Abstract
Arthrogryposis multiplex congenita (AMC) is a developmental condition characterized by multiple joint contractures resulting from reduced or absent fetal movements. Through linkage analysis, homozygosity mapping, and exome sequencing in four unrelated families affected by lethal AMC, we identified biallelic mutations in GLDN in the affected individuals. GLDN encodes gliomedin, a secreted cell adhesion molecule involved in the formation of the nodes of Ranvier. Transmission electron microscopy of the sciatic nerve from one of the affected individuals showed a marked lengthening defect of the nodes. The GLDN mutations found in the affected individuals abolish the cell surface localization of gliomedin and its interaction with its axonal partner, neurofascin-186 (NF186), in a cell-based assay. The axoglial contact between gliomedin and NF186 is essential for the initial clustering of Na+ channels at developing nodes. These results indicate a major role of gliomedin in node formation and the development of the peripheral nervous system in humans. These data indicate that mutations of GLDN or CNTNAP1 (MIM: 616286), encoding essential components of the nodes of Ranvier and paranodes, respectively, lead to inherited nodopathies, a distinct disease entity among peripheral neuropathies.
Main Text
Arthrogryposis multiplex congenita (AMC) has an overall incidence of 1 in 3,000.1, 2 Some non-genetic factors, such as extrinsic limitation of fetal movements or maternal autoimmune myasthenia, can cause AMC. Non-syndromic or isolated AMC are the direct consequence of reduced fetal movements, which could lead, in addition to AMC, to pterygia, pulmonary hypoplasia, diaphragmatic defect, or cleft palate. Non-syndromic AMCs are genetically heterogeneous and include a large spectrum of diseases that arise as a result of mutations in genes encoding components required for the formation or the function of neuromuscular junctions and skeletal muscle, survival of motor neurons, or myelination of peripheral nerves.3 However, many affected individuals remain without a genetic diagnosis, suggesting the involvement of other pathogenic mechanisms. Here, we have further explored genetic alterations in a group of families affected by genetically undiagnosed AMC.
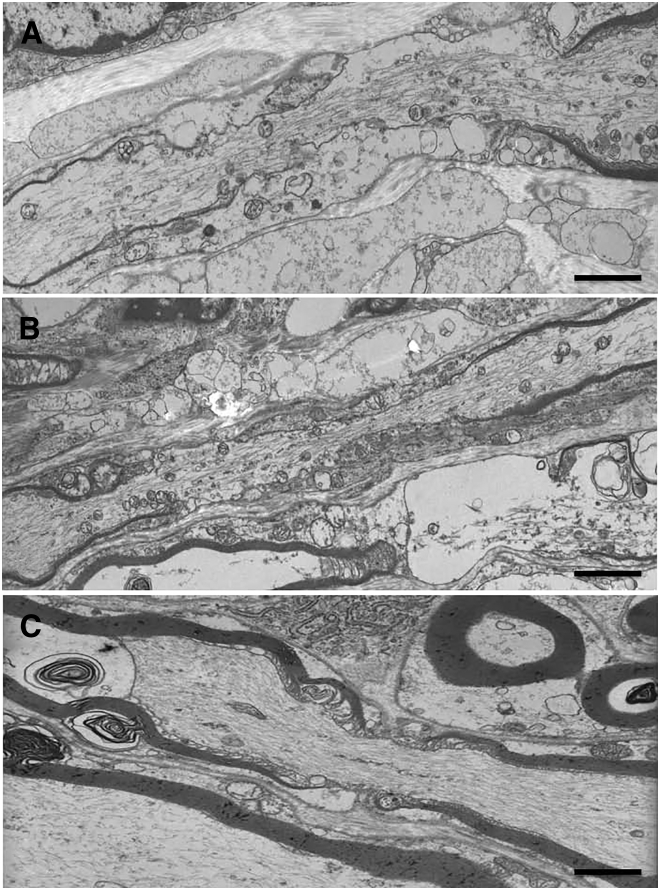
The parents of all affected individuals provided written informed consent for genetic analyses of their children or fetuses and themselves in accordance with the ethical standards of our institutional review boards. In family 1, two affected fetuses were born to unrelated healthy parents. According to ultrasound examination, the first fetus displayed akinesia associated with polyhydramnios from 32 weeks of gestation (w.g.). Fetal death occurred at 33 w.g. Postmortem examination revealed marked extension of lower limbs associated with extension contractures of wrists and pulmonary hypoplasia (Figure S1). For the second fetus, ultrasound examination revealed an identical phenotype and the pregnancy was terminated at 33 w.g. at the request of the parents. Karyotype, SMN1 (MIM: 600354), and DMPK (MIM: 605377) analyses were normal. Histological examination of the spinal cord and skeletal muscle was normal. Genetic mapping of disease loci was carried out with an Affymetrix GeneChip Human Mapping 250K SNP microarray. Multipoint linkage analysis of SNP data was performed with the Alohomora4 and Merlin softwares.5 We performed whole-exome sequencing (WES) on DNA of the index case of family 1 by using the Exome Capture Agilent SureSelect XT V5 kit for library preparation and exome enrichment as previously described.6 Sequencing was performed on a Genome Analyzer IIx Illumina instrument in paired-end mode with a read length of 2 × 100 bp. The median coverage of the WES was 65. Reads were aligned to the human reference genome sequence (UCSC Genome Browser hg19, NCBI build 37.3) via the Burrows-Wheeler Aligner.7 Variants were selected with SAMtools8 then annotated with Annovar softwares.9 Variants in coding regions (including non-synonymous and nonsense mutations), intron-exon junctions, or short coding insertions or deletions were selected when the minor-allele frequency (MAF) was less than 0.0030. Prediction of pathogenicity of non-synonymous or splice mutations was performed with PolyPhen-210 and the Human Splicing Finder, respectively.11 By combining genetic mapping of disease loci under the hypothesis of autosomal-recessive inheritance and WES, we identified compound heterozygous mutations in GLDN in the affected individuals of family 1 (Figure 1A). A heterozygous frameshift deletion (c.758delC [GenBank: NM_181789.2]) resulting in premature stop codon p.Pro253Leufs∗51 was identified. This variant was not annotated in the 1000 Genomes or ExAC Browser databases. The second mutation was a missense mutation (c.1423G>C) predicted to be pathogenic with a high score (p.Ala475Pro; PolyPhen-2 score of 1). This mutation is annotated in dbSNP 146 (rs764239923) and has a very low MAF in the ExAC database (0.00000824). Both mutations were confirmed by Sanger sequencing using primers flanking the mutations (Table S1) in both affected fetuses and were allelic (Figure 1A and Figure S2). Transmission electron microscopy of the sciatic nerve from the index case revealed a reduced number of myelinated fibers. In a comparable field of view (5,000 μm2), 47 axons were myelinated as compared to 77 in two age-matched control fetuses (data not shown). The nodal length was significantly increased in the affected individual (5.96 ± 2.33 μm) as compared to nodal length in the control fetus (1.12 ± 0.98 μm) (Student’s t test = 0.0004, n = 7 in each group, Figure 2).
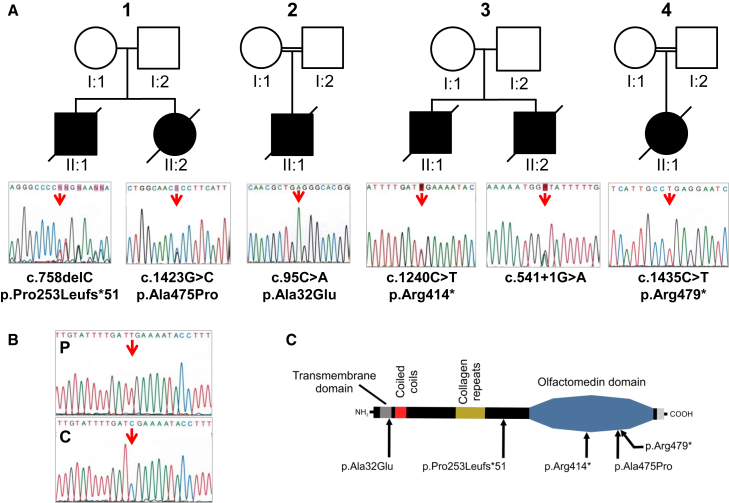
Figure 1.
Mutations Identified in GLDN in AMC-Affected Families
(A) Pedigrees for families 1, 2, 3, and 4 are shown. Arrows indicate mutant nucleotide positions. The affected individuals carry either compound heterozygous (families 1 and 3) or homozygous (families 2 and 4) mutations. The nucleotide and amino acid changes are indicated with respect to the GenBank: NM_181789.2 and Genbank: NP_861454.2 reference sequences, respectively. Open symbols, unaffected; filled symbols, affected.
(B) Sequencing of RT-PCR product from the RNA of the family 3 affected individual, II:1, (P) and a control individual (C) is shown. RT-PCR analysis was performed from RNA after reverse transcription with random hexamers. Long-range PCR amplification was carried out with Advantage GC Genomic LA polymerase (Clontech), then Sanger sequencing was performed from the RT-PCR products with primers GLDN-Ex1Fq and GLDN-Ex10Rq (Table S1). Sequence analysis revealed the nonsense mutation, only indicating that the allelic splice donor mutation leads to either a retention of large intronic sequence or instability of the corresponding transcript.
(C) Location of mutations relative to the predicted protein domains of gliomedin is shown.
Figure 2.
Transmission Electron Microscopy Analysis of Nerve in the Family 1 Affected Individual II:1 Carrying Deleterious GLDN Mutations
Tissue samples were fixed in a 2% glutaraldehyde fixative solution, post-fixed with osmium tetroxide, and embedded in resin epoxy. Semi-thin sections were stained with toluidine blue. Ultra-thin sections were contrasted with uranyl acetate and lead citrate and examined under a PHILIPS CM10 transmission electron microscope. Longitudinal sciatic nerve sections of the affected fetus, displaying increased length of the node of Ranvier (A and B), are shown, as is that of an age-matched (33 w.g.) control indiviudal (C). Scale bars represent 3 μm.
Targeted exome sequencing (TES) was performed on the DNA samples of additional affected individuals with arthrogryposis and/or reduced fetal mobility of unknown origin (n = 106). TES was performed with the Agilent SureSelectXT Custom Kit (targeting 500 Kb, including all known AMC-associated genes and candidate genes, including GLDN) for library preparation and exome enrichment. Sequencing was performed on an Illumina MiSeq System with paired-end 150 bp reads and with the MiSeq Reagent Micro Kit v.2, according to Illumina’s protocol. The average of the median coverage was 92. Variants were selected according to the same criteria as those used for WES data. TES allowed for the identification of three additional families carrying biallelic mutations in GLDN. In family 2, the affected individual was born to consanguineous parents at 30 w.g. with marked polyhydramnios, intrauterine growth retardation, and AMC characterized by flexion contractures of the elbows, extension of the knees, camptodactyly, and retrognathia. Ultrasound examinations performed at 14 and 23 w.g. were normal. The male neonate died at day 1. By combining homozygosity by descent using 250K SNP microarray and TES, we identified a homozygous missense mutation in GLDN (c.95C>A; p.Ala32Glu), predicted to be pathogenic with a PolyPhen-2 score of 0.8, in the affected individual (Figure 1A). This mutation is annotated in dbSNP146 (rs779432560) and has a very low MAF in the ExAC database (0.00002829). The mutation was confirmed by Sanger sequencing, and both parents were heterozygous carriers (Figure 1A, Figure S2, and Table S1). In family 3, two affected fetuses were born to unrelated healthy parents. The first affected individual died at day 1 and presented with akinesia, polyhydramnios, AMC involving the fingers, wrists, thumbs, and knees, and pulmonary hypoplasia. Ultrasound examinations during pregnancy were normal at 22 w.g., then detected a marked polyhydramnios at 28 w.g. with bilateral flexion of fingers. Karyotype, SMN1, and DMPK analyses were normal. Pathological examination of the brain and spinal cord was normal. For the second male fetus, ultrasound examinations at 12 and 22 w.g. were normal. At 31 w.g., ultrasound examination revealed a similar abnormal phenotype characterized by polyhydramnios, marked extension of lower limbs associated with flexion of fingers and marked reduced mobility. On the basis of ultrasound findings, the parents elected termination of pregnancy, which was achieved at 31 w.g. Postmortem examination confirmed AMC associated with microretrognathia and pulmonary hypoplasia. TES was performed on the DNA sample of the first affected individual and identified compound heterozygous mutations in GLDN: a heterozygous nonsense mutation (c.1240C>T; p.Arg414∗) annotated in dbSNP146 (rs539703340) that has a very low MAF in both the 1000 Genomes (0.0002) and ExAC databases (0.000008) and a splice donor mutation (c.541+1G>A) not annotated in the same databases. Both mutations were confirmed by Sanger sequencing in both fetuses and were allelic (Figure 1A, Figure S2, and Table S1). Sequencing of RT-PCR products of GLDN from muscle RNA of the affected individual II:1 revealed the nonsense mutation only, indicating that the allelic splice donor mutation leads to either a retention of large intronic sequence or instability of the corresponding transcript (Figure 1B and Table S1). In family 4, the affected individual was born to consanguineous parents. Ultrasound examinations performed at 13 and 21 w.g. were normal. At 27 w.g., ultrasound examination revealed reduced fetal mobility with marked polyhydramnios, and at 29 w.g., fetal immobility. Karyotype, SMN1, and DMPK analyses were normal. Pregnancy was terminated at 30 w.g. at the request of the parents. Postmortem examination revealed distal arthrogryposis of the hands, bilateral clubfoot, and pulmonary hypoplasia. Pathological examination of the brain and spinal cord was normal. In the affected individual, TES identified a homozygous nonsense mutation in GLDN (c.1435C>T; p.Arg479∗). The mutation is annotated in dbSNP 146 (rs368085516) and has a very low MAF in the ExAC database (0.00001647). The mutation was confirmed by Sanger sequencing, and both parents were heterozygous carriers (Figure 1A, Figure S2, and Table S1).
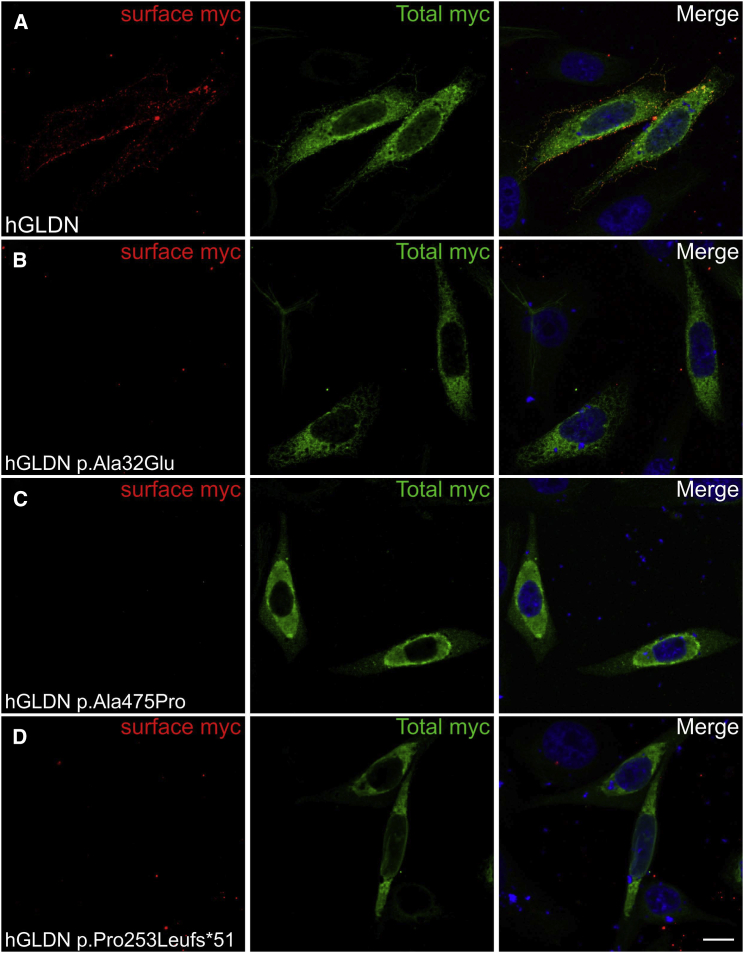
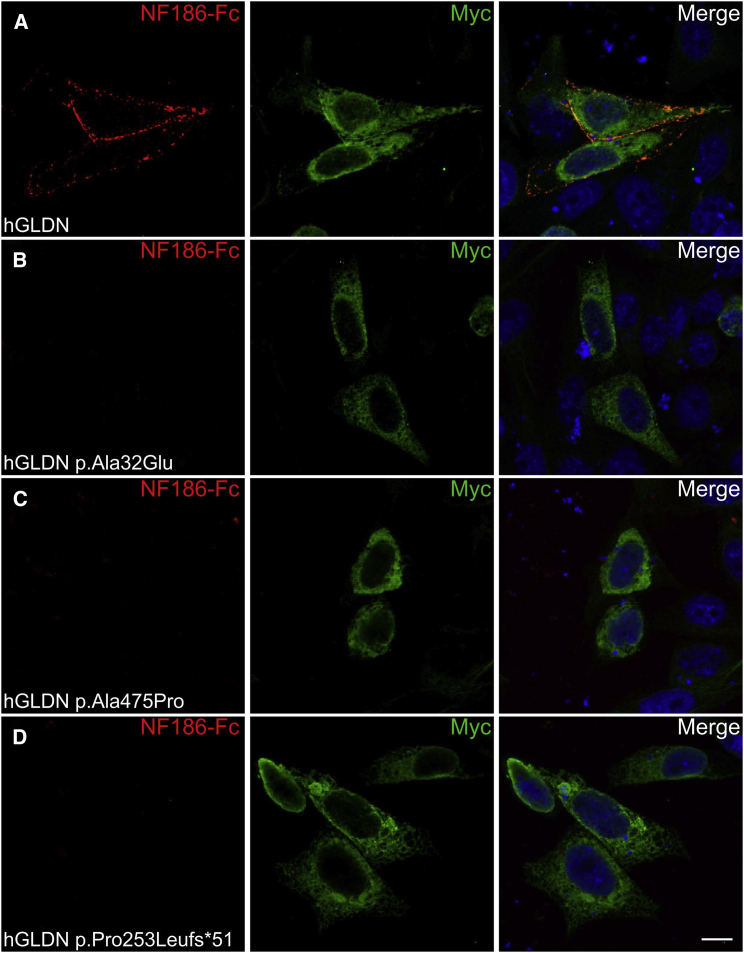
The p.Ala475Pro variant found in family 1 is located within the olfactomedin domain (aa 300–550, Figure 1C) of gliomedin.12 The p.Pro253Leufs∗51, p.Arg414∗, and p.Arg479∗ variants found in families 1, 3, and 4, respectively, are also suspected to affect this domain. The olfactomedin domain mediates the interaction between gliomedin and NrCAM, as well as neurofascin-186 (NF186), two cell adhesion molecules expressed at the nodes of Ranvier.12, 13, 14, 15 Thus, these variants might impact the formation of the NrCAM-NF186-gliomedin complex at nodes. In family 2, the p.Ala32Glu variant is localized within the transmembrane domain of gliomedin (aa 16–38, Figure 1C).12, 13, 14 Gliomedin exists as a transmembrane and a secreted form, and the latter form only is found at nodes.12, 13 However, variants within the transmembrane domain could also affect gliomedin secretion. To determine whether these variants had deleterious effects on gliomedin surface localization and NF186 binding, a myc-tag epitope was inserted at the extracellular C terminus of wild-type (WT) and mutant p.Ala32Glu, p.Ala475Pro, or p.Pro253Leufs∗51 gliomedin constructs. These constructs were then transfected into CHO cells, and living cells were incubated with anti-myc antibodies to detect surface gliomedin or incubated with soluble NF186 (NF186-Fc). An antibody to myc tag was then used to reveal the total WT and mutant gliomedin after fixation and permeabilization. WT gliomedin was readily localized at the cell surface (Figure 3) and bound NF186-Fc (Figure 4). In contrast, p.Ala32Glu, p.Ala475Pro, or p.Pro253Leufs∗51 gliomedin variants strongly affected the surface localization of gliomedin (Figure 3) and resulted in the loss of NF186-Fc binding to cells (Figure 4). Western blotting experiments were performed to characterize the protein amounts of variants (p.Ala32Glu, p.Ala475Pro, or p.Pro253Leufs∗51) as compared to those of WT gliomedin in transfected CHO cells (Figure S3). The variants found in affected individuals resulted in either normal, increased, or mild reduction of protein amount, indicating that a defect of gliomedin localization and binding to NF186 did not result from the lack of gliomedin (Figure S3). These data indicate that GLDN mutations found in the affected individuals had major deleterious effects on gliomedin transport to the cell surface and its binding to NF186. The p.Arg414∗ and p.Arg479∗ variants identified in families 3 and 4, respectively, are located within the olfactomedin domain and are expected to lead to similar effects.
Figure 3.
Mutations in GLDN Affect the Surface Localization of the Protein
Human gliomedin cDNA was amplified by PCR from a human placental cDNA library and sub-cloned into pcDNA3.1 (Thermofisher) at KpnI and XbaI sites, and a myc-tag epitope was inserted at the extracellular C terminus. Mutations found in the affected individuals (c.1423G>C, c.758delC, and c.95C>A) were inserted into cDNA with PrimeSTAR HS DNA polymerase (Clontech). All constructs were sequenced through the entire coding region. CHO cells transfected with myc-tagged human gliomedin (hGLDN) were incubated with mouse antibodies against myc before fixation to label surface myc (red) and after fixation and permeabilization to label total myc (green). Nuclei are stained with DAPI (blue). WT gliomedin (A) is readily localized at the cell surface. By contrast, p.Ala32Glu (B), p.Ala475Pro (C), and p.Pro253Leufs∗51 (D) variants strongly impacted the surface localization of gliomedin. Scale bars represent 10 μm.
Figure 4.
NF186 Binding is Disrupted by GLDN Mutations
CHO cells transfected with myc-tagged human gliomedin (hGLDN, green) were incubated with the extracellular domain of NF186 fused to human IgG Fc (NF186-Fc, red). NF186-Fc was expressed in human embryonic kidney cells and purified with Protein A-Sepharose beads (Sigma-Aldrich). Nuclei are stained with DAPI (blue). NF186-Fc bound to cells transfected with WT gliomedin (A), but did not bind to p.Ala32Glu (B), p.Ala475Pro (C), or p.Pro253Leufs∗51 (D) mutants. Scale bars represent 10 μm.
In the peripheral nervous system (PNS), the axoglial contact mediated by the binding of glial gliomedin and NrCAM to axonal NF186 is crucial for the initial clustering of voltage-gated Na+ channels at the nodes of Ranvier.12, 13, 14 The concentration of these channels allows saltatory conduction along myelinated axons. In the mature peripheral nerve, the secreted form of gliomedin is localized at the node of Ranvier and maintains the position of the Schwann cell microvilli toward the nodal axolemma.12, 13, 14 Here, our approach allowed for the identification of biallelic GLDN mutations in four unrelated families affected by severe hypokinesia and AMC. The parents of the affected individuals were healthy. The p.Ala475Pro and p.Pro253Leufs∗51 variants affected the olfactomedin domain of gliomedin and resulted in defects in both surface localization of gliomedin and interaction with NF186. This suggests that the affected individuals carrying these mutations on both alleles might exhibit alterations of the axoglial contact at nodes. In keeping with this suggestion, one of these affected individuals showed important node lengthening and a myelination defect in the sciatic nerve. Interestingly, the p.Ala32Glu mutation also resulted in defects in surface localization of gliomedin and thus might affect gliomedin aggregation at nodes. Surprisingly, mice carrying homozygous deletions of exons 9 and 10 of Gldn, which encodes for the olfactomedin domain of gliomedin, display no overt neurological abnormalities.16 The mutations found in the affected individuals might have stronger effects because they impact the gliomedin-NrCAM-NF186 complex. Indeed, altered gliomedin proteins might affect the localization or transport of glial partner proteins. Our data add to the recent report of four unrelated AMC-affected families showing homozygous frameshift mutations in CNTNAP117 (MIM: 616286). CNTNAP1 encodes CASPR-1, an essential component of the paranodal axoglial junction composed by caspr-1, contactin-1, and neurofascin-155 (NF155). This junction is believed to form a barrier against the lateral diffusion of nodal Na+ channels.18, 19 Altogether, these results indicate that mutations of GLDN or CNTNAP, encoding essential components of the nodes of Ranvier and paranodes, respectively, lead to severe developmental defects of the PNS, leading to reduced fetal mobility and AMC. These inherited nodopathies represent a distinct disease entity among peripheral neuropathies.
Acknowledgments
This work was supported by grants from the French Ministry of Health (PHRC 2010, AOM10181), the Association Française contre les Myopathies (DAJ1891), the University Paris Saclay (ERM2013), and INSERM to J. Melki. This work was supported by the Agence Nationale pour la Recherche under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases, and the Association Française contre les Myopathies (MNM1 2012-14580) to J.D. The authors would like to thank Damien Gentil for technical assistance, the Clinical Research Unit of Bicêtre Hospital, and the Direction of Clinical Research and Development (AP-HP).
Published: September 8, 2016
Footnotes
Supplemental Data include three figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.021.
Web Resources
1000 Genomes, http://www.1000genomes.org
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Human Splicing Finder, http://www.umd.be/HSF3/HSF.html
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Supplemental Data
References
- 1.Hall J.G. Genetic aspects of arthrogryposis. Clin. Orthop. Relat. Res. 1985;(194):44–53. [PubMed] [Google Scholar]
- 2.Fahy M.J., Hall J.G. A retrospective study of pregnancy complications among 828 cases of arthrogryposis. Genet. Couns. 1990;1:3–11. [PubMed] [Google Scholar]
- 3.Hall J.G. Arthrogryposis (multiple congenital contractures): diagnostic approach to etiology, classification, genetics, and general principles. Eur. J. Med. Genet. 2014;57:464–472. doi: 10.1016/j.ejmg.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Rüschendorf F., Nürnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 5.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J., Tawk M., Tiziano F.D., Veillet J., Bayes M., Nolent F., Garcia V., Servidei S., Bertini E., Castro-Giner F. Spinal muscular atrophy associated with progressive myoclonic epilepsy is caused by mutations in ASAH1. Am. J. Hum. Genet. 2012;91:5–14. doi: 10.1016/j.ajhg.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eshed Y., Feinberg K., Poliak S., Sabanay H., Sarig-Nadir O., Spiegel I., Bermingham J.R., Jr., Peles E. Gliomedin mediates Schwann cell-axon interaction and the molecular assembly of the nodes of Ranvier. Neuron. 2005;47:215–229. doi: 10.1016/j.neuron.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Eshed Y., Feinberg K., Carey D.J., Peles E. Secreted gliomedin is a perinodal matrix component of peripheral nerves. J. Cell Biol. 2007;177:551–562. doi: 10.1083/jcb.200612139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maertens B., Hopkins D., Franzke C.W., Keene D.R., Bruckner-Tuderman L., Greenspan D.S., Koch M. Cleavage and oligomerization of gliomedin, a transmembrane collagen required for node of ranvier formation. J. Biol. Chem. 2007;282:10647–10659. doi: 10.1074/jbc.M611339200. [DOI] [PubMed] [Google Scholar]
- 15.Labasque M., Devaux J.J., Lévêque C., Faivre-Sarrailh C. Fibronectin type III-like domains of neurofascin-186 protein mediate gliomedin binding and its clustering at the developing nodes of Ranvier. J. Biol. Chem. 2011;286:42426–42434. doi: 10.1074/jbc.M111.266353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg K., Eshed-Eisenbach Y., Frechter S., Amor V., Salomon D., Sabanay H., Dupree J.L., Grumet M., Brophy P.J., Shrager P., Peles E. A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron. 2010;65:490–502. doi: 10.1016/j.neuron.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laquérriere A., Maluenda J., Camus A., Fontenas L., Dieterich K., Nolent F., Zhou J., Monnier N., Latour P., Gentil D. Mutations in CNTNAP1 and ADCY6 are responsible for severe arthrogryposis multiplex congenita with axoglial defects. Hum. Mol. Genet. 2014;23:2279–2289. doi: 10.1093/hmg/ddt618. [DOI] [PubMed] [Google Scholar]
- 18.Peles E., Nativ M., Lustig M., Grumet M., Schilling J., Martinez R., Plowman G.D., Schlessinger J. Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rios J.C., Melendez-Vasquez C.V., Einheber S., Lustig M., Grumet M., Hemperly J., Peles E., Salzer J.L. Contactin-associated protein (Caspr) and contactin form a complex that is targeted to the paranodal junctions during myelination. J. Neurosci. 2000;20:8354–8364. doi: 10.1523/JNEUROSCI.20-22-08354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.