Abstract
Tubulinopathies constitute a family of neurodevelopmental/neurodegenerative disorders caused by mutations in several genes encoding tubulin isoforms. Loss-of-function mutations in TBCE, encoding one of the five tubulin-specific chaperones involved in tubulin folding and polymerization, cause two rare neurodevelopmental syndromes, hypoparathyroidism-retardation-dysmorphism and Kenny-Caffey syndrome. Although a missense mutation in Tbce has been associated with progressive distal motor neuronopathy in the pmn/pmn mice, no similar degenerative phenotype has been recognized in humans. We report on the identification of an early-onset and progressive neurodegenerative encephalopathy with distal spinal muscular atrophy resembling the phenotype of pmn/pmn mice and caused by biallelic TBCE mutations, with the c.464T>A (p.Ile155Asn) change occurring at the heterozygous/homozygous state in six affected subjects from four unrelated families originated from the same geographical area in Southern Italy. Western blot analysis of patient fibroblasts documented a reduced amount of TBCE, suggestive of rapid degradation of the mutant protein, similarly to what was observed in pmn/pmn fibroblasts. The impact of TBCE mutations on microtubule polymerization was determined using biochemical fractionation and analyzing the nucleation and growth of microtubules at the centrosome and extracentrosomal sites after treatment with nocodazole. Primary fibroblasts obtained from affected subjects displayed a reduced level of polymerized α-tubulin, similarly to tail fibroblasts of pmn/pmn mice. Moreover, markedly delayed microtubule re-polymerization and abnormal mitotic spindles with disorganized microtubule arrangement were also documented. Although loss of function of TBCE has been documented to impact multiple developmental processes, the present findings provide evidence that hypomorphic TBCE mutations primarily drive neurodegeneration.
Main Text
Tubulinopathies are a family of severe neurodevelopmental disorders caused by mutations in multiple genes encoding tubulin isoforms (e.g., TUBA1A [MIM: 602529], TUBA8 [MIM: 605742], TUBB2B [MIM: 612850], TUBB3 [MIM: 602661], and TUBB5 [MIM: 191130]), with defective neuronal migration and cerebral cortex malformation representing their major features.1 Inactivating mutations in TBCE (tubulin folding cofactor E [MIM: 604934]), encoding one of the five tubulin-specific chaperones involved in tubulin folding and polymerization, are known to cause two rare neurodevelopmental syndromes, hypoparathyroidism-retardation-dysmorphism syndrome (HRDS [MIM: 241410]), which is characterized by congenital hypoparathyroidism, intellectual disability, facial dysmorphism, and extreme growth failure, and Kenny-Caffey syndrome type 1 (KCS1 [MIM: 244460]), which overlaps HRDS but exhibits osteosclerosis and recurrent bacterial infections as additional features.2 All reported children with HRDS and KCS1 but one originate from inbred families of Middle Eastern origin (Saudi and Israeli pedigrees) and carry a recurrent homozygous in-frame deletion (c.155_166delGCCACGAAGGGA [p.Ser52_Gly55del]) in TBCE (GenBank: NM_001079515.2) resulting in virtually absent protein level.2, 3 Two additional mutations, both truncating (c.66_67delAG [p.Val23fs48∗] and c.T1113A [p.Cys371∗]), have been reported in a compound heterozygous state in a Belgian pedigree with two siblings manifesting features of HRDS.2 More recently, microtubule-disrupting mutations in tubulin genes have also been shown to trigger neurodegeneration, as illustrated by TUBA4A (MIM: 191110) and TUBB4A (MIM: 602662) mutations underlying amyotrophic lateral sclerosis (MIM: 616208) and hypomyelinating leukodystrophy associated with progressive spastic dystonic tetraplegia (MIM: 612438), respectively.4, 5 Similarly, a missense Tbce mutation predicting the p.Trp524Gly amino acid substitution has been associated with progressive distal motor neuronopathy in pmn/pmn mice,6, 7 but no similar phenotype has so far been reported in humans. Here, we report on the identification of an early-onset neurodegenerative encephalopathy resembling the phenotype of pmn/pmn mice, caused by biallelic TBCE mutations, with the c.464T>A missense change (p.Ile155Asn) occurring at the heterozygous/homozygous state in all affected subjects. We also show that the mutation results in a reduced amount of TBCE and that defective TBCE function in primary fibroblasts is associated with a lowered amount of polymerized α-tubulin, markedly delayed microtubule re-polymerization, and abnormal mitotic spindle morphology. Consistent with what we had previously been observed in pmn/pmn mice, the present findings demonstrate that hypomorphic TBCE function primarily drives neurodegeneration.
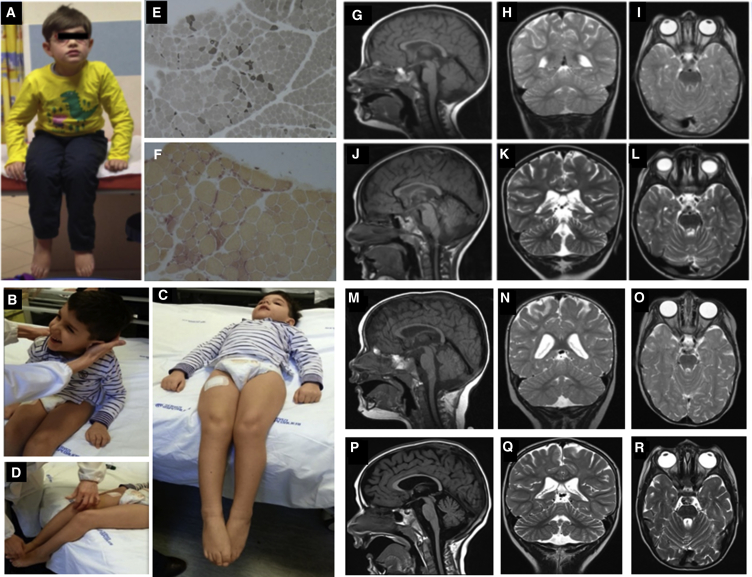
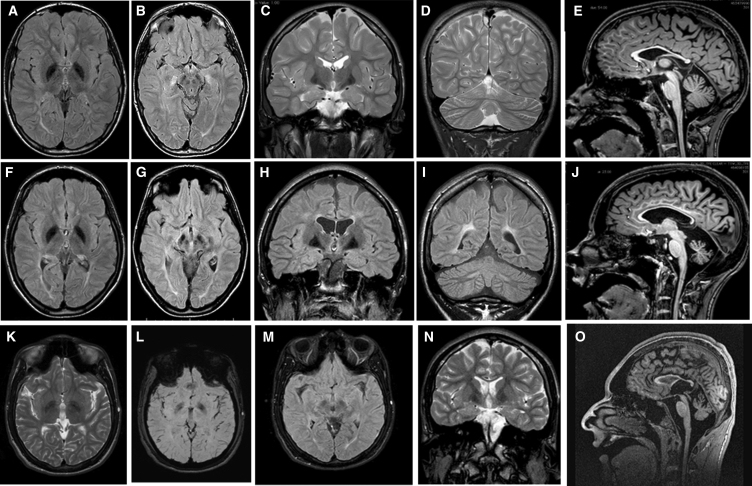
Six individuals from four apparently unrelated families originating from the same geographical area (metropolitan area of Naples in Campania, Southern Italy, and Ischia Island, just off the coast) were included in the study. Clinical features are schematically listed in Table 1 and summarized in the Supplemental Case Reports. Clinical data and biological material collection and storage were attained from the participating families after written informed consent was obtained, and the study was approved by the Ethical Committees of the participating centers. Genomic DNA and total RNA was isolated from peripheral blood leukocytes, using standard protocols. We originally performed whole-exome sequencing (WES) on genomic DNA obtained from subjects 1544334 and 2518864 (Figure 1). Both subjects had been followed at the Ospedale Pediatrico Bambino Gesù, Rome, and shared a neurodegenerative disorder characterized by early-onset slowly progressive distal motor neuropathy resembling distal spinal muscular atrophy (SMA) with bilateral foot drop associated to spasticity and cerebellar ataxia. They did not exhibit dysmorphia or growth defects. Muscle biopsy confirmed neurogenic muscle atrophy, and brain magnetic resonance imaging (MRI) documented cerebellar atrophy (Figure 1). In both cases, we used a trio-based (proband and unaffected parents) strategy. Exome capture was carried out using SureSelect Human All Exon v.4 (Agilent), and WES data analysis was performed using an in-house implemented pipeline, as previously reported.8, 9, 10 Paired-end reads were aligned to the human genome (UCSC GRCh37/hg19) with the Burrows-Wheeler Aligner (BWA v.0.7.10). Presumed PCR duplicates were discarded using Picard tools’ MarkDuplicates. GATK tools were used for realignment of sequences encompassing indels and for base quality recalibration.11 SNPs and small indels were identified by means of the GATK’s HaplotypeCaller used in gVCF mode, followed by family-level joint genotyping and phasing, according to GATK’s latest best practices. Called variants were filtered to retain variants with quality > 100 and quality-by-depth score > 1.5. High-quality variants were then filtered against public databases (dbSNP142 and ExAC v.0.3) to retain novel and clinically associated variants, and annotated variants with unknown frequency or having MAF < 0.1% and occurring with a frequency < 2% in an in-house database including frequency data from approximately 600 population-matched WES. SnpEff toolbox (v.4.1)12 was used to predict the functional impact of variants, which were filtered to retain only those located in exons with any effect on the coding sequence, and splice site regions (−3 to +8 with respect to exon-intron junctions). Functional annotation of variants was performed with SnpEff v.4.1 and dbNSFP v.2.8,12, 13 and their functional impact was analyzed by Combined Annotation Dependent Depletion (CADD) v.1.3 and dbNSFP Support Vector Machine (SVM) v.3.0 algorithms.14, 15 For sequencing statistics, see Table S1. Data annotation predicted 13,541 and 13,484 high-quality variants having functional impact (i.e., non-synonymous and splice site changes) in subjects 1544334 and 2518864, respectively. Among them, 356 and 350 clinically associated, private, and low/unknown frequency variants were retained for further analyses. Because family 1 had documented consanguinity, filtering was first directed to identify genes shared by the probands having variants compatible with a recessive inheritance models. We identified only one gene, TBCE, satisfying the filtering criteria. Specifically, subject 1544334 (family 1) was homozygous for the c.464T>A missense change (p.Ile155Asn) and subject 2518864 was a compound heterozygote for the same nucleotide substitution and the truncating c.1076delC (p.Leu360Ter) change. Sanger sequencing confirmed both variants and segregation in each family (Figure S1). No gene with a putative de novo change was shared by the two probands, and none of the genes with putative de novo variants in each individual was functionally linked to processes relevant to neurodegeneration (Table S1), strongly pointing to TBCE as the gene implicated in the disorder. Mutation scan of the TBCE coding sequence was successively performed on two monozygotic twins (subjects 00997847 and 00997844) of a third family originating from the same geographical area. Both individuals exhibited MRI profiles and an overall clinical phenotype overlapping that of the two previous cases, and Sanger sequencing allowed us to identify homozygosity for the recurrent c.464T>A change (Figure S1). Finally, interrogation of the Telethon Institute of Genetics and Medicine WES variant database allowed us to identify an additional sibling pair homozygous for the c.464T>A missense substitution (Figure S1). In that family, WES had been performed to identify the genetic cause of an unsolved apparently recessive neurodegenerative disorder characterized by spastic tetraparesis, optic atrophy, and abnormal brain MRI (cerebellar and brainstem atrophy, corpus callosum hypotrophy, and white matter hyperintensity with iron deposition in the globus pallidus and substantia nigra) resembling a pattern of neurodegeneration with brain iron accumulation (NBIA [MIM: 234200]) (Figure 2). Clinical reassessment of the two affected subjects revealed clinical features matching the disorder characterizing the previously identified subjects homozygous/compound heterozygous for the TBCE c.464T>A change. Remarkably, similar to the other case subjects, the family was from Pozzuoli and Mergellina, located in the Naples metropolitan area, along the coast near Ischia Island. Parents were not consanguineous. WES data statistics are reported in Table S1.
Table 1.
Clinical Features of Subjects with Biallelic Mutations in TBCE
| Affected Subjects | 1544334 | 2518864 | 00997847 | 00997844 | VN_X3359 | VN_3360 |
|---|---|---|---|---|---|---|
| Family | 1 | 2 | 3 | 3 | 4 | 4 |
| Gender | male | male | male | male | female | male |
| Year of birth | 2007 | 2009 | 2000 | 1996 | 2001 | |
| Inheritance | simplex case | simplex case | simplex case | familial | familial | |
| Consanguinity | yes | no | no | no | no | |
| Pregnancy, delivery, neonatal period | normal, weight at birth 3,360 g at 39th week of gestation | normal, weight at birth 3,490 g at 38th week of gestation | normal, weight 3,030 g at 37th week gestation | normal, weight 2,570 g at 37th week gestation | normal, weight at birth 2,600 g at 38th week of gestation | normal, weight at birth 3,250 g at 39th week of gestation |
| Initial motor & cognitive development | neonatal period normal | abnormal | neonatal period normal | neonatal period normal | normal until 8 months | |
| Age at presentation | around 4–5 months of age | at birth | around 4–5 months of age | 14 months | 8 months | |
| Signs of presentation | hypotonia and developmental delay in the first year of life | hypotonia and developmental delay in the first year of life | hypotonia and developmental delay in the first year of life | spasticity and developmental delay in the first year of life | spasticity and developmental delay in the first year of life | |
| Signs of regression | distal amyotrophy, ataxia, spasticity | distal amyotrophy, ataxia, spasticity | distal amyotrophy, ataxia, spasticity | distal amyotrophy, spastic tetraparesis, optic atrophy | distal amyotrophy, spastic paraparesis, optic atrophy | |
| Outcome | ||||||
| Age (years) | 6 | 6 | 15 | 15 | 20 | 15 |
| Motor function | able to sit | never able to sit | lost the ability to sit | lost the ability to sit | lost the ability to sit | lost the ability to sit |
| Cognitive level | mildly affected | moderately affected | moderately affected | moderately affected | severely affected | severely affected |
| Speech and language | able to speak, dysarthria | not able to speak | able to speak, severe dysarthria | able to speak, severe dysarthria | not able to speak | severe dysarthria |
| TBCE mutationsa | c.463T>A (p.Ile155Asn), c.463T>A (p.Ile155Asn) | c.463T>A (p.Ile155Asn), c.1076delC (p.Leu360Ter) | c.463T>A (p.Ile155Asn), c.463T>A (p.Ile155Asn) | c.463T>A (p.Ile155Asn), c.463T>A (p.Ile155Asn) | c.463T>A (p.Ile155Asn), c.463T>A (p.Ile155Asn) | |
Both variants have been reported with low recurrence in public databases (c.464T>A, rs780472451, 0.0000082/1, ExAC; c.1076delC, rs750781063, 0.00002/3, ExAC). Mutations are annotated according to GenBank: NM_001079515.2 (NP_001072983.1).
Figure 1.
TBCE Mutations Cause Early-Onset Encephalopathy with Distal Spinal Muscular Atrophy
Clinical features of subjects 1544334 (A) and 2518864 (B–D); muscle biopsy of 1544334 stained with ATPase 4.3 showing fiber type grouping and scattered atrophic angulated fibers (E) and atrophic angulated fibers that are denervated because positively stained with non-specific esterase (F); brain MRIs of subjects 1544334 (G–I, at 2 years; J–L, at 3 years) and 2518864 (M–O, at 1 year; P–R, at 2 years). Note the hypoplasia of corpus callosum and the cerebellar atrophy with prominent folia (K, Q).
Figure 2.
TBCE Mutations Cause a Pattern of Neurodegeneration with Brain Iron Accumulation in the Second Decade of Life of Affected Individuals
(A–J) Brain MRIs of subjects VN_X5360 (A–E, performed at age 13 years) and VN_X3359 (F–J, performed at age 17 years). Axial FLAIR-weighted images shown in (A), (B), (F), and (G); coronal T2-weighted images shown in (C) and (D); T1-weighted sagittal images shown in (E) and (J); (A), (B), (F), (G), (H), and (I) are FLAIR-weighted images. Note T2 or FLAIR hypointense areas corresponding to globi pallidi, substantia nigra, and mesencephalic red nuclei in both siblings, indicating abnormal iron deposits.
(K–O) Brain MRIs of subject 00997847 performed at age 16 years. Axial T2-weighted image shown in (K); a T2 coronal weighted image shown in (N); two axial T2∗ relaxation-weighted images documenting the presence of iron shown in (L) and (M); and a sagittal T1-weighted image shown in (O). The hypointense areas detected in T2-weighted images correspond to the increased iron for age accumulated at the level of the globi pallidi and substantia nigra.
All affected subjects were homozygous or compound heterozygous for the c.464T>A missense change and were from the same geographical area, strongly suggesting a founder effect. The variant has recently been reported in the ExAC database (rs780472451) with an overall estimated frequency of 0.000008 (1/121,411, information on the ethnic origin of the carrier not available). Genotyping of 400 unrelated and apparently unaffected individuals from the Naples metropolitan area by ARMS-PCR (primers and PCR conditions available on request) failed in identifying any allele carrying the variant (0–0.0025, 95% confidence interval). Based on the informative variants identified by WES to flank the c.464T>A change, the genomic region encompassing the disease-causing variant shared by the three families was estimated to cover approximately 1.3 Mb (chr1: 235,357,303–236,687,580). To more precisely define the size of the shared fragment, eight microsatellite markers covering the TBCE locus were selected, and the four trios were genotyped using an ABI PRISM 3500 Sequencer and the GeneMapper 4.0 software (Applied Biosystems). Reconstruction of the haplotypes allowed us to refine the length of the shared genomic region, which spanned approximately 0.5 Mb (Table S2), confirming the identity by descent of the c.464T>A missense variant.
All individuals homozygous or compound heterozygous for the c.464T>A variant in TBCE shared a strikingly homogeneous phenotype with clinical signs of progressive neurodegeneration, associated with normal calcium/phosphate metabolism and absence of repeated infections (Table 1 and Supplemental Case Reports), clearly differing from HRDS and KCS. By contrast, their clinical features were reminiscent of the pmn/pmn mouse,16 in which the progressive motor neuronopathy, hindlimb atrophy, and reduced brain size is caused by a missense substitution in the same gene (p.Trp524Gly) documented to partially impair TBCE stability and function.6, 7
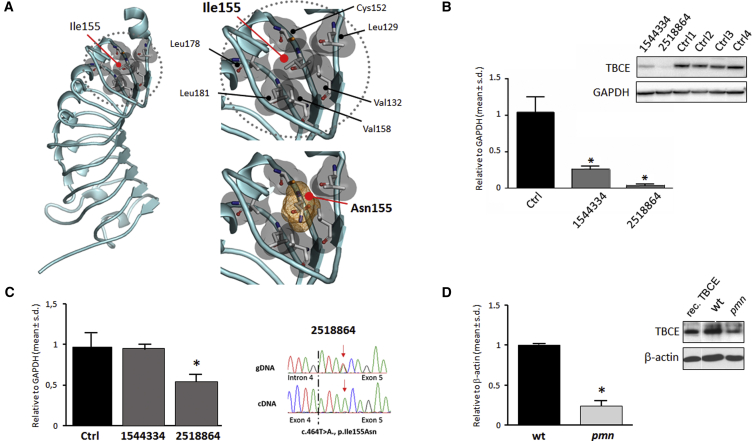
TBCE is a 527-amino-acid-long protein characterized by an N-terminal CAP-Gly domain, a motif mediating protein binding to α-tubulin, followed by nine tandemly arranged leucine-rich repeat (LRR) motifs, and a C-terminal ubiquitin-like domain possibly implicated in α-tubulin degradation via the proteasome (Figure S1).17, 18 Ile155 is a highly conserved residue across orthologs (Figure S1) and is located within the LRR domain. A homology model of the protein region spanning the nine LRRs (residues 123–348) was generated on the basis of the available LRR domain structure of internalin H (PDB: 1H6U), using the program Swiss-PdbViewer 4.1;19 loops were modeled with MODELER v.9.13.20 The generated model fitted well with the established structure of these motifs21 and localized the affected residue in the hydrophobic core stabilizing the second LRR motif (Figure 3A). At the same position, the introduction of the hydrophilic asparagine was predicted to perturb dramatically the structure of the motif and the overall folding of the LRR domain. Since LRR domains are functional modules that mediate protein-protein interaction, this amino acid substitution was expected to affect TBCE function or the overall TBCE structure and protein stability. To verify the latter hypothesis, the level of TBCE was evaluated by western blot analysis in skin fibroblasts obtained from affected subjects 1544334 and 2518864 and four healthy control subjects. Fibroblasts were lysed with RIPA buffer containing protease inhibitors cocktail (Roche), and 30 μg of total proteins were separated on a 4%–12% Bis-Tris gel (Invitrogen) and transferred to polyvinylidene difluoride membrane (Amersham). After blocking, membranes were incubated with anti-TBCE (Novus) (1:1,000) and anti-GAPDH (Sigma) (1:10,000) antibodies, the latter used to normalize samples loading. After washing, membranes were incubated with the secondary antibodies (Jackson ImmunoResearch), and immunoreactive bands were visualized using Lite Ablot Extend Long Lasting Chemiluminescent Substrate (Euroclone). Western blot analysis documented a significantly reduced amount of TBCE in fibroblasts from affected individuals compared to control cells (Figure 3B), suggestive of rapid degradation of the TBCE mutant. Of note, reduction in TBCE levels was more accentuated in fibroblasts from subject 2518864, who was compound heterozygote for the p.Ile155Asn and p.Leu360Ter changes. To compare the levels of TBCE mRNA among samples, total RNA from cell lysates was isolated using TRIzol solution (Invitrogen) and 1 μg RNA was reverse transcribed with the SuperScript First-Strand Synthesis system (Invitrogen), using random hexamers as primers. TBCE mRNA expression levels were determined by quantitative real-time PCR using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) and Power SYBR Green I dye chemistry. Assays were performed in triplicate, with GAPDH as internal controls, and documented a reduction to half of normal of TBCE mRNA in fibroblasts from subject 2518864 compared to what was observed in individual 1544334 and control cells (Figure 3C). Consistent with this finding, the frameshift variant was undetectable at the mRNA level, as determined by cDNA Sanger sequencing (Figure 3C), indicating nonsense-mediated RNA decay. A similar reduction in Tbce levels was documented in fibroblasts (Figure 3D) of pmn/pmn mice. Overall these data indicated that TBCE levels in fibroblasts of subjects with isolated neurodegenerative disease is preserved, in part, in sharp contrast with the extremely reduced or completely absent level of the protein documented in HRDS and KCS.3, 22
Figure 3.
Predicted Structural Impact of the p.Ile155Asn Substitution, and Consequence of TBCE Mutations on mRNA and Protein Levels
(A) Homology model of the TBCE leucine-rich repeat (LRR) domain (residues 123–348, GenBank: NP_001072983.1). Ile155 with surrounding hydrophobic residues contributes to the hydrophobic core required for proper folding of the domain. Introduction of Asn is predicted to disrupt the intramolecular binding network stabilizing the LRR structure.
(B) Western blot (WB) analysis documenting reduced TBCE levels in fibroblasts from affected subjects. Data are presented as means ± SD, ∗p < 0.05 (Mann-Whitney test).
(C) Real-time qRT-PCR showing reduced expression level of TBCE mRNA in fibroblasts from subject 2518864. ∗p < 0.05 (Mann-Whitney test). Such reduced level of the transcript is caused by transcript decay of the allele carrying the truncating c.1076delC variant, as documented by Sanger sequencing of TBCE cDNA showing solely the transcript with the c.464T>A missense change.
(D) WB analysis demonstrating reduced levels of Tbce in tail fibroblasts from the pmn/pmn mouse (TbceTrp524Gly); recombinant protein (rec. TBCE) is also reported. Data are presented as means ± SD, ∗p < 0.01 (Mann-Whitney test).
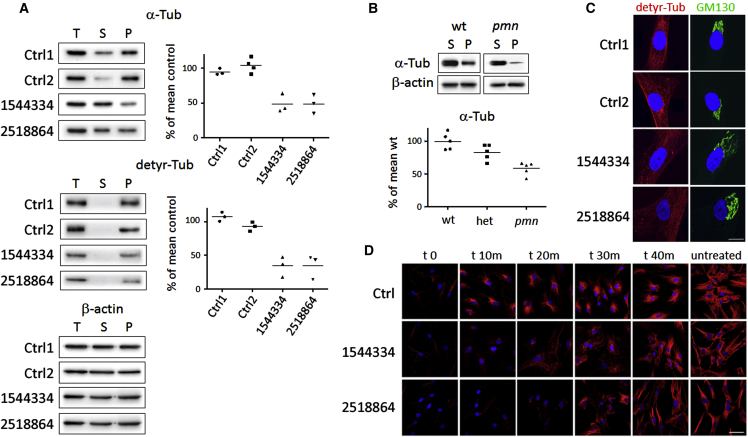
TBCE is required for the folding of α-tubulin, α/β-tubulin dimerization, and subsequent heterodimer polymerization into microtubules, which are major components of the cytoskeleton.23, 24 To explore the impact of the two TBCE mutations on microtubule polymerization, we first determined the levels of total, soluble, and polymerized α-tubulin in fibroblasts of affected and control subjects, using biochemical fractionation. Total, soluble, and polymerized tubulins from fibroblast lysates were separated by centrifugation (16,000 × g, 30 min, room temperature) as previously described.25 Proteins were solubilized by heating in Laemmli buffer, separated on 10% SDS-PAGE, and transferred on membranes, which were immunoblotted with antibodies against α-tubulin (Sigma) (1:5,000), detyrosinated tubulin (a kind gift of Dr. A. Andrieux, INSERM, Grenoble, France) (1:10,000), and β-actin (TuJ1, Babco) (1:10,000). The level of total α-tubulin did not significantly differ among cells (Figure 4A). By contrast, the amount of polymerized α-tubulin was significantly reduced in fibroblasts from affected subjects compared to control cells (Figure 4A), in line with what was observed in tail fibroblasts of pmn/pmn mice (Figure 4B). Moreover, significantly reduced levels of detyrosinated α-tubulin, a marker of stable and long-lived microtubules in neurons, were also noted (Figure 4A), indicating again a reduced microtubule stability in fibroblasts with biallelic mutations in TBCE.
Figure 4.
Defective Microtubule Polymerization in Subjects with Homozygous/Heterozygous TBCE c.464T>A Missense Change and in pmn/pmn Mice
(A) Western blot (WB) analyses showing total (T), soluble (S), and polymerized (P) α-tubulin (above), detyrosinated tubulin (middle), and β-actin (below) in fibroblasts from affected individuals (1544334, 2518864) and in control cells (Ctrl1, Ctrl2). Diagrams (right) show reduced fractions of polymerized α-tubulin and detyrosinated tubulin in fibroblasts of affected subjects. In both comparisons, differences were statistically significant by Mann-Whitney test (p < 0.01).
(B) WB analyses showing reduced amount of polymerized α-tubulin (P) in fibroblasts from pmn/pmn mice compared to control fibroblasts (wt). Differences were statistically significant by Mann-Whitney test (p < 0.01).
(C) Immunofluorescence analysis documenting loss of microtubules containing detyrosinated tubulin (detyr-Tub) in fibroblasts of affected subjects. Note that detyrosinated microtubules (red) in control fibroblasts are closely associated with the GM130-stained Golgi apparatus (green), while the Golgi apparatus in fibroblasts carrying mutated TBCE alleles appears to have lost compaction. Nuclei are DAPI stained (blue). Scale bar represents 10 μm.
(D) Immunofluorescence analyses showing reduced re-polymerization of microtubules (labeled for α-tubulin, red) after nocodazole treatment in fibroblasts from subjects 1544334 and 2518864, as compared to control cells (Ctrl). Nuclei are labeled with DAPI (blue). Scale bar represents 20 μm.
We hypothesized that defective TBCE function might alter microtubule nucleation and dynamics. To verify this hypothesis, synchronized fibroblasts from individuals with mutated TBCE alleles and unaffected controls were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, seeded on coverslips, and allowed to attach for 24 hr. Cells were treated with the microtubule-disrupting drug nocodazole (Sigma) (10 μM, 35 min at 37°C), and microtubule regrowth was analyzed after drug washout by confocal microscopy analysis, in time-course experiments, after fibroblasts were fixed, and immunostained for α-tubulin (Sigma) and with DAPI (Life Technologies). Cultures were visualized with a confocal microscope Fluoview FV1000 (Olympus) and images were acquired with the software FV10-ASW v.2.0. In control fibroblasts, microtubules began to polymerize efficiently as early as 10 min after nocodazole washout, and extended polymerized structures were well formed at 30 min (Figure 4C). By contrast, microtubule re-polymerization appeared markedly delayed in fibroblasts from affected subjects; microtubules were also less abundant and strongly disorganized in both early and late stages of re-polymerization. Such altered dynamics appeared particularly pronounced in fibroblasts with compound heterozygosity for the p.Ile155Asn and p.Leu360Ter changes, consistent with the more profound reduction of TBCE levels.
TBCE is concentrated at the Golgi membrane in an Arf1-regulated manner and facilitates both nucleation rates and polymerization speed of Golgi-derived microtubules.26 Immunofluorescence of synchronized primary fibroblasts was performed to visualize tubulin polymerization using antibodies against detyrosinated tubulin (1:1,000), as a marker of microtubules, and the Golgi membrane, using an anti-GM130 antibody (Becton Dickinson) (1:300). A decreased level of detyrosinated tubulin was observed in fibroblasts from affected subjects (Figure 4D), in line with the previously collected data and the reported loss of Golgi-derived microtubules in motor neurons of pmn/pmn mice.26 Of note, such reduced nucleation of microtubules was associated with a decreased compaction of the Golgi apparatus (Figure 4D).
Microtubules are a key cytoskeletal component of the mitotic apparatus, and their centrosomal nucleation coordinates formation and dynamics of the mitotic spindle during mitosis. To explore the occurrence of altered microtubule spindle organization, confocal microscopy analysis of synchronized fibroblasts from subjects with mutated TBCE alleles and unaffected individuals was performed. Fixed cells were stained with pericentrin (Abcam), γ-tubulin (Sigma), and α-tubulin (Abcam) antibodies, followed by the appropriate secondary antibodies (Invitrogen) and DAPI to visualize chromosomes. An abnormal mitotic morphology and disorganized mitotic microtubules were observed; spindles appeared asymmetric and unrefined and exhibited an irregular aster structure (Figure S2), documenting the impact of the hypomorphic defect in TBCE on mitotic spindle formation and organization.
The level of α/β-tubulin heterodimers as well as their polymerization and depolymerization at the microtubule ends are tightly controlled in cells to guarantee proper dynamics of this cytoskeletal component. Since microtubules control multiple neuronal processes, including cell division, migration, function, and survival,23 neurons are particularly vulnerable to altered microtubule dynamics. In this report, we provide evidence that defective TBCE function underlies a recessive early-onset neurodegenerative disorder and is associated with perturbed microtubule dynamics characterized by decreased and delayed microtubule polymerization. TBCE is a tubulin-binding cofactor required for the formation of α/β-tubulin heterodimers, microtubule polymerization, and also tubulin scavenging. Virtually complete loss of TBCE function has previously been identified to underlie two clinically related developmental disorders, HRDS and KCS1.2, 3 In contrast, hypomorphic Tbce function has specifically been reported to cause a neurodegenerative disorder in pmn/pmn mice due to retrograde dying back of motor axons and degeneration of motor neuron cell bodies.6, 7, 16, 27 The present data provide evidence of a similar neurodegenerative phenotype in humans resulting from defective TBCE function.
Consistent with the presently observed reduced nucleation of microtubules at the Golgi apparatus and decreased compaction of the Golgi apparatus, previous observations in pmn/pmn mice and TBCE-depleted motor neurons have documented that defective TBCE function impairs microtubule polymerization at the Golgi apparatus, leading to defective trafficking of COPI vesicles and Golgi fragmentation, which interestingly represents one of the earliest pathological features of degenerating motor neurons in amyotrophic lateral sclerosis.28, 29 In sharp contrast, HRDS and KCS are syndromic conditions mainly affecting multiple developmental processes and growth, with no sign of early-onset and progressive neurodegeneration, and in particular no sign of functional alterations involving upper and lower motor neurons. At the moment we have no explanation for this remarkable phenotypic difference. This picture has similarities with the phenotypic heterogeneity associated with mutations in ASAH1 (MIM: 613468), encoding N-acylsphingosine amidohydrolase 1, in which hypomorphic defects have been linked to childhood SMA associated with progressive myoclonic epilepsy (SMA-PME [MIM: 159950]), a rare autosomal-recessive phenotype restricted to the central nervous system generally starting with lower motor neuron disease,29 while a different class of mutations dramatically affecting catalytic activity of the enzyme results in Farber disease (MIM: 228000), a lysosomal storage disease characterized by severe motor and intellectual disability, early-onset subcutaneous lipogranulomata, painful and progressively deformed joints, and hoarseness of the voice.
Another remarkable clinical aspect of TBCE-related neurodegeneration is the MRI pattern resembling NBIA that was documented in the four older affected sibs in their second decade (subjects VN_X3359, VN_X5360, 00997847, and 00997844). We suppose that this pattern might represent a feature evolving with age, possibly appearing during the second decade of life. This pattern is unexpected because iron accumulation has not been reported among the neuropathological features of the pmn/pmn mouse.16 Although additional observations are needed to confirm this unexpected association, this finding suggests a critical role of cytoskeletal components in the complex cellular processes implicated in iron homeostasis in the brain.30
Overall, we describe a distal motor neuropathy, spastic ataxia, and juvenile-onset brain iron accumulation caused by biallelic mutations in TBCE overlapping the phenotype of the pmn/pmn mouse, a well-known model of motor neuron disease. Expanding beyond the previously documented impact of abolished TBCE function on developmental processes and growth, our findings indicate a stringent requirement of TBCE specifically for neuronal survival and function.
Acknowledgments
We are grateful to the participating families. This work was supported in part by grants from Fondazione Bambino Gesù (Vite Coraggiose to M.T.), Bulgari (GeneRare to B.D.), Ministero della Salute (RC2015 and RC2016 to M.N., M.T., and E. Bertini), and Agence Nationale de la Recherche (to G.H.).
Published: September 22, 2016
Footnotes
Supplemental Data include case reports, two figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.08.006.
Contributor Information
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
Enrico Bertini, Email: bertini@opbg.net.
Web Resources
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
Chimera, http://www.cgl.ucsf.edu/chimera
dbNSFP v.2.0, https://sites.google.com/site/jpopgen/dbNSFP
ExAC Browser, http://exac.broadinstitute.org/
GATK Best Practices, https://www.broadinstitute.org/gatk/guide/best-practices
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Data
References
- 1.Bahi-Buisson N., Poirier K., Fourniol F., Saillour Y., Valence S., Lebrun N., Hully M., Bianco C.F., Boddaert N., Elie C., LIS-Tubulinopathies Consortium The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain. 2014;137:1676–1700. doi: 10.1093/brain/awu082. [DOI] [PubMed] [Google Scholar]
- 2.Parvari R., Hershkovitz E., Grossman N., Gorodischer R., Loeys B., Zecic A., Mortier G., Gregory S., Sharony R., Kambouris M., HRD/Autosomal Recessive Kenny-Caffey Syndrome Consortium Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat. Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 3.Courtens W., Wuyts W., Poot M., Szuhai K., Wauters J., Reyniers E., Eleveld M., Diaz G., Nöthen M.M., Parvari R. Hypoparathyroidism-retardation-dysmorphism syndrome in a girl: A new variant not caused by a TBCE mutation--clinical report and review. Am. J. Med. Genet. A. 2006;140:611–617. doi: 10.1002/ajmg.a.31122. [DOI] [PubMed] [Google Scholar]
- 4.Smith B.N., Ticozzi N., Fallini C., Gkazi A.S., Topp S., Kenna K.P., Scotter E.L., Kost J., Keagle P., Miller J.W., SLAGEN Consortium Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–331. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simons C., Wolf N.I., McNeil N., Caldovic L., Devaney J.M., Takanohashi A., Crawford J., Ru K., Grimmond S.M., Miller D. A de novo mutation in the β-tubulin gene TUBB4A results in the leukoencephalopathy hypomyelination with atrophy of the basal ganglia and cerebellum. Am. J. Hum. Genet. 2013;92:767–773. doi: 10.1016/j.ajhg.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin N., Jaubert J., Gounon P., Salido E., Haase G., Szatanik M., Guénet J.L. A missense mutation in Tbce causes progressive motor neuronopathy in mice. Nat. Genet. 2002;32:443–447. doi: 10.1038/ng1016. [DOI] [PubMed] [Google Scholar]
- 7.Bommel H., Xie G., Rossoll W., Wiese S., Jablonka S., Boehm T., Sendtner M. Missense mutation in the tubulin-specific chaperone E (Tbce) gene in the mouse mutant progressive motor neuronopathy, a model of human motoneuron disease. J. Cell Biol. 2002;159:563–569. doi: 10.1083/jcb.200208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordeddu V., Redeker B., Stellacci E., Jongejan A., Fragale A., Bradley T.E., Anselmi M., Ciolfi A., Cecchetti S., Muto V. Mutations in ZBTB20 cause Primrose syndrome. Nat. Genet. 2014;46:815–817. doi: 10.1038/ng.3035. [DOI] [PubMed] [Google Scholar]
- 9.Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 10.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations impairing GSK3-mediated MAF phosphorylation cause cataract, deafness, intellectual disability, seizures, and a Down syndrome-like facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmalbruch H., Jensen H.J., Bjaerg M., Kamieniecka Z., Kurland L. A new mouse mutant with progressive motor neuronopathy. J. Neuropathol. Exp. Neurol. 1991;50:192–204. doi: 10.1097/00005072-199105000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Serna M., Carranza G., Martín-Benito J., Janowski R., Canals A., Coll M., Zabala J.C., Valpuesta J.M. The structure of the complex between α-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism. J. Cell Sci. 2015;128:1824–1834. doi: 10.1242/jcs.167387. [DOI] [PubMed] [Google Scholar]
- 18.Voloshin O., Gocheva Y., Gutnick M., Movshovich N., Bakhrat A., Baranes-Bachar K., Bar-Zvi D., Parvari R., Gheber L., Raveh D. Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress. Cell. Mol. Life Sci. 2010;67:2025–2038. doi: 10.1007/s00018-010-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 20.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 21.Bella J., Hindle K.L., McEwan P.A., Lovell S.C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian G., Huang M.C., Parvari R., Diaz G.A., Cowan N.J. Cryptic out-of-frame translational initiation of TBCE rescues tubulin formation in compound heterozygous HRD. Proc. Natl. Acad. Sci. USA. 2006;103:13491–13496. doi: 10.1073/pnas.0602798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapitein L.C., Hoogenraad C.C. Building the neuronal microtubule cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 24.Sanders A.A., Kaverina I. Nucleation and dynamics of Golgi-derived microtubules. Front. Neurosci. 2015;9:431. doi: 10.3389/fnins.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minotti A.M., Barlow S.B., Cabral F. Resistance to antimitotic drugs in Chinese hamster ovary cells correlates with changes in the level of polymerized tubulin. J. Biol. Chem. 1991;266:3987–3994. [PubMed] [Google Scholar]
- 26.Bellouze S., Schäfer M.K., Buttigieg D., Baillat G., Rabouille C., Haase G. Golgi fragmentation in pmn mice is due to a defective ARF1/TBCE cross-talk that coordinates COPI vesicle formation and tubulin polymerization. Hum. Mol. Genet. 2014;23:5961–5975. doi: 10.1093/hmg/ddu320. [DOI] [PubMed] [Google Scholar]
- 27.Schaefer M.K., Schmalbruch H., Buhler E., Lopez C., Martin N., Guenet J.L., Haase G. Progressive motor neuronopathy: a critical role of the tubulin chaperone TBCE in axonal tubulin routing from the Golgi apparatus. J. Neurosci. 2007;27:8779–8789. doi: 10.1523/JNEUROSCI.1599-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonatas N.K., Stieber A., Mourelatos Z., Chen Y., Gonatas J.O., Appel S.H., Hays A.P., Hickey W.F., Hauw J.J. Fragmentation of the Golgi apparatus of motor neurons in amyotrophic lateral sclerosis. Am. J. Pathol. 1992;140:731–737. [PMC free article] [PubMed] [Google Scholar]
- 29.Mourelatos Z., Gonatas N.K., Stieber A., Gurney M.E., Dal Canto M.C. The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu,Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc. Natl. Acad. Sci. USA. 1996;93:5472–5477. doi: 10.1073/pnas.93.11.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arber C.E., Li A., Houlden H., Wray S. Review: Insights into molecular mechanisms of disease in neurodegeneration with brain iron accumulation: unifying theories. Neuropathol. Appl. Neurobiol. 2016;42:220–241. doi: 10.1111/nan.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.