Abstract
Disruption of the establishment of left-right (L-R) asymmetry leads to situs anomalies ranging from situs inversus totalis (SIT) to situs ambiguus (heterotaxy). The genetic causes of laterality defects in humans are highly heterogeneous. Via whole-exome sequencing (WES), we identified homozygous mutations in PKD1L1 from three affected individuals in two unrelated families. PKD1L1 encodes a polycystin-1-like protein and its loss of function is known to cause laterality defects in mouse and medaka fish models. Family 1 had one fetus and one deceased child with heterotaxy and complex congenital heart malformations. WES identified a homozygous splicing mutation, c.6473+2_6473+3delTG, which disrupts the invariant splice donor site in intron 42, in both affected individuals. In the second family, a homozygous c.5072G>C (p.Cys1691Ser) missense mutation was detected in an individual with SIT and congenital heart disease. The p.Cys1691Ser substitution affects a highly conserved cysteine residue and is predicted by molecular modeling to disrupt a disulfide bridge essential for the proper folding of the G protein-coupled receptor proteolytic site (GPS) motif. Damaging effects associated with substitutions of this conserved cysteine residue in the GPS motif have also been reported in other genes, namely GPR56, BAI3, and PKD1 in human and lat-1 in C. elegans, further supporting the likely pathogenicity of p.Cys1691Ser in PKD1L1. The identification of bi-allelic PKD1L1 mutations recapitulates previous findings regarding phenotypic consequences of loss of function of the orthologous genes in mice and medaka fish and further expands our understanding of genetic contributions to laterality defects in humans.
Main Text
Although vertebrates may appear symmetrical when viewed externally, there is marked left-right (L-R) asymmetry in the positioning of almost all the internal organs. The normal arrangement of internal organs is known as situs solitus. Failure to establish normal organ asymmetry along the left-right axis can result in situs inversus totalis (SIT) or situs ambiguous (also known as heterotaxy). SIT is the complete mirror image of situs solitus whereas heterotaxy refers to discordant organ arrangement with reversal of at least one organ relative to the others.1 In vertebrates, the establishment of L-R asymmetry occurrs during early embryonic development and involves complex signaling transduction cascades and nodal cilia.2, 3, 4 Whereas the activation of NODAL in the left lateral plate mesoderm results in normal position of internal organs (situs solitus), the expression of NODAL in the right side of the embryo leads to SIT.4 In SIT, organ concordance is preserved and congenital organ malformation is rare. In contrast, heterotaxy (randomization of organ positioning and organ discordance) arises from bilateral or absent NODAL activation.5 Approximately 80% of individuals with heterotaxy have complex congenital heart disease (CHD).6 Overall, situs anomalies have a prevalence of 1 in 10,000 live births and account for approximately 3% of complex CHD.5, 6, 7
The genetic causes of laterality defects in humans are highly heterogeneous. SIT and heterotaxy with complex CHD can be caused by defects in a number of genes including ZIC3 (MIM: 300265), CFC1 (MIM: 605194), MMP21 (MIM: 608416), GDF1 (MIM: 602880), NODAL (MIM: 601265), LEFTY2 (MIM: 601877), ACVR2B (MIM: 602730), and FOXH1 (MIM: 603621); those genes encode components or modifiers of nodal signaling, a signal transduction pathway that plays an important role in mesoderm and endoderm formation and subsequent organization of L-R axial structures.2, 3, 5, 8, 9, 10, 11, 12, 13, 14 Additionally, rare variants in other genes including CCDC11 (MIM: 614759), CRELD1 (MIM: 607170), MED13L (MIM: 608771), SHROOM3 (MIM: 6045700), MEGF8 (MIM: 604267), NKX2-5 (MIM: 600584), NPHP4 (MIM: 607215), and PKD2 (MIM: 173910) have also been associated with L-R patterning defects.5, 15, 16, 17, 18, 19, 20 Furthermore, approximately 50% of individuals with primary ciliary dyskinesia (PCD) have SIT and approximately 6% have heterotaxy.21 Despite these previous discoveries, our understanding of the genetic basis of this group of developmental disorders remains relatively limited.
Here, we describe the identification of homozygous mutations in the polycystic kidney disease 1 like 1 gene (PKD1L1 [MIM: 609721]) by WES and Sanger sequencing. The gene has previously been characterized as an important determinant of L-R axis in mouse and medaka fish.22, 23, 24, 25 Three subjects affected with laterality defects from two unrelated families were studied. Subject 1 from family 1 was ascertained from approximately 6,900 consecutive clinical exome case subjects referred to the Exome Laboratory at the Baylor Genetics from November 2011 to December 2015. The similarly affected sibling (subject 2) in family 1 was later recruited and studied by Sanger sequencing. Subject 3 from family 2 was enrolled in the Baylor Hopkins Center for Mendelian Genomics (BHCMG) research study. Written informed consent for all subjects was obtained in accordance with protocols approved by the appropriate human subject ethics committees at Baylor College of Medicine.
WES was completed in the Exome Laboratory at Baylor Genetics (subject 1, father, and mother) and at Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) (subject 3) (Table S1). Sequencing and data analyses were conducted as previously described, targeting approximately 20,000 genes, including the coding and the untranslated region (UTR) exons.26, 27 Samples were also analyzed by cSNP array (Illumina HumanExome-12 v1 array) for quality control assessment of exome data, as well as for detecting large copy-number variants (CNVs) and regions of absence of heterozygosity (AOH).28
Family 1 is a non-consanguineous family of Northern European ancestry with one fetus (subject 1, II-3) and one deceased child (subject 2, II-2), both affected with heterotaxy and complex CHD, and one healthy child (II-1) (Table 1, Figure 1). The family was referred for prenatal genetic assessment after an ultrasound examination performed at 21 weeks and 6 days of gestation revealed complex congenital malformations of the fetus. The fetus presented with situs ambiguus with atrial situs solitus, unbalanced atrioventricular septal defect with left ventricular hypoplasia, double outlet right ventricle with malposed great arteries, right-sided stomach, left-sided liver, and right-sided spleen (Table 1). The diagnosis was confirmed after birth. The subject expired at 3 weeks of age due to complications of CHD. The family history indicated that a previous child (subject 2) was also diagnosed with heterotaxy. He had atrial situs ambiguus, unbalanced atrioventricular septal defect with left ventricular hypoplasia, pulmonary atresia, right-sided stomach, left-sided liver, and right-sided spleen (Table 1, Figure 1). This child died in the neonatal period due to complications of severe CHD.
Table 1.
Summary of Phenotypic and Molecular Data
| Family 1; Subject 1 | Family 1; Subject 2 | Family 2; Subject 3 | |
|---|---|---|---|
| Age of diagnosis | prenatal | prenatal | 3 weeks |
| Sex | male | male | female |
| Cardiac situs | ND | levocardia | dextrocardia |
| Atrial situs | solitus: normal systemic and pulmonary venous return | ambiguus: IVC, hepatics to L-sided atrium; R-SVC, PV to R-sided atrium | inversus: normal systemic and pulmonary venous connections with mirror image situs |
| AV valves and ventricles | U-AVSD; LV hypoplasia | U-AVSD; LV hypoplasia | CCTGA; LV hypoplasia; VSD |
| Great arteries | DORV with malposed great arteries | PA with MAPCAS; R-aortic arch | PA |
| Abdominal situs | inversus: R-sided stomach, L-sided liver, R-sided spleen | inversus: R-sided stomach, L-sided liver, R-sided spleen | inversus, R-sided stomach, L-sided liver, R-sided spleen |
| Nucleotide change (Hom) | c.6473+2_6473+3delTG | c.5072G>C | |
| Effect | splice donor site at exon 42/intron 42 boundary | p.Cys1691Ser | |
| Coordinate (Hg19) | chr7: 47,870,812–47,870,813 | chr7: 47,886,558 | |
| ExAC (Het) (African) | 3/10,394 | ND | |
| ExAC (Het) (Latinos) | 1/11,560 | ND | |
| ExAC (Het) (European) | 41/66,146 | ND | |
| In silico predictions | elimination of the splice donor site (see Figure S1) | SIFT (damaging); MutationTaster (disease-causing); PolyPhen-2 (damaging) | |
Abbreviations are as follows: CCTGA, congenitally corrected transposition of the great arteries (ventricular inversion); DORV, double outlet right ventricle; ExAC, Exome Aggregation Consortium; Hom, homozygous; Het, heterozygous; IVC, inferior vena cava; L, left; LV, left ventricle; MAPCAS, major aortopulmonary collaterals; ND, no data; PA, pulmonary atresia; PV, pulmonary veins; R, right; SVC, superior vena cava; U-AVSD, unbalanced atrioventricular septal defect; VSD, ventricular septal defect.
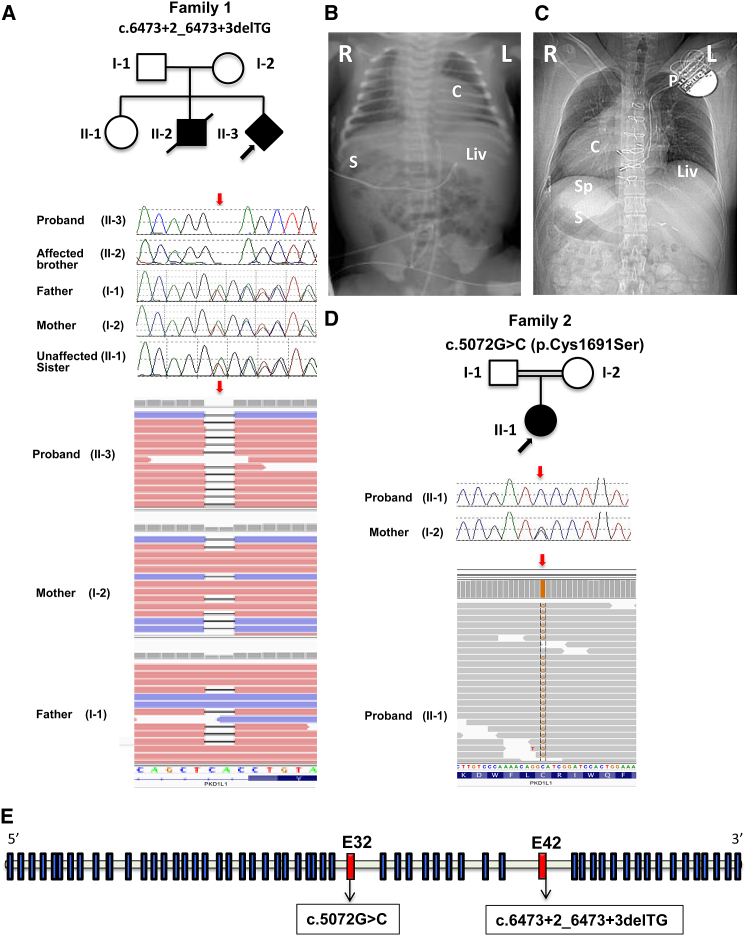
Figure 1.
Segregation of PKD1L1 Mutations with Laterality Defects in Families 1 and 2
(A) NGS reads in Integrative Genomic Viewer (IGV) and Sanger chromatograms show the c.6473+2_6473+3delTG deletion in family 1: proband (II-3, subject 1), affected brother (II-2, subject 2), unaffected sister (II-1), father (I-1), and mother (I-2). The c.6473+2_6473+3delTG deletion is homozygous in the proband and the affected brother and heterozygous in the parents. The mutation is indicated by red arrows.
(B) Plain radiograph of the chest and abdomen of the affected brother in family 1 (subject 2) at birth shows situs ambiguous with levocardia and leftward apex (white “C”), left-sided liver (Liv), right-sided stomach (S); R = right, L = left.
(C) CT localizer radiograph of the proband in family 2 (subject 3) demonstrates situs inversus totalis with dextrocardia and rightward apex (C), left-sided liver (Liv), right-sided stomach (S), and right-sided spleen (Sp). Median sternotomy wires are seen as well as a subcutaneous pacemaker (P) with intravascular leads terminating in the left-sided right atrium and subpulmonary ventricle; R = right, L = left.
(D) NGS reads in IGV and Sanger chromatograms show the c.5072G>C (p.Cys1691Ser) missense change in family 2: proband (II-1, subject 3) and mother (I-2). The c.5072G>C (p.Cys1691Ser) change is homozygous in the proband and heterozygous in the mother. The mutation is indicated by red arrows.
(E) Schematic representation of genomic structure of human PKD1L1, where solid blue rectangles indicate exons and the horizontal bars introns. The exons 32 and 42 are colored in solid red and the mutations with their relative position are shown.
Subject 3 (II-1 from family 2) is a 46-year-old female born to consanguineous parents of Iranian origin. She was diagnosed in the first weeks of life with SIT and congenital heart disease including congenitally corrected transposition of the great arteries (ventricular inversion) with a small left ventricle, pulmonary atresia, and ventricular septal defect (Table 1, Figure 1). She underwent placement of a left ventricle to pulmonary artery conduit and ventricular septal defect closure. She has paroxysmal atrial flutter and a dual-chamber pacemaker (Figure 1).
Trio exome-sequencing analysis was performed on DNA extracted from cultured fetal amniocytes from subject 1 and blood samples from the parents in family 1. WES analysis did not detect any pathogenic variants in genes previously associated with laterality defects.5, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 However, WES identified a homozygous c.6473+2_6373+3delTG variant affecting the invariant splice donor site at the exon 42/intron 42 junction of PKD1L1 (GenBank: NM_138295.3) in subject 1; WES also showed that the variant is present in the heterozygous state in both parents, consistent with autosomal-recessive Mendelian expectations (Figure 1, Table 1). The next-generation sequencing (NGS) data revealed the presence of this mutation in 97 out of 97 (97:0) reads in the proband, and 45:63 and 30:58 of the mutation versus the wild-type reads in the mother and the father, respectively (Figure 1, Table 1). In silico programs predicted that the c.6473+2_6473+3delTG variant abolishes the canonical splice donor site in intron 42 (Figure S1), as is usually expected for changes affecting invariant splice sites at the exon-intron boundaries. The minor allele frequency (MAF) of this variant is 0.06% (41/66,146 alleles) in the European non-Finnish population, 0.028% (3/10,394 alleles) in Africans, and 0.008% (1/11,560) in Latinos in the database from the Exome Aggregation Consortium (ExAC; n = 61,486 exomes; accessed February 2016) (Table 1). No homozygotes have been reported for this variant in ExAC. Subsequent Sanger sequencing confirmed the homozygous mutation in the proband, validated the segregation of alleles in both the parents, and showed that the deceased brother was homozygous and the healthy sister was heterozygous for this change (Figure 1). The detection of this variant in homozygous configuration in the proband and the affected brother shows that the homozygous genotype segregates with the congenital malformations in the family and supports a causative role of this variant in the affected individuals.
For family 2, proband WES analysis performed on the DNA sample from subject 3 detected a homozygous c.5072G>C (p.Cys1691Ser) missense mutation in the G protein-coupled receptor proteolytic site (GPS) motif of PKD1L1 (GenBank: NM_138295.3) (Figure 1, Table 1). The NGS data revealed the presence of this mutation in 110 out of 110 (110:0) reads in the proband; no wild-type reads were identified (Figure 1). This variant is located in exon 32 and affects a cysteine residue that is highly conserved during evolution (Figure 2). The p.Cys1691Ser substitution is predicted to be deleterious by in silico prediction tools (Table 1). This variant is absent from ExAC (n = 61,486 exomes; accessed February 2016). Sanger sequencing confirmed that the variant is homozygous in the proband and also showed that the unaffected mother is heterozygous for the change. The father was not available for study (Figure 1). However, it is likely that the father is heterozygous for the variant since this change is located within a copy-neutral AOH region of approximately 16 Mb (chr7: 40,538,810–56,534,467) in subject 3.
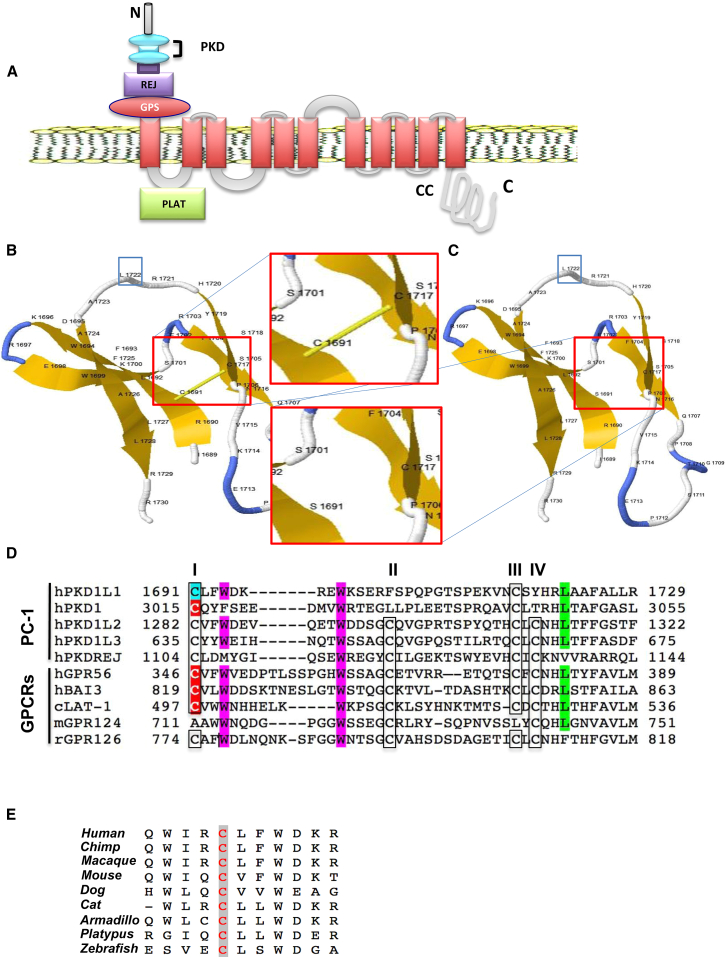
Figure 2.
Alignment and Molecular Modeling of the Impact of p.Cys1691Ser Variant on the GPS Motif
(A) Schematic representation of the human PKD1L1 structural domains. PKD1L1 has two Ig-like PKD domains, a REJ domain and a GPS motif in the N-terminal extracellular region, an LH2/Plat domain in the first intracellular loop, and a coiled-coil domain at the C-terminal (CC).
(B and C) 3D models of the GPS motif in PDK1L1 in the wild-type (B) and p.Cys1691Ser mutant (C) by Phyre2 based on the GPS motif in the GAIN and HormR domains of human brain angiogenesis inhibitor 3 (BAI3). The five β strands are shown with numbered amino acids corresponding to the coding sequence of PKD1L1. Cys to Ser substitution at the position 1691 (magnification boxed in red) eliminates the highly conserved disulfide bridge between cysteine residues 1691 and 1717 represented by the yellow solid bar. The conserved Leu1722 residue at the putative cleavage site is boxed in blue.
(D) Alignment of the GPS motif in PKD1L1, other members of PC-1 family, and G protein couple receptors (GPCRs). The GPS motif contains four conserved cysteines arranged in a specific fashion (C-x2-W-x6-16- W-x4-C-x-10-22-C-x-C) just before the first transmembrane domain. The conserved cysteine (C) residues are boxed; the conserved tryptophan (W) residues are highlighted in magenta. The first cysteine residues are numbered with respect to the protein sequence. PKD1L1 and PKD1 do not have the second and fourth (II and IV) cysteine residues in the conserved positions. The putative cleavage site leucine residues that are located in the turn between the last two β strands of the GPS motif are highlighted in green in the alignment. Highlighted in cyan is the mutant site of PKD1L1 GPS in subject 3 (II-1 from family 2) of this study. Highlighted in red are the previously reported mutant sites in the GPS motif in hPKD1, hBAI3, and hGPR56 and cLAT-1. Abbreviations are as follows: h, human; r, rat; m, mouse; c, C. elegans.
(E) ClustalW multiple alignment analysis shows high level evolutionary conservation of the human Cys1691 residue in PKD1L1 across multiple species (highlighted in gray).
Human PKD1L1, which belongs to the polycystin cation channel family 1 (PC-1), is a paralog of PKD1 (polycystin-1 [MIM: 601313]) and consists of 58 exons spanning 187 kb of genomic DNA. The 2,849-amino acid protein contains in the N-terminal extracellular region two immunoglobulin (Ig)-like polycystic kidney disease (PKD) domains, a small receptor for egg jelly (REJ) domain, a GPS motif, 11 putative transmembrane segments, a polycystin-1, lipoxygenase, alpha-toxin (PLAT) domain, and a C-terminal intracellular coiled-coil (CC) (Figure 2); all the domains can also be found in PKD1. RNA dot-blot analysis of multiple human tissues has demonstrated PKD1L1 expression in adult and fetal heart and in testis.29
Several independent animal models provide evidence for the involvement of PKD1L1 in the L-R patterning in vertebrates. First, in a Pkd1l1−/− mouse model, approximately one-third of the homozygous mutant mice showed situs inversus (SI) without other phenotype or lesions suggestive of underlying primary ciliary dyskinesia (PCD) such as impaired mucociliary clearance or reproductive defects.23 The Pkd1l1−/− mice showed markedly reduced viability during development or early postnatal period. The author hypothesized that undiagnosed CHD associated with laterality defects might be the potential cause, although the hearts and great vessels of these mice were not examined. Second, the Pkd1l1rks/rks mutant mice carrying a homozygous p.Asp411Gly substitution, which disrupts a highly conserved PKD domain of PKD1L1, also displayed L-R laterality defects including randomized stomach situs, randomized heart apex, and right lung isomerism. Mutant Pkd1l1rks/rks mice showed embryonic lethality by 15.5 dpc.24 In addition to internal organ discordance, the authors also showed that Pkd1l1rks/rks mutants failed to activate the asymmetric gene expression at the node or in the lateral plate.24 Furthermore, Field and collaborators showed that Pkd1l1 was exclusively expressed in the node with a spatiotemporal pattern corresponding to the establishment of L-R asymmetry. Ciliary morphology and motility of nodal cells in Pkd1l1 and Pkd2 mutants were both normal, suggesting a functional role for the two genes downstream of nodal flow.24 Additionally, they demonstrated that PKD1L1 interacts through the C-terminal coiled coil (CC) domain with the C-terminal region of PKD2 in the node and hypothesized that PKD1L1 and PKD2 form complexes within cilia that sense nodal flow and are involved in left-sided activation of the Nodal signaling cascade.24 In a third study,22 Grimes and collaborators demonstrated that (1) PKD1L1 is required to restrict Nodal activation to the left side downstream of nodal flow; (2) PKD1L1 can mediate fluid flow-induced Ca+ signal response in vitro, suggesting an analogous role in vivo in the elicitation of nodal flow response; and (3) importantly, the complete loss-of-function Pkd1l1tm1/tm1 mutant mice presented with randomized laterality of heart and stomach at E13.5. In addition, 43% of the mutants surviving until adulthood exhibited reversed situs. Finally, keeping with the role of Pkd1l1 in laterality defects in mice, a previous study showed that medaka fish mutant abecobe (abc is the homolog of PKD1L1 in humans and Pkd1l1 in mouse) displays L-R developmental patterning defects, which included randomized direction of heart looping and randomized liver and gallbladder positioning.25
Collective data based on animal models, the severity of the variant, and the co-segregation of disease and genotype suggested that the c.6473+2_6473+3delTG PKD1L1 mutation in subjects 1 and 2 from family 1 is the most parsimonious molecular explanation for these individuals’ phenotypic features. In order to further determine the effect of the c.5072G>C (p.Cys1691Ser) variant in family 2, a structural prediction of the extracellular region containing the amino acid change in PKD1L1 was determined using the in silico protein modeling bioinformatics tools Phyre2 and I-TASSER.30, 31, 32, 33 The crystal structures of the GPS motif in the GPCR-autoproteolysis inducing (GAIN) and hormone receptor (HormR) domains of human brain angiogenesis inhibitor 3 (BAI3) and the GAIN and HormR domains of rat CIRL/latrophilin 1 (CL1) were identified by the prediction programs as top-ranked structural analogs and were used as templates for subsequent analyses (Figures 2, S2, and S3). The GPS motif serves as an autoproteolytic cleavage site located just before the first trans-membrane domain; this motif is shared by some G protein-coupled receptors (GPCRs) and the polycystin-1 (PC-1) family members (Figure 2).34 Previous studies showed that the GPS motif is structurally characterized by five β strands that are integrated into the approximately 320-residue GAIN domain.34, 35 The GAIN domain and the ∼40-residue GPS motif form a module that is evolutionarily conserved from slime molds to mammals; the module is involved in receptor activation, signaling activity, and intracellular trafficking functions of the GPCRs and PC-1 proteins.34, 35 Three key elements appear to be fundamental for the proper folding and activity of the GPS motif: (1) disulfide bonds between neighboring β strands that enable the formation of the sharp loop between the last two strands containing the conserved leucine at the cleavage site (Figure 2); (2) hydrophobic interaction between the last β strands; and (3) the trapping of the highly conserved leucine within a conserved hydrophobic pocket.34 Of note, all the members of the PC-1 family share these structural features with the GPCR proteins, although PKD1 and PKD1L1 have only one disulfide bond at the penultimate β strand (Figure 2). GPS autoproteolytic cleavage is essential for protein maturation, intracellular trafficking, and other normal functions of the GPS motif-containing proteins.34, 35, 36, 37, 38 Mutations that alter the GPS structural integrity without affecting autoproteolysis can also abrogate the protein function.35 Our analysis revealed that p.Cys1691Ser in PKD1L1 abolishes the essential disulfide bridge between the first and the penultimate β strand (Figures 2 and S3) and is therefore predicted to destabilize the tridimensional structure of the GPS motif with potential detrimental effects on protein function and signaling activity as recently shown.35 Moreover, the predicted alteration of the loop containing the conserved critical leucine residue Leu1722 (Figure 2) may in turn affect GPS cleavage and the trafficking of the protein to the cytoplasm.34
Intriguingly, changes affecting the same conserved residue in the GPS motif corresponding to Cys1691 in PKD1L1 (residue highlighted in cyan in Figure 2) have been reported in at least three other human genes—GPR56 (MIM: 604110), BAI3 (MIM: 602684), and PKD1 (MIM: 601313)—as well as latrophilin-1 (lat-1) in C. elegans, as identified by Domain Mapping of Disease Mutation (DMDM) analysis and literature review (summarized below and in Table 2).35, 36, 37, 38, 39, 40 In one study, a homozygous p.Cys346Ser substitution in GPR56 corresponding to the identical substitution detected in PKD1L1 has been reported in two subjects with bilateral frontoparietal polymicrogyria (BFPP [MIM: 606854]) (Figure 2); the molecular studies have demonstrated that this change produces a GPR56 protein with dramatically impaired cleavage that fails to traffic beyond the endoplasmic reticulum, with consequent loss of function of the protein.36, 37 Another study showed that the conserved Cys819 in the putative tumor-suppressor gene BAI3 (Figure 2) is mutated to Tyr in cancer cells and the change is predicted to affect the activity of the protein.39 Furthermore, it has been shown that in vitro site-specific mutagenesis of Cys to Ser at the position 3015 in PKD1 (Figure 2) affects the cleavage of the protein at the GPS motif.38 Lastly, a recent study in C. elegans showed that the corresponding substitution p.Cys497Ser in the LAT-1 protein (Figure 2) affects the GPS structural integrity by disrupting the normal disulfide bond patterns and results in an inactive receptor.35 Based on the predictions from molecular modeling that shows that p.Cys1691Ser in PKD1L1 abolishes an essential disulfide bridge, together with the evidence of the damaging effects associated with changes of the conserved cysteine in other genes, we conclude that the homozygous PKD1L1 c.5072G>C (p.Cys1691Ser) mutation is most likely the cause of the laterality defects in subject 3 from family 2.
Table 2.
Reported Mutations Affecting the Conserved Cysteine in the GPS Motif
| Gene | Residue Position | Variant | Associated Phenotype/Effect |
|---|---|---|---|
| PKD1L1 | c.5072G>C | p.Cys1691Ser | SIT (subject 3, family 2 of this study) |
| GPR56 | c.1036T>A | p.Cys346Ser | bilateral frontoparietal polymicrogyria (BFPP)36, 37 |
| BAI3 | c.2904G>A | p.Cys819Tyr | mutated in non-small cell lung cancer39 |
| PKD1 | c.9043T>A | p.Cys3015Ser | affects PKD1 cleavage activity in vitro38 |
| lat-1 | c.1489T>A | p.Cys497Ser | disrupts disulfide bond and affects LAT-1 receptor activity in C. elegans35 |
PDKD1L1 interacts with PKD2,22, 24, 25, 41 which is also involved in L-R patterning in mouse, medaka fish, and zebrafish models.42, 43 Additionally, Pkd1l1−/− and Pkd2−/− mouse mutants phenocopy strongly. However, in humans, PKD2 heterozygous mutations cause polycystic kidney disease 2 (MIM: 613095), a form of ciliopathy arising from abnormalities of the renal primary cilium. Heterozygous mutations in PKD2 have also been reported in individuals with laterality defects but the occurrence is rare.17 Further studies are needed in order to elucidate the role of PKD1L1 and PKD2 in the establishment of left-right body asymmetry in humans and to determine whether additional PKD1L1 partners might be potentially involved in human laterality disorders.
In summary, we identified two homozygous mutations in PKD1L1 in three individuals who presented with laterality defects. Our findings recapitulate those in mouse and medaka fish and further expand the genetic heterogeneity of laterality defects in humans.
Conflicts of Interest
J.R.L. is a paid consultant for Regeneron Pharmaceuticals, holds stock ownership in 23andMe and Lasergen, Inc., is on the Scientific Advisory Board of Baylor Genetics, and is a co-inventor on United States and European patents related to molecular diagnostics. J.W.B. is currently employed by Illumina, Inc. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered at the Baylor Genetics.
Acknowledgments
We thank the families for their participation and collaboration. We thank Irene Miloslavskaya, Anh Dang from Baylor Genetics, and Theodore Chiang from Human Genome Center for their technical support. The study was supported in part by the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) grant no. U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (J.R.L.). T.H. is supported by the NIH/NIGMS T32 GM07526 Medical Genetics Research Fellowship Program.
Published: September 8, 2016
Footnotes
Supplemental Data include three figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.011.
Accession Numbers
The ClinVar accession numbers for the DNA variant data reported in this paper are SCV000280028 and SCV000280029.
Web Resources
1000 Genomes, http://www.1000genomes.org
Domain Mapping of Disease Mutation, http://bioinf.umbc.edu/DMDM/generatelogo.php?accession=smart00303
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Phyre2, http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Raya A., Izpisúa Belmonte J.C. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat. Rev. Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X., Sasaki H., Lowe L., Hogan B.L., Kuehn M.R. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 3.Collignon J., Varlet I., Robertson E.J. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhu L., Belmont J.W., Ware S.M. Genetics of human heterotaxias. Eur. J. Hum. Genet. 2006;14:17–25. doi: 10.1038/sj.ejhg.5201506. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland M.J., Ware S.M. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- 6.Peeters H., Devriendt K. Human laterality disorders. Eur. J. Med. Genet. 2006;49:349–362. doi: 10.1016/j.ejmg.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Kathiriya I.S., Srivastava D. Left-right asymmetry and cardiac looping: implications for cardiac development and congenital heart disease. Am. J. Med. Genet. 2000;97:271–279. doi: 10.1002/1096-8628(200024)97:4<271::aid-ajmg1277>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 8.Perles Z., Moon S., Ta-Shma A., Yaacov B., Francescatto L., Edvardson S., Rein A.J., Elpeleg O., Katsanis N. A human laterality disorder caused by a homozygous deleterious mutation in MMP21. J. Med. Genet. 2015;52:840–847. doi: 10.1136/jmedgenet-2015-103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guimier A., Gabriel G.C., Bajolle F., Tsang M., Liu H., Noll A., Schwartz M., El Malti R., Smith L.D., Klena N.T. MMP21 is mutated in human heterotaxy and is required for normal left-right asymmetry in vertebrates. Nat. Genet. 2015;47:1260–1263. doi: 10.1038/ng.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiraishi I., Ichikawa H. Human heterotaxy syndrome – from molecular genetics to clinical features, management, and prognosis –. Circ. J. 2012;76:2066–2075. doi: 10.1253/circj.cj-12-0957. [DOI] [PubMed] [Google Scholar]
- 11.Hamada H., Meno C., Watanabe D., Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- 12.Ware S.M., Harutyunyan K.G., Belmont J.W. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev. Dyn. 2006;235:1631–1637. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- 13.Kaasinen E., Aittomäki K., Eronen M., Vahteristo P., Karhu A., Mecklin J.P., Kajantie E., Aaltonen L.A., Lehtonen R. Recessively inherited right atrial isomerism caused by mutations in growth/differentiation factor 1 (GDF1) Hum. Mol. Genet. 2010;19:2747–2753. doi: 10.1093/hmg/ddq164. [DOI] [PubMed] [Google Scholar]
- 14.Mohapatra B., Casey B., Li H., Ho-Dawson T., Smith L., Fernbach S.D., Molinari L., Niesh S.R., Jefferies J.L., Craigen W.J. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum. Mol. Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tariq M., Belmont J.W., Lalani S., Smolarek T., Ware S.M. SHROOM3 is a novel candidate for heterotaxy identified by whole exome sequencing. Genome Biol. 2011;12:R91. doi: 10.1186/gb-2011-12-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen T.A., Troelsen Kde.L., Larsen L.A. Of mice and men: molecular genetics of congenital heart disease. Cell. Mol. Life Sci. 2014;71:1327–1352. doi: 10.1007/s00018-013-1430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bataille S., Demoulin N., Devuyst O., Audrézet M.P., Dahan K., Godin M., Fontès M., Pirson Y., Burtey S. Association of PKD2 (polycystin 2) mutations with left-right laterality defects. Am. J. Kidney Dis. 2011;58:456–460. doi: 10.1053/j.ajkd.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Izumi K., Noon S., Wilkens A., Krantz I.D. NKX2.5 mutation identification on exome sequencing in a patient with heterotaxy. Eur. J. Med. Genet. 2014;57:558–561. doi: 10.1016/j.ejmg.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Narasimhan V., Hjeij R., Vij S., Loges N.T., Wallmeier J., Koerner-Rettberg C., Werner C., Thamilselvam S.K., Boey A., Choksi S.P. Mutations in CCDC11, which encodes a coiled-coil containing ciliary protein, causes situs inversus due to dysmotility of monocilia in the left-right organizer. Hum. Mutat. 2015;36:307–318. doi: 10.1002/humu.22738. [DOI] [PubMed] [Google Scholar]
- 20.Twigg S.R., Lloyd D., Jenkins D., Elçioglu N.E., Cooper C.D., Al-Sannaa N., Annagür A., Gillessen-Kaesbach G., Hüning I., Knight S.J. Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization. Am. J. Hum. Genet. 2012;91:897–905. doi: 10.1016/j.ajhg.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles M.R., Daniels L.A., Davis S.D., Zariwala M.A., Leigh M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimes D.T., Keynton J.L., Buenavista M.T., Jin X., Patel S.H., Kyosuke S., Vibert J., Williams D.J., Hamada H., Hussain R. Genetic analysis reveals a hierarchy of interactions between polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genet. 2016;12:e1006070. doi: 10.1371/journal.pgen.1006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel P., Read R., Hansen G.M., Freay L.C., Zambrowicz B.P., Sands A.T. Situs inversus in Dpcd/Poll-/-, Nme7-/-, and Pkd1l1-/- mice. Vet. Pathol. 2010;47:120–131. doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- 24.Field S., Riley K.L., Grimes D.T., Hilton H., Simon M., Powles-Glover N., Siggers P., Bogani D., Greenfield A., Norris D.P. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development. 2011;138:1131–1142. doi: 10.1242/dev.058149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamura K., Kobayashi D., Uehara Y., Koshida S., Iijima N., Kudo A., Yokoyama T., Takeda H. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development. 2011;138:1121–1129. doi: 10.1242/dev.058271. [DOI] [PubMed] [Google Scholar]
- 26.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalani S.R., Liu P., Rosenfeld J.A., Watkin L.B., Chiang T., Leduc M.S., Zhu W., Ding Y., Pan S., Vetrini F. Recurrent muscle weakness with rhabdomyolysis, metabolic crises, and cardiac arrhythmia due to bi-allelic TANGO2 mutations. Am. J. Hum. Genet. 2016;98:347–357. doi: 10.1016/j.ajhg.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuasa T., Venugopal B., Weremowicz S., Morton C.C., Guo L., Zhou J. The sequence, expression, and chromosomal localization of a novel polycystic kidney disease 1-like gene, PKD1L1, in human. Genomics. 2002;79:376–386. doi: 10.1006/geno.2002.6719. [DOI] [PubMed] [Google Scholar]
- 30.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J., Zhang Y. Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics. 2015;52:1–15. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araç D., Boucard A.A., Bolliger M.F., Nguyen J., Soltis S.M., Südhof T.C., Brunger A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prömel S., Frickenhaus M., Hughes S., Mestek L., Staunton D., Woollard A., Vakonakis I., Schöneberg T., Schnabel R., Russ A.P., Langenhan T. The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep. 2012;2:321–331. doi: 10.1016/j.celrep.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Z., Tietjen I., Bu L., Liu-Yesucevitz L., Gaur S.K., Walsh C.A., Piao X. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum. Mol. Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 37.Piao X., Hill R.S., Bodell A., Chang B.S., Basel-Vanagaite L., Straussberg R., Dobyns W.B., Qasrawi B., Winter R.M., Innes A.M. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 38.Qian F., Boletta A., Bhunia A.K., Xu H., Liu L., Ahrabi A.K., Watnick T.J., Zhou F., Germino G.G. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc. Natl. Acad. Sci. USA. 2002;99:16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M., Yue P., Haverty P.M., Bourgon R., Zheng J. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 40.Peterson T.A., Adadey A., Santana-Cruz I., Sun Y., Winder A., Kann M.G. DMDM: domain mapping of disease mutations. Bioinformatics. 2010;26:2458–2459. doi: 10.1093/bioinformatics/btq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian F., Germino F.J., Cai Y., Zhang X., Somlo S., Germino G.G. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat. Genet. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 42.Pennekamp P., Karcher C., Fischer A., Schweickert A., Skryabin B., Horst J., Blum M., Dworniczak B. The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 2002;12:938–943. doi: 10.1016/s0960-9822(02)00869-2. [DOI] [PubMed] [Google Scholar]
- 43.Bisgrove B.W., Snarr B.S., Emrazian A., Yost H.J. Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer’s vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 2005;287:274–288. doi: 10.1016/j.ydbio.2005.08.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.