2. RRBS library preparation and sequencing
The RRBS library was made at The Australian Genome Research Facility (AGRF) from 100 ng of purified DNA using the Ovation RRBS Methyl-Seq System (NuGEN), according to the manufacturers recommendations, which includes use of the Qiagen Epitect kit for bisulfite conversion. Once made, the same library was sequenced on both the HiSeq2500 at AGRF and the NextSeq500 at WEHI. For the HiSeq2500, we performed 100 bp paired-end sequencing. For the NextSeq500, 75 bp paired-end sequencing was performed. For sequencing on each instrument read 1 (R1) was obtained with the MetSeq Primer 1 (NuGEN), read 2 (R2) with the Illumina Reverse Read primer (Illumina) and the index with the Illumina Index Read primer (Illumina). We obtained 95 M reads from the HiSeq2500 and 65 M from the NextSeq500.
3.3. Data processing
Sequencing reads obtained from both instruments were processed the same way. Quality control (QC) of reads was performed using FastQC [7] and found to be comparable between instruments. Trimming of adapters and low quality base calls was performed with trim_galore [8] in paired-end mode. Reads derived from the NextSeq500 required the additional trimming of the 6 bp N6 unique molecular identifier (UMI) sequence from the 5′ end of R2, discussed in more detail below. Trimmed reads were then filtered for true RRBS reads (which contain an MspI cut site at the 5′ end of both read 1 and read 2) with trimRRBSdiversityAdaptCustomers.py (NuGEN), which is recommended in the Ovation RRBS Methyl-Seq System. To eliminate potential mapping bias between the alleles, sequencing reads were aligned to a bisulfite converted custom version of the mouse genome (mm10), where SNPs that exist between Mus musculus domesticus and Mus musculus castaneus have been N-masked, as described previously [9], using Bismark [10]. Methylation bias in the reads was determined using the bismark_methylation_extractor using the—mbias_only option, and methylation calls were made, excluding the 13 5′ bases of reads due to mbias, using bismark_methylation_extractor.
To assess the nucleotide composition of reads derived from the two sequencing platforms, all read files were converted to uniform FASTA format using seqtk (https://github.com/lh3/seqtk), and parsed to CGAT [11] to calculate per read nucleotide composition. Statistical summaries were produced using datamash (https://www.gnu.org/software/datamash/). Boxplots were produced in R [12]. Profiling of methylation over genes was performed using Seqmonk [13] by quantifying the genome using windows of 2 kb sliding by 1 kb, with only informative windows containing more than 100 reads considered. Analysis of methylation at CGIs was also performed in Seqmonk with only CGIs containing greater than 100 reads considered.
4. Results
We created an RRBS library using the Ovation RRBS Methyl-Seq System, from one E9.5 mouse embryo. The library was then run on both the HiSeq2500 and NextSeq500 platforms, to obtain paired-end reads for each. The quality of the reads was high from each platform, however we found that very few reads (2.45%) obtained from the NextSeq500 passed our quality control, despite an appropriate number of HiSeq2500 reads passing. Reads were predominantly being lost following the RRBS specific diversity trimming quality control step, which ensures that reads begin with the expected MspI restriction enzyme cut site. For this process we used a script supplied for use with the Ovation RRBS Methyl-Seq System, but it can also be performed using the—rrbs option with trim_galore [8], however we found this made no improvement to the mapping rate. The failure to map was later determined to be due to the addition of the N6 UMI sequence to the 5′ end of R2, meaning the MspI site appeared further in to the read than expected, and in turn caused the read to fail QC. The addition of the N6 sequence to R2, which should have appeared on the index read, was a peculiarity of the demultiplexing process. Further information on the N6 sequence can be obtained from the Ovation RRBS Methyl-Seq System protocol [14]. The N6 sequence can be trimmed post sequencing with little effect on data quality, which is what we did here, however a better approach is to mask the UMI sequence at the time of demultiplexing if it is not required. Once the 5′ most 6 bases were trimmed from R2 of the NextSeq500 dataset, we re-ran the data processing pipeline and obtained a mapping rate that was comparable to that obtained from the HiSeq2500.
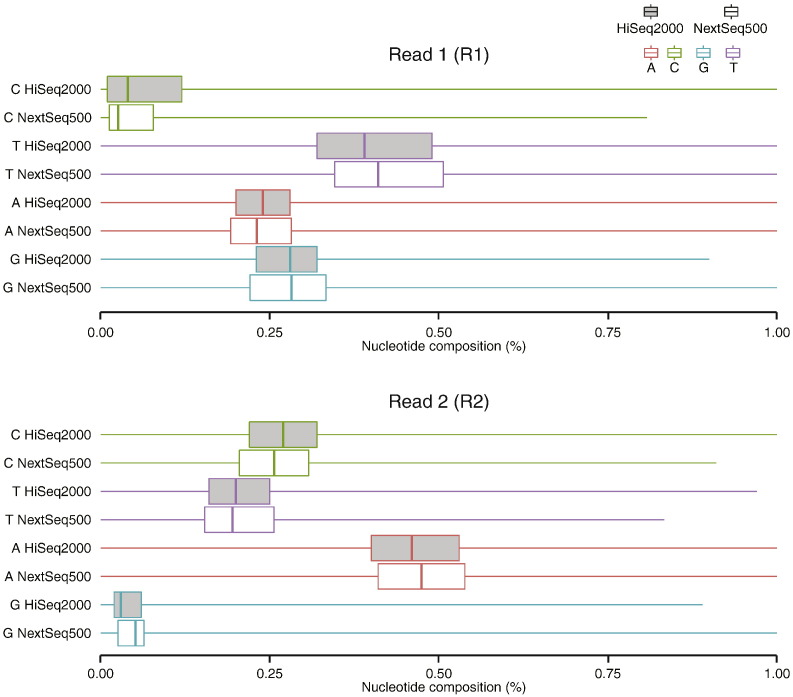
RRBS libraries have a highly skewed base composition due to the bisulfite conversion of unmethylated cytosine to uracil, which is then read as a thymine by the sequencer. As data from the HiSeq2500, which uses a 4-colour system to detect DNA residues, was performing well, we wondered whether the 2-colour system employed by the NextSeq500 was also capable of accurately calling bases from a library with highly skewed base composition. To assess this possibility we looked at the frequency of each base within both R1 and R2, obtained from the NextSeq500 and the HiSeq2500 in the unprocessed fastq files (Fig. 1). As expected, our RRBS libraries had a highly skewed base composition, with decreased C′s and increased T's in R1 and decreased G's and increased A's in R2. Interestingly, we found no substantial difference in base composition between the platforms, suggesting that both the 2- and 4-colour systems for detecting DNA residues are capable of accurately reading libraries with skewed base composition.
Fig. 1.
Box plots showing the percentage of each base within all reads of the raw fastq files produced on either the NextSeq500 or HiSeq2500. Separate plots are shown for R1 and R2. The box indicates the 25th to 75th percentile range and median and the whiskers indicate the maximum and minimum values.
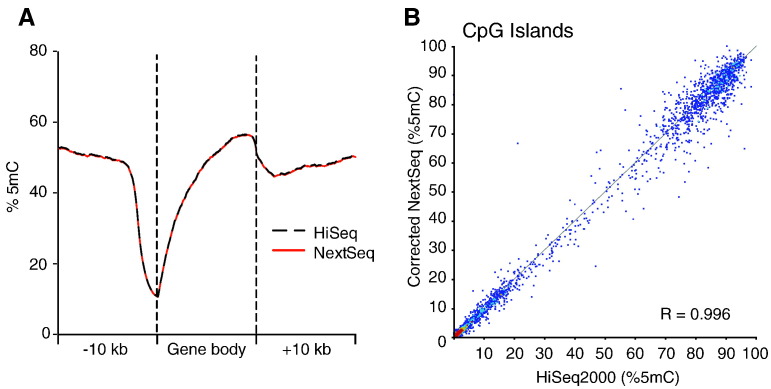
Given that the read quality from each instrument appeared comparable, we next looked at the methylation profiles obtained from the HiSeq2500 and the NextSeq500 data. Calls obtained by both methods showed the expected pattern of high 5mC at gene bodies and intergenic regions, and low 5mC at promoters (Fig. 2A). Encouragingly, the NextSeq500 5mC profile matched the HiSeq2500 profile very tightly. We next quantified 5mC over CGIs and found that the methylation calls obtained from the HiSeq2500 data were highly correlated to those of the NextSeq500 data (R = 0.996, Fig. 2B). These results suggest that RRBS data obtained from both the HiSeq2500 and the NextSeq500 are equivalent and reliable.
Fig. 2.
A) Quantification of 5mC across the average of all gene bodies ± 10 kb for data obtained from the HiSeq2500 and NextSeq500. B) Scatter plot showing the correlation between HiSeq2500 and NextSeq500 methylation calls at CpG islands. The Pearson Correlation Coefficient (R) is also shown.
5. Conclusion
We conclude that the 2-colour detection system employed by the NextSeq500 is capable of producing reads of a similarly high quality to that of the HiSeq2500 from RRBS libraries, despite the highly skewed base composition of bisulfite treated DNA. Moreover the methylation calls produced are highly concordant between instruments.
Acknowledgements
We thank The Dyson Bequest and The DHB Foundation for philanthropic funding to MEB. This work was supported by grants from the Australian National Health and Medical Research Council (GNT1045936) to MEB. MEB and MER are RD Wright Biomedical Research fellows of the Australian National Health and Medical Research Council (GNT1110206 and GNT1104924 respectively). We thank the AGRF for producing the RRBS library and sequencing on the HiSeq2500 platform, and Dr. Stephen Wilcox (WEHI) for sequencing on the Nextseq500. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian National Health and Medical Research Council Research Institute Infrastructure Support Scheme.
Contributor Information
Marnie E. Blewitt, Email: blewitt@wehi.edu.au.
Andrew Keniry, Email: keniry@wehi.edu.au.
References
- 1.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 2.Bird A., Taggart M., Frommer M., Miller O.J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 3.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark S.J., Harrison J., Paul C.L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle P., Clement K., Gu H., Smith Z.D., Ziller M., Fostel J.L., Holmes L., Meldrim J., Kelley F., Gnirke A., Meissner A. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Available from.
- 8.Krueger F. trim_galore: a wrapper tool around Cutadapt and FastQC. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (Available at:)
- 9.Keniry A., Gearing L.J., Jansz N., Liu J., Holik A.Z., Hickey P.F., Kinkel S.A., Moore D.L., Breslin K., Chen K. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenetics Chromatin. 2016;9:16. doi: 10.1186/s13072-016-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims D., Ilott N.E., Sansom S.N., Sudbery I.M., Johnson J.S., Fawcett K.A., Berlanga-Taylor A.J., Luna-Valero S., Ponting C.P., Heger A. CGAT: computational genomics analysis toolkit. Bioinformatics. 2014;30:1290–1291. doi: 10.1093/bioinformatics/btt756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Core Team R: A Language and Environment for Statistical Computing. 2016. http://www.R-project.org/: (R Foundation for Statistical Computing, Vienna, Austria)
- 13.Andrews S. Seqmonk: a tool to visualise and analyse high throughput mapped sequence data. http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ (Available at:)
- 14.NuGEN http://www.nugen.com/sites/default/files/M01394v2_UG_Ovation_RRBS_Methyl-Seq.pdf