Abstract
Microtubules are dynamic cytoskeletal elements coordinating and supporting a variety of neuronal processes, including cell division, migration, polarity, intracellular trafficking, and signal transduction. Mutations in genes encoding tubulins and microtubule-associated proteins are known to cause neurodevelopmental and neurodegenerative disorders. Growing evidence suggests that altered microtubule dynamics may also underlie or contribute to neurodevelopmental disorders and neurodegeneration. We report that biallelic mutations in TBCD, encoding one of the five co-chaperones required for assembly and disassembly of the αβ-tubulin heterodimer, the structural unit of microtubules, cause a disease with neurodevelopmental and neurodegenerative features characterized by early-onset cortical atrophy, secondary hypomyelination, microcephaly, thin corpus callosum, developmental delay, intellectual disability, seizures, optic atrophy, and spastic quadriplegia. Molecular dynamics simulations predicted long-range and/or local structural perturbations associated with the disease-causing mutations. Biochemical analyses documented variably reduced levels of TBCD, indicating relative instability of mutant proteins, and defective β-tubulin binding in a subset of the tested mutants. Reduced or defective TBCD function resulted in decreased soluble α/β-tubulin levels and accelerated microtubule polymerization in fibroblasts from affected subjects, demonstrating an overall shift toward a more rapidly growing and stable microtubule population. These cells displayed an aberrant mitotic spindle with disorganized, tangle-shaped microtubules and reduced aster formation, which however did not alter appreciably the rate of cell proliferation. Our findings establish that defective TBCD function underlies a recognizable encephalopathy and drives accelerated microtubule polymerization and enhanced microtubule stability, underscoring an additional cause of altered microtubule dynamics with impact on neuronal function and survival in the developing brain.
Main Text
Microtubules are dynamic cytoskeletal elements required for many cellular and developmental processes. Their function is particularly critical in neurons, where they coordinate proliferation and migration, maintain neuronal polarity, support intracellular trafficking, and contribute to signal transduction.1 In neurons, their dynamic remodeling also contributes to the finely controlled events required for neuronal differentiation, proper morphological and functional maturation, and the establishment and maintenance of synaptic connections and plasticity.1, 2 The structural unit of microtubules is the αβ-tubulin heterodimer, whose assembly relies upon the CCT/TriC complex, five tubulin cofactor (TBC) proteins (i.e., TBCA to TBCE), and the small GTPase, ARL2.3, 4 Mutations in genes encoding tubulins and microtubule-associated proteins have been reported to cause a wide spectrum of neurodevelopmental and neurodegenerative disorders.5, 6 These mutations commonly affect tubulin dimer formation and/or polymerization and reduce microtubule stability. Growing evidence, however, suggests that altered microtubule dynamics can also lead to neurodegeneration.7 Here we report that mutations in TBCD, encoding one of the five co-chaperones required for assembly and disassembly of the αβ-tubulin heterodimer, underlie a previously unrecognized encephalopathy inherited as a recessive trait and exhibiting both neurodevelopmental and neurodegenerative features, including developmental delay and profound intellectual disability, seizures, progressive spasticity, optic atrophy, early-onset cortical atrophy associated with secondary hypomyelination, and thin corpus callosum. We demonstrate that disease-causing mutations affect TBCD synthesis, stability, and/or function and perturb microtubule dynamics, with an overall shift toward a more rapidly growing and stable microtubule population.
Seven subjects from five unrelated families were included in the study. Clinical data and biological material collection, use, and storage were obtained from the participating families after written informed consent was provided, and studies for each family were approved by the respective institutional review boards. To identify the genetic cause of a severe encephalopathy affecting two siblings (F118_346 and F118_347) exhibiting early-onset cortical atrophy, thinned corpus callosum, developmental delay, intellectual disability, epilepsy, and spastic tetraplegia (Figure 1 and Table S1), we performed whole-exome sequencing (WES). Exome capture was conducted using SureSelect Human All Exon v.4 (Agilent Technologies) on genomic DNA extracted from circulating leukocytes of the two siblings and their unaffected parents. Sequencing data processing, annotation, and filtering were performed using an in-house implemented pipeline which mainly takes advantage of the Genome Analysis Toolkit (GATK v.3.4) framework,8 following the GATK’s latest best practices, as previously reported.9, 10, 11, 12 For sequencing statistics, see Table S2. High-quality variants were filtered against public databases (dbSNP142 and ExAC v.0.3) to retain private and clinically associated variants, and annotated variants with unknown frequency or having MAF < 0.1%, and occurring with a frequency < 2% in an in-house WES variant database (approx. 600 population-matched exomes). Functional annotation of variants was performed using SnpEff v.4.1 and dbNSFP v.2.8.13, 14 Autosomal-recessive transmission was considered as the most likely inheritance model, and variant filtering and prioritization revealed a homozygous missense change, c.3365C>T (p.Pro1122Leu), in TBCD (tubulin-specific chaperone D) (MIM: 604649, GenBank: NM_005993.4) as the only strong candidate underlying the disease. Sanger sequencing confirmed homozygosity for the variant in each sibling (Figure S1). Biallelic TBCD variants were independently identified by WES in five additional subjects with similar clinical features from four unrelated families (Figure 1 and Table S1). In one family, matching was facilitated by GeneMatcher.15 The oldest sibling, CMH444, presented with focal epilepsy associated with congenital spastic tetraplegia and cognitive deficits. Brain MRI documented a thinned corpus callosum, cortical brain atrophy, and white matter volume loss. His sister, CMH445, presented with a similar constellation of symptoms, but with a relatively less severe phenotype. DNA was prepared utilizing the KAPA Biosystems library preparation kit (KAPA Biosystems) followed by TruSeqExome enrichment v.4 (Illumina). Read alignment and variant calling, filtering, and prioritization were performed as previously published,16, 17, 18 using custom-developed software, RUNES and VIKING. Variants were filtered to retain those with predicted impact on transcript processing and protein-coding sequence having allele frequency < 1% in public and in-house databases. Among the identified genes with variants compatible with a recessive mode of inheritance, TBCD emerged as the best candidate (Table S2). Similarly, matching was facilitated by GeneMatcher for subject 1455707. This child could not stand or sit independently. She had absent speech, minimal movement, and some response to noises and voices. Developmental delay and microcephaly were first noted at 4 months. MRI at 6 and 12 months showed a thin corpus callosum with hypoplasia of the rostrum and splenium and delayed myelination. Exome sequencing was performed by GeneDx on exon targets isolated by capture using the Clinical Research Exome kit (Agilent). Reads were aligned to the human genome build hg19 and analyzed and interpreted using a custom-developed analysis tool (Xome Analyzer) as previously described.19 General assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (submitter code: 26957). WES data analysis did not identify any de novo variant, and filtering and prioritization of the seven genes with biallelic variants pointed out to c.686T>G (p.Leu229Arg) and c.3365C>T (p.Pro1122Leu) in TBCD as the most promising disease-causing candidates (Table S2). The fourth subject, 3641284, presented with generalized hypotonia, developmental delay, seizures, and marked cerebral atrophy at 8 months. At 3 years, he exhibited severe intellectual disability with no speech or voluntary movement and a hypotonic tetraplegia dominated by a severe pure motor neuropathy. Finally, subject 6215546 was characterized by a relatively more attenuated phenotype, exhibiting developmental delay, only incidental seizures during infancy, moderate cognitive deficits, dysarthria, very mild pyramidal dysfunction of the arms, and marked hypertonic spastic paraplegia of the legs. MRI indicated minor cortical atrophy and thin corpus callosum. For both individuals, capture of target sequences was performed using the SureSelect Human All Exon v4 enrichment kit (Agilent Technologies) and sequencing was outsourced (BGI). WES data were processed, annotated, and filtered using an in-house developed pipeline as previously reported.20 Candidates were prioritized taking into account both the predicted functional impact of filtered variants and the biological relevance of individual genes. In both individuals, compound heterozygosity for mutations in TBCD emerged as the most likely events underlying the trait (Table S2).
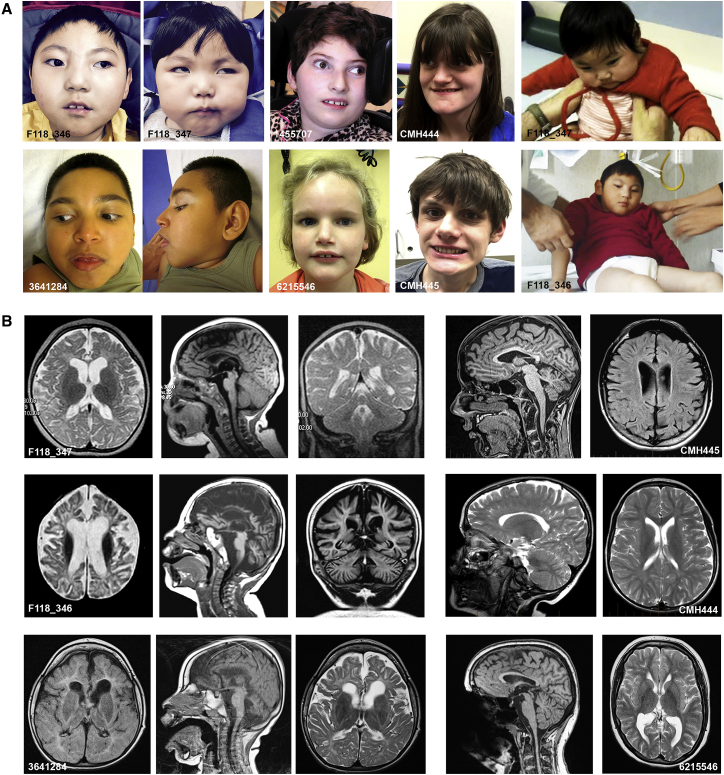
Figure 1.
Clinical Features of Subjects with Biallelic TBCD Mutations
(A) Features of affected subjects. Note microcephaly in subjects F118_346, F118_347, 3641284, and 1455707 and tetraplegia in F118_346 and F18_347.
(B) Brain MRIs of individuals F118_347 (8 months old; top left three panels) and F118_346 (2 years old; middle left three panels) showing axial, sagittal, and coronal views (from the left). F118_347 exhibits diffuse prominence of the cortical sulci with thinned corpus callosum and white matter volume loss with hypomyelination; the eldest sib exhibits more severe MRI features, with diffusely widened cortical sulci and cerebellar foliae and enlargement of lateral and third ventricles. MRIs of subjects CMH445 (16 years old; top right two panels) and CMH444 (13 years old; middle two right panels). In the former, diffuse prominence of the cortical sulci and cerebellar foliae with mild enlargement of the lateral and third ventricles and thinned corpus callosum are documented. In the latter, a milder pattern is observed, documenting a thin corpus callosum and partial white matter hypomyelination and mild cortical atrophy. MRIs of subjects 3641284 (3.5 years old; bottom three left panels) and 6215546 (17 years old; bottom right two panels). In the former, enlarged ventricles and generalized atrophy of gray matter, thin corpus callosum, cerebellar vermis atrophy, and abnormal white matter T2 and FLAIR signal hyperintensity due to severe secondary hypomyelination are observed. In the latter, slightly enlarged posterior lateral ventricles, mild cortical atrophy, hypoplastic splenium of the corpus callosum, and ectopic neuropituitary are noted.
All putative disease-causing variants were confirmed by Sanger sequencing (Figure S1). With the exception of one splice site change, all variants were missense nucleotide substitutions affecting conserved residues among orthologs (Figure S2). Functional impact of variants was analyzed by Combined Annotation Dependent Depletion (CADD) v.1.3 and Database for Nonsynonymous SNPs’ Functional Predictions (dbNSFP) Support Vector Machine (SVM) v.3.0 algorithms.21, 22 All variants were predicted to be deleterious and were either rare or not present in reference population databases (Table 1).
Table 1.
List of the Identified TBCD Mutations Causing Early-Onset Encephalopathy
| Subject(s) | Genomic Coordinate (hg19) | Nucleotide Changea | Coding Exon | Annotation | Status | Amino Acid Changea | Funct. Impactb(CADD Score, metaSVM Score) |
|---|---|---|---|---|---|---|---|
| F118_346 and F118_347 | 17: 80,896,008 | c.3365C>T | 36 | rs755177846 ExAc: 3.3 × 10−5 | hom. | p.Pro1122Leu | 26.0, −0.37 |
| CMH444 and CMH445 | 17: 80,887,366, 17: 80,895,956 | c.2981C>T, c.3313G>A | 32, 33 | novel rs764003906 ExAc: 1.6 × 10−5 | het. (P), het. (M) | p.Thr994Met, p.Val1105Met | 29.2, −0.35; 20.8, −1.11 |
| 3641284 | 17: 80,863,883, 17: 80,765,526 | c.1876G>A, c.1130G>A | 20, 11 | novel rs764085684 ExAc: 8.2 × 10−7 | het. (P), het. (M) | p.Ala626Thr, p.Arg377Gln | 27.2, −0.04; 32.0, −0.94 |
| 6215546 | 17: 80,739,598, 17: 80,765,517 | c.771+1_771+10del, c.1121C>T | int7, 11 | novel, novel | het. (M), het. (P) | ND, p.Thr374Met | 24.0, ND; 29.0, −0.64 |
| 1455707 | 17: 80,739,512, 17: 80,896,008 | c.686T>G, c.3365C>T | 7, 36 | rs778417127 ExAc: 2.5 × 10−5, rs755177846 ExAc: 3.3 × 10−5 | het. (P), het. (M) | p.Leu229Arg, p.Pro1122Leu | 26.2, 0.25; 26.0, −0.37 |
Abbreviations are as follows: ND, not determined; hom., homozygous change; het., heterozygous change; P, paternally inherited; M, maternally inherited.
Nucleotide and amino acid positions refer to GenBank: NM_005993.4 (cDNA) and NP_005984.3 (protein).
Prediction based on CADD v.1.3 and dbSNP v.3.0 SVM.
Major recurrent features in individuals with biallelic TBCD mutations included developmental delay and profound intellectual disability, seizures, progressive spasticity, optic atrophy, cortical atrophy, and thin corpus callosum. Microcephaly occurred in four subjects, and in three individuals it was present at birth and correlated with the time of onset and progression of atrophy. Quadriplegia was documented in all individuals. Four subjects were never able to walk, and three of them never achieved sitting or head control; those that attained walking subsequently lost this ability because of severe and progressive spasticity. Five individuals had epileptic seizures; peripheral motor axonal neuropathy was recorded in one subject. MRI showed signs of cortical atrophy and a thin corpus callosum in all subjects; this was associated with secondary hypomyelination and cerebellar atrophy in the most severely involved individuals. The clinical phenotype was relatively homogeneous, allowing validation of the causative role of the identified TBCD missense changes, even though variability in the onset and progression was observed. Of note, subject 6215546 presented an attenuated phenotype, which correlated with the milder perturbing effect of the p.Thr374Met substitution, while the particularly severe condition recorded in individuals F118_346 and F118_347 was associated with an extremely reduced TBCD level (discussed below). Overall, we consider this recessive condition caused by TBCD mutations as a disorder characterized by both neurodevelopmental and neurodegenerative features, in which the less-severe phenotypes appear to be characterized by early-onset neurodegeneration. The subjects’ clinical history, including delayed motor milestones, the progressive nature of the disorder, brain MRI evidence of cortical atrophy and hypomyelination, followed later by cerebellar atrophy, all seem to indicate that this condition is a neurodevelopmental disorder with evidence of slowly progressive neurodegeneration. This is further evidenced by the presence of microcephaly in most affected individuals. Of note, none of the affected individuals exhibited defects of cortical development associated with altered neuronal proliferation and migration, which is a common finding among tubulinopathies.23
TBCD encodes one of the five co-chaperones participating in the assembly of α- and β-tubulin in tubulin heterodimers to be incorporated in microtubules.24 It is a large (133 kDa) protein predicted to be predominantly α-helical in structure and containing armadillo/HEAT repeats, which mediate protein-protein interaction.25 The identified disease-associated variants mapped at different regions of the protein, and to explore in silico their impact on TBCD structure and function, we performed molecular dynamics (MD) simulations on the modeled structure of native and mutant TBCD obtained by homology modeling using SWISS-MODEL server (Figure S3). The starting model was refined by geometry optimization followed by MD simulations with Gromacs v.5.0.4 package, using the gromos54a7.ff force field.26, 27 The starting structure was embedded in a dodecahedron box filled with SPC water molecules,28 which extended up to 12 Å from the solute, and counter ions were added to neutralize the overall charge with Gromacs genion tool. After a long energy minimization (10,000 iterations), the system was slowly relaxed for 5 ns applying positional restraints (1,000 kJ mol−1 nm−2) to the protein atoms, and an uncontrolled MD simulation was protracted for 400 ns with a time step of 2 fs. V-rescale temperature coupling was employed to keep the temperature constant at 300 K.29 The simulation was performed enforcing periodic boundary condition, and the Particle-Mesh Ewald method was used for the treatment of long-range electrostatic interactions.30 Starting models for the mutant systems were generated by introducing mutations in silico into a wel-equilibrated (after ∼50 ns) snapshot of the wild-type (WT) protein, using UCSF-Chimera software.31 In all cases, the side chain orientation was chosen based on the best rotamer that had the least/no steric clashes with the neighboring residues. During the whole simulation time, the secondary structure of the WT protein appeared very stable; by contrast, all amino acid substitutions led to substantial long-range and/or local structural perturbations. In most cases (i.e., TBCDp.Thr374Met, TBCDp.Arg377Gln, TBCDp.Ala626Thr, TBCDp.Thr994Met, and TBCDp.Pro1122Leu), a considerable decrease in structured residues due to transition from α helix to coil was observed, while reduced flexibility at residues 836–910 occurred in all modeled mutants (particularly evident in TBCDp.Leu229Arg and TBCDp.Val1105Met) (Figure S3). Of note, all mutants exhibited a statistically significant reduction of the solvent accessibility surface (SAS) (Figure S4). Maximum deviation was observed for the modeled protein carrying the p.Pro1122Leu substitution, which was associated with the more severe clinical phenotype. Remarkably, visualization of positive and negative electrostatic potential isosurfaces showed variable perturbations in all mutants (Figures S5 and S6), with more dramatic changes involving proteins carrying the p.Leu229Arg, p.Arg377Gln, p.Thr994Met, and p.Val1105Met substitutions. Overall, these in silico simulations consistently predicted a variable but relevant impact of all amino acid substitutions on protein structure and folding.
To analyze the functional impact of variants directly, we first evaluated whether they affected the level of the TBCD protein in skin fibroblasts obtained from three unrelated subjects (F118_346, 3641284, and 6215546) cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (EuroClone). Western blot (WB) analysis performed on cell lysates, using a previously validated anti-TBCD antibody,32 followed by visualization of the immunoreactive proteins by enhanced chemiluminescence (Millipore) documented variably reduced levels of TBCD protein, with a dramatic decrease particularly occurring in fibroblasts homozygous for the p.Pro1122Leu substitution (Figure 2A). To confirm these findings, the single-nucleotide changes resulting in the p.Thr374Met, p.Arg377Gln, p.Ala626Thr, and p.Pro1122Leu substitutions were introduced by site-directed mutagenesis in a C-terminal Myc-DDK-tagged human TBCD cDNA cloned in pCMV6-Entry vector (RC200381, Origene), and each construct was transiently transfected (Fugene 6, Promega) in HEK293T. At 48 hr after transfection, cells were lysed and the level of TBCD was assessed by immunoblotting with an anti-Myc antibody (Cell Signaling), which reproduced this variable destabilization of each of these mutants (Figure 2A). Similar results were obtained in transiently transfected HeLa cells. Treatment of transfected cells with cycloheximide (20 μg/mL), a protein synthesis inhibitor, documented a pronounced decay of protein level for the TBCDp.Pro1122Leu mutant (Figure S7), confirming the impact of mutation on protein stability.
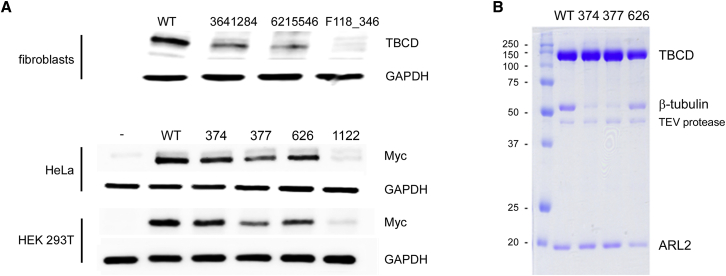
Figure 2.
Disease-Causing TBCD Mutations Affect TBCD Stability and Function
(A) Western blot (WB) analysis showing decreased level of TBCD in fibroblasts from affected individuals 3641284, 6215546, and F118_346 compared to control cells (WT) (above). Levels of Myc-tagged TBCDp.Thr374Met (374), TBCDp.Arg377Gln (377), TBCDp.Ala626Thr (626), and TBCDp.Pro1122Leu (1,122) mutants were confirmed to be variably reduced compared to wild-type TBCD (WT) by transient expression experiments in HeLa and HEK293T cells (below).
(B) TBCDp.Thr374Met (374) and TBCDp.Arg377Gln (377) mutants display defective TBCD:β-tubulin:ARL2 complex formation. Constructs encoding the GST-tagged TBCD proteins were co-expressed with ARL2 in HEK293T cells and affinity purified, followed by GST cleavage. Proteins were resolved by SDS-PAGE and stained with Coomassie blue.
TBCD localizes at the centrosome, the major microtubule-organizing center,32, 33 and confocal immunofluorescence analysis was performed to evaluate possible altered subcellular localization of TBCD mutants. Fibroblasts were seeded at the density of 20 × 103 in 24-well cluster plates onto 12-mm cover glasses and treated with thymidine and nocodazole (Sigma). Fixed cells were stained with anti-TBCD and anti-γ-tubulin (Sigma) antibodies, the respective fluorochrome-conjugated secondary antibodies (Invitrogen), and DAPI. Cells stained only with the fluorochrome-conjugated secondary antibodies were used to set up acquisition parameters, and signals from the different fluorescent probes were taken in sequential scanning mode, using a TCS SP2 AOBS apparatus (Leica Microsystems). In all tested cells from affected subjects, co-localization of TBCD mutants with γ-tubulin, used as a centrosome marker, was documented (Figure S8), indicating the retention of this feature of TBCD function.
TBCD is known to bind to β-tubulin and ARL.34 Based on the recessive transmission of the disorder and the variable reduced level of individual TBCD mutants, we hypothesized a hypomorphic/loss-of-function role for the identified disease-causing variants possibly affecting proper interaction of TBCD with its binding partners. To test this hypothesis, we co-expressed each disease-associated TBCD cDNA encoding the TBCDp.Thr374Met, TBCDp.Arg377Gln, and TBCDp.Ala626Thr mutants as GST-fusion proteins,35 along with the binding partner ARL2, in HEK293T cells to allow ready purification and analysis of complex formation. Soluble proteins were obtained by centrifugation of cell lysates and complexes were purified using Glutathione Sepharose 4B (GE 17-0756-01) beads prior to removal of the GST, by addition of purified TEV protease. SDS-PAGE and Coomassie blue staining showed that WT TBCD and TBCDp.Ala626Thr purified as complexes of TBCD:β-tubulin:ARL2 in the expected 1:1:1 ratio (Figure 2B). In contrast, TBCDp.Thr374Met and TBCDp.Arg377Gln co-purified with substantially reduced amounts of β-tubulin, despite retention of ARL2 binding, indicating specific loss in β-tubulin binding. All purified complexes were then analyzed using a dye binding (ThermoFluor) assay to determine thermal stability. Thermal denaturation curves were generated using StepOnePlus Real-Time temperature block by mixing TBCD (20 μL, final concentration 1 mg/mL protein) with 2 μL 1:100 dilution of SYPRO Orange dye (S5692, Sigma). Fluorescence emission (603 nm) was determined using excitation at 488 nm every 2 min for 2 hr over the range of 25°C–98°C, and the melting and plateau phases of collected data were analyzed with GraphPad Prism 6 software. The curves were fit to a Boltzmann sigmoidal equation to determine the Tm (inflection point) of each sample. Data obtained from four replicates of each protein, from two independent preparations of each, indicated that the TBCDp.Thr374Met and TBCDp.Arg377Gln proteins displayed a clearly decreased stability, as evident by the statistically significant lower Tm compared to the WT protein (Table S3). This decreased stability is predicted to be a consequence of the paucity of β-tubulin in each of these preparations, and consistently supported a loss-of-function role of disease-causing TBCD variants. Overall, the structural and biochemical characterization of disease-causing TBCD mutations consistently predicted a substantial impact on protein structure and folding, resulting in variable reduced stability and defective function of TBCD mutants.
Perturbed TBCD levels have been documented to affect assembly and disassembly of αβ-tubulin polymers.36, 37 Because proper microtubule dynamics is required for correct plasticity of cytoskeletal components38 and depends upon a tightly controlled balance of assembly/disassembly of αβ-tubulin polymers, we then examined the impact of TBCD mutations on the microtubule array. The amounts of total, soluble, and polymerized tubulin pools were determined in fibroblasts from affected individuals and control subjects. Tubulin pools were extracted as described.39 After 12% SDS-PAGE, equal volumes of each pool were analyzed by WB analysis using monoclonal anti-α-tubulin (Sigma cat# T5168, RRID: AB_477579) antibodies and revealed by a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Rockland). Analyses revealed that total α-tubulin levels (Figure 3A) were not significantly altered in fibroblasts from affected individuals, though soluble pools comprised a smaller tubulin fraction, indicating an enhanced microtubule stability and a reduction of the pool of tubulin dimers available for polymerization. In addition to its role in building the core structure of microtubules, TBCD has been proposed to modulate microtubule dynamics by promoting tubulin disassembly.37 To further explore the impact of mutations on microtubule dynamics, the kinetics of tubulin polymerization was assessed in primary fibroblasts from affected individuals and control subjects treated with 10 μM nocodazole (30 min, 37°C) to completely depolymerize the microtubules. After drug washout, recovery was allowed for 5, 10, 20, and 30 min. Immunofluorescence analysis was performed on fixed cells after staining with α-tubulin antibody (Abcam) followed by the appropriate secondary antibody (Invitrogen) and DAPI. Compared to what observed in treated control fibroblasts, microtubule re-polymerization was accelerated in cells from subjects with biallelic TBCD mutations (Figure 3B), consistent with these mutations being loss of function or hypomorphic, as TBCD overexpression has been demonstrated to prevent the growth of microtubules in the same assay.37 Together, these findings indicated that disease-associated defective TBCD function results in a shift toward a more rapidly growing and perhaps more stable microtubule population.
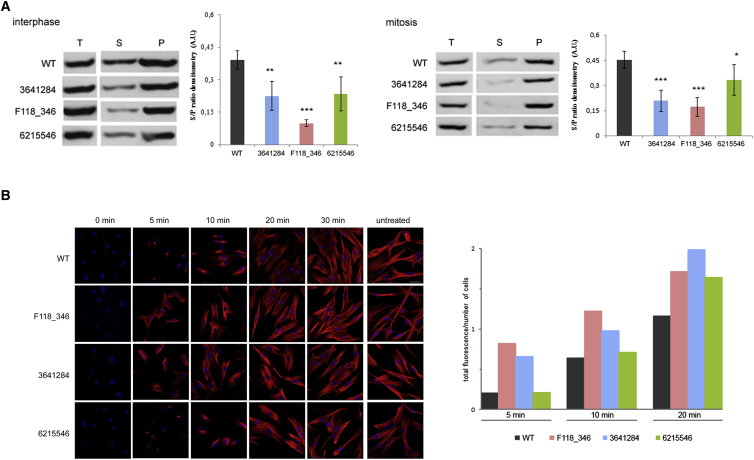
Figure 3.
Defective TBCD Function Impacts Microtubule Dynamics
(A) Increased levels of polymerized α-tubulin in fibroblasts with biallelic mutations in TBCD. Representative western blots (WB) of total (T), soluble (S), and polymerized (P) pools of α-tubulin in untreated cells during interphase (left) and synchronized cells during mitosis (right). Equal amounts of proteins from both fractions were loaded onto SDS-PAGE and analyzed by WB probing with anti-α-tubulin antibody. Densitometry is reported. Data are expressed as means ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) Accelerated microtubule re-polymerization in primary fibroblasts obtained from subjects with biallelic TBCD mutations. Representative confocal microscopy analysis of fibroblasts from affected subjects after nocodazole treatment showing accelerated microtubule re-polymerization compared to control cells (left). Fixed cells were stained with anti-α-tubulin antibody (red) and DAPI (blue). The scale bar (47 μm) is the same for all panels. Quantification of total fluorescence normalized on cell number, referred to the exponential phase of re-polymerization (5 to 20 min), is also shown (right).
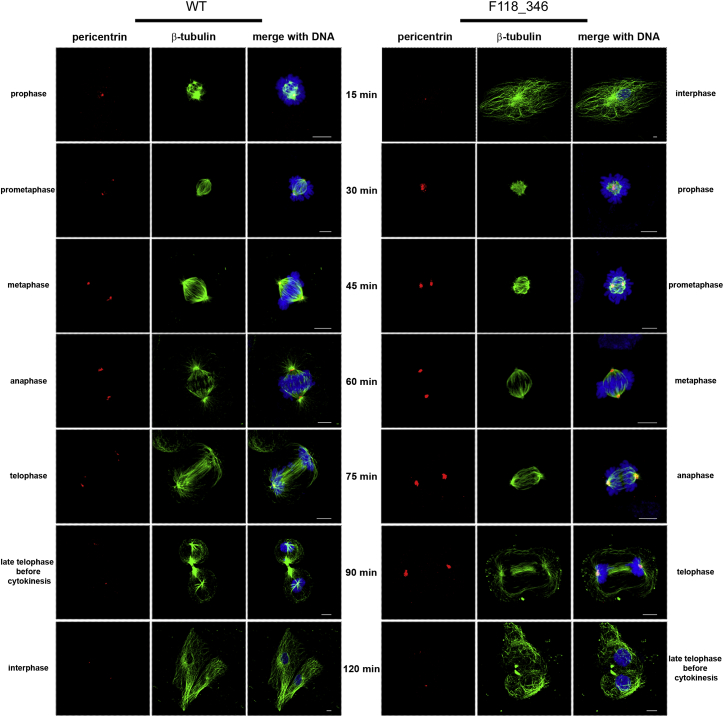
At the centrosome, TBCD is required for initiation of microtubule growth and organization of the mitotic spindle.32, 33 To further examine the impact of defective TBCD function on microtubule rearrangement and dynamics, confocal analysis was directed to evaluate possible perturbation on mitosis and microtubule spindle configuration in fibroblasts from affected subjects. After 24 hr of culture in complete medium, cells were treated with 2 mM thymidine (24 hr), washed with PBS, and treated with 100 ng/mL nocodazole (12 hr). After recovery (120 min), cells were fixing every 15 min using PHEMO buffer for 10 min at room temperature24 and stained with pericentrin (Abcam), γ-tubulin (Sigma), β-tubulin (Sigma), and/or α-tubulin (Abcam) antibodies, followed by the appropriate secondary antibodies (Invitrogen) and DAPI. These time-course experiments on synchronized cells reveled altered spindle structure in all fibroblast lines expressing biallelic TBCD mutations (Figure 4). Specifically, these cells exhibited disorganized, tangle-shaped mitotic microtubules, with markedly reduced aster formation. Moreover, progression of mitosis appeared delayed, possibly due to a transient blockade occurring during spindle formation. Aberrant spindle morphology was associated with significantly larger centrosomes and enhanced γ-tubulin and pericentrin signals (Figure S9). Of note, the observed spindle anomalies did not alter significantly the rate of fibroblast proliferation, as demonstrated by XTT-based cell proliferation assay (Roche Molecular Biochemicals) (Figure S10).
Figure 4.
Defective TBCD Function Affects the Mitotic Spindle
Confocal microscopy analysis was performed in synchronized skin fibroblasts from subjects with biallelic TBCD mutations and control cells. Images are representative of each stage of the cell cycle. Cells were stained using antibodies against pericentrin (red, centrosome marker) and β-tubulin (green, marker for microtubules and mitotic spindle); chromosomes are DAPI stained (blue). Scale bars represent 6 μm.
Here we established that biallelic hypomorphic/inactivating mutations in TBCD profoundly impact microtubule dynamics and cause an early-onset and severe encephalopathy. TBCD is one of the five tubulin-specific chaperones that function downstream of the cytosolic chaperonin complex, mediating the reversible assembly of the α/β-tubulin heterodimer.34, 40 The protein also modulates microtubule dynamics by sequestering β-tubulin from GTP-bound α/β-tubulin heterodimers.37 More recently, TBCD has been reported to localize at centrosomes and Fleming bodies and to be required for the assembly and maintenance of the mitotic spindle, microtubule retraction during cytokinesis, and cell abscission.32, 33 Moreover, TBCD has been shown to participate in centriolar and ciliary basal body assembly, strongly suggesting a more wider role in processes linked to microtubule nucleation.41, 42 In contrast with the involvement of this protein in such diverse fundamental cellular processes with relevance for diverse cell lineages, the major clinical features associated with TBCD mutations appear to be almost restricted to neurons, which suggests a specific and stringent dependence of these cells for proper TBCD function. Consistent with the present findings, a recent study provided evidence that loss of TBCD in Drosophila results in ectopic arborization of dendrites and axonal degeneration.43 Of note, overexpression of TBCD was also associated with microtubule disruption and ectopic dendrite arborization, indicating that proper TBCD function is crucial for in vivo neuronal morphogenesis. TBCD was also shown to physically interacts with the intracellular domain of Down syndrome cell adhesion molecule (Dscam),43 a neuronal adhesion molecule that is highly expressed in the central nervous system during development and is implicated in dendritogenesis, axonal outgrowth, neuron-to-neuron recognition events, and neural circuit formation.44
Although mutations affecting TBCA (MIM: 610058), TBCB (MIM: 601303), and TBCC (MIM: 602971) have not been associated with human disease thus far, biallelic inactivating mutations in TBCE (MIM: 604934) have been shown to underlie hyperparathyroidism-retardation-dysmorphism syndrome (MIM: 241410) and Kenny-Caffey syndrome (MIM: 244460).45 Interestingly, substitution at the last residue of Tbce (p.Trp524Gly) has been documented to cause a nonsyndromic motor neuropathy in mice resulting from decreased stability of the Tbce mutant protein and associated with a reduced number of microtubules suggested to originate from defective stabilization.46 In line with this finding, we recently identified a different hypomorphic missense mutation in TBCE (p.Ile1155Asn) occurring at the compound heterozygous/homozygous state in subjects exhibiting early-onset neurodegenerative encephalopathy with distal spinal muscular atrophy.66 In these individuals, clinical features resemble the phenotype of mice homozygous for the p.Trp524Gly substitution and are associated with reduced TBCE levels and altered tubulin polymerization. These data indicate that, differently from the truncating TBCE mutations described to perturb developmental processes, partial retention of TBCE function results in neurodegeneration also in humans, which is consistent with the present findings indicating both neurodevelopmental and neurodegenerative components associated with mutations in TBCD. Based on these findings, we expect that complete loss of TBCD function might result in a particularly severe neurodevelopmental disorder. Consistently, the present data suggest the occurrence of genotype-phenotype correlations, with the onset, progression, and overall severity of the phenotype linked to the relative functional impact of mutations. Co-existence of neurodevelopmental and neurodegenerative components has previously been documented in other encephalopathies, such as pontocerebellar hypoplasia (PCH) caused by mutations in EXOSC3 (MIM: 606489), TSEN54 (MIM: 608755), TSEN34 (MIM: 608754), TSEN15 (MIM: 608756), and TSEN2 (MIM: 608753) that are early-onset neurodevelopmental disorders but are also associated with signs of neurodegeneration after birth.47, 48, 49 Similarly to what was observed in subjects with biallelic TBCD mutations, MRI profiles in these disorders do not show signs of brain malformations, such as cortical dysplasia or simplified gyral pattern, but include brain cortical and cerebellar atrophy associated to PCH as predominant features. Hypomyelination was also observed to occur in subjects with TBCD mutations, which does not appear to be a primary event since the early signs of brain cortical and cerebellar atrophy suggest a primary neuronal involvement driving secondary white matter hypomyelination and loss. Regarding this specific aspect, the condition appears similar to aspartate-glutamate carrier 1 deficiency (MIM: 612949), a recessive encephalopathy caused by mutations in SLC25A12 (MIM: 603667),50 which was originally considered as a primary hypomyelination disorder but later recognized as being characterized by a primary neuronal cortical involvement with secondary hypomyelination.51 Another surrogate argument that would exclude primary hypomyelination in TBCD mutation-related encephalopathy is the finding of low levels of N-acetylaspartate (NAA) by MRS in CMH445 and 3641284, the only subjects included in this study for whom this information was available, since NAA is generally characterized by normal to increased levels by MRS in primary hypomyelination disorders due to abnormal raised levels of N-acetylaspartylglutamate.52, 53 Additional individuals with TBCD mutations will be necessary to characterize more accurately the nature of this disorder as well as to more precisely appreciate the extent of clinical variability associated with defective TBCD function.
Aberrant microtubule organization has been documented to underlie neurodevelopmental disorders.54, 55 For example, mutations in TUBB3 causing malformations in cortical development impair proper α/β-tubulin heterodimer formation and destabilize microtubules.56 Emerging evidence supports the view that altered microtubule dynamics can also underlie or contribute to neurodegenerative disorders.7 Increased microtubule depolymerization and defective microtubule assembly as a result of altered function of the microtubule-associated protein Tau, encoded by MAPT (MIM: 157140), has been proposed to play a relevant role in Alzheimer disease.57 Similarly, neurite defects occurring in Parkinson disease have been attributed to increased microtubule depolymerization,39, 58 and toxins that lead to parkinsonism alter microtubule dynamics, causing a decrease in length and number of microtubules.59, 60 Accumulating evidence also indicates that disruption of presynaptic microtubules generally precede synapse degeneration.61 Of note, similarly to what was observed for the presently identified phenotype resulting from biallelic mutations in TBCD, hyper-stabilization of microtubules has been shown to result in neurodegeneration, as in the case of autosomal-dominant spastic paraplegia type 4 (SPG4 [MIM: 182601]), a disease characterized by the degeneration of corticospinal tracts and caused by heterozygous mutations in SPG4, encoding the microtubule-severing enzyme spastin.62 In this progressive axonal degeneration disorder, defective spastin function results in a local accumulation of stable microtubules enriched in detyrosinated α-tubulin.63 Remarkably, the degenerative phenotype could be partly rescued in a SPG4 mouse model by treatment with nocodazole,64 strongly indicating hyperstability and reduced dynamics as the event triggering neuronal degeneration also in this disorder65 and providing insights on possible therapeutic strategies.
In summary, we have recognized a previously unappreciated neurodevelopmental disorder with evidence of acquired hypomyelination and neurodegenerative features, which is caused by defective TBCD function and associated with aberrant microtubule dynamics. These findings further emphasize the relevant role of TBCD in modulating stability and polymerization of microtubules and disclose a novel mechanism driving accelerated polymerization and enhanced stability of this cytoskeletal component with dramatic impact on neuronal function and survival in developing brain.
Acknowledgments
We are grateful to the participating individuals and their families. We thank colleagues in the Center for Pediatric Genomic Medicine, Children’s Mercy Kansas City, for WES sequencing data, and Megan Truitt Cho, GeneDx, for WES data analysis. This work was supported, in part, by the following grants: Fondazione Bambino Gesù (Vite Coraggiose to M.T.); Bulgari (GeneRare to B.D.); Ministero della Salute (RC2016 to M.N., M.L.D., E.B., and M.T.); CINECA (computational resources to M.T.); Marion Merrell Dow Foundation, W.T. Kemper Foundation, Pat & Gil Clements Foundation, Claire Giannini Foundation, and Black & Veatch (to I.T., E. Farrow, and C.J.S.); and NIH (R01GM090158 to R.A.K. and F31CA189672 to J.W.F.).
Published: September 22, 2016
Footnotes
Supplemental Data include ten figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.08.003.
Web Resources
Chimera, http://www.cgl.ucsf.edu/chimera
dbNSFP v.2.0, https://sites.google.com/site/jpopgen/dbNSFP
GATK Best Practices, https://www.broadinstitute.org/gatk/guide/best-practices
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://genematcher.org/
Gromacs, http://www.gromacs.org
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
OMIM, http://www.omim.org/
SWISS-MODEL, http://swissmodel.expasy.org/
Supplemental Data
References
- 1.Kapitein L.C., Hoogenraad C.C. Building the neuronal microtubule cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 2.Jaworski J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou C., Cunningham L., Marcus A.I., Li Y., Kahn R.A. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuss M., Keays D.A. Microtubules and neurodevelopmental disease: the movers and the makers. Adv. Exp. Med. Biol. 2014;800:75–96. doi: 10.1007/978-94-007-7687-6_5. [DOI] [PubMed] [Google Scholar]
- 6.Tischfield M.A., Cederquist G.Y., Gupta M.L., Jr., Engle E.C. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr. Opin. Genet. Dev. 2011;21:286–294. doi: 10.1016/j.gde.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubey J., Ratnakaran N., Koushika S.P. Neurodegeneration and microtubule dynamics: death by a thousand cuts. Front. Cell. Neurosci. 2015;9:343. doi: 10.3389/fncel.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordeddu V., Redeker B., Stellacci E., Jongejan A., Fragale A., Bradley T.E., Anselmi M., Ciolfi A., Cecchetti S., Muto V. Mutations in ZBTB20 cause Primrose syndrome. Nat. Genet. 2014;46:815–817. doi: 10.1038/ng.3035. [DOI] [PubMed] [Google Scholar]
- 10.Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 11.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations impairing GSK3-mediated MAF phosphorylation cause cataract, deafness, intellectual disability, seizures, and a Down syndrome-like Facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong J.X., Caputo V., Phelps I.G., Stella L., Worgan L., Dempsey J.C., Nguyen A., Leuzzi V., Webster R., Pizzuti A., University of Washington Center for Mendelian Genomics Recessive inactivating mutations in TBCK, encoding a Rab GTPase-activating protein, cause severe infantile syndromic encephalopathy. Am. J. Hum. Genet. 2016;98:772–781. doi: 10.1016/j.ajhg.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saunders C., Smith L., Wibrand F., Ravn K., Bross P., Thiffault I., Christensen M., Atherton A., Farrow E., Miller N. CLPB variants associated with autosomal-recessive mitochondrial disorder with cataract, neutropenia, epilepsy, and methylglutaconic aciduria. Am. J. Hum. Genet. 2015;96:258–265. doi: 10.1016/j.ajhg.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders C.J., Moon S.H., Liu X., Thiffault I., Coffman K., LePichon J.B., Taboada E., Smith L.D., Farrow E.G., Miller N. Loss of function variants in human PNPLA8 encoding calcium-independent phospholipase A2 γ recapitulate the mitochondriopathy of the homologous null mouse. Hum. Mutat. 2015;36:301–306. doi: 10.1002/humu.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neveling K., Feenstra I., Gilissen C., Hoefsloot L.H., Kamsteeg E.J., Mensenkamp A.R., Rodenburg R.J., Yntema H.G., Spruijt L., Vermeer S. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum. Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- 21.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romaniello R., Arrigoni F., Bassi M.T., Borgatti R. Mutations in α- and β-tubulin encoding genes: implications in brain malformations. Brain Dev. 2015;37:273–280. doi: 10.1016/j.braindev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Tian G., Huang Y., Rommelaere H., Vandekerckhove J., Ampe C., Cowan N.J. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 25.Grynberg M., Jaroszewski L., Godzik A. Domain analysis of the tubulin cofactor system: a model for tubulin folding and dimerization. BMC Bioinformatics. 2003;4:46. doi: 10.1186/1471-2105-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 28.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 29.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 30.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 31.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham L.A., Kahn R.A. Cofactor D functions as a centrosomal protein and is required for the recruitment of the gamma-tubulin ring complex at centrosomes and organization of the mitotic spindle. J. Biol. Chem. 2008;283:7155–7165. doi: 10.1074/jbc.M706753200. [DOI] [PubMed] [Google Scholar]
- 33.Fanarraga M.L., Bellido J., Jaén C., Villegas J.C., Zabala J.C. TBCD links centriologenesis, spindle microtubule dynamics, and midbody abscission in human cells. PLoS ONE. 2010;5:e8846. doi: 10.1371/journal.pone.0008846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Fanarraga M., Avila J., Guasch A., Coll M., Zabala J.C. Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J. Struct. Biol. 2001;135:219–229. doi: 10.1006/jsbi.2001.4386. [DOI] [PubMed] [Google Scholar]
- 35.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 36.Tian G., Thomas S., Cowan N.J. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton (Hoboken) 2010;67:706–714. doi: 10.1002/cm.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín L., Fanarraga M.L., Aloria K., Zabala J.C. Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett. 2000;470:93–95. doi: 10.1016/s0014-5793(00)01293-x. [DOI] [PubMed] [Google Scholar]
- 38.Howard J., Hyman A.A. Growth, fluctuation and switching at microtubule plus ends. Nat. Rev. Mol. Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 39.Choi W.S., Palmiter R.D., Xia Z. Loss of mitochondrial complex I activity potentiates dopamine neuron death induced by microtubule dysfunction in a Parkinson’s disease model. J. Cell Biol. 2011;192:873–882. doi: 10.1083/jcb.201009132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nithianantham S., Le S., Seto E., Jia W., Leary J., Corbett K.D., Moore J.K., Al-Bassam J. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble αβ-tubulin pool for microtubule dynamics. eLife. 2015;4:e08811. doi: 10.7554/eLife.08811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lüders J., Stearns T. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 42.Fanarraga M.L., Carranza G., Castaño R., Jiménez V., Villegas J.C., Zabala J.C. Emerging roles for tubulin folding cofactors at the centrosome. Commun. Integr. Biol. 2010;3:306–308. doi: 10.4161/cib.3.4.11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okumura M., Sakuma C., Miura M., Chihara T. Linking cell surface receptors to microtubules: tubulin folding cofactor D mediates Dscam functions during neuronal morphogenesis. J. Neurosci. 2015;35:1979–1990. doi: 10.1523/JNEUROSCI.0973-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montesinos M.L. Roles for DSCAM and DSCAML1 in central nervous system development and disease. Adv. Neurobiol. 2014;8:249–270. doi: 10.1007/978-1-4614-8090-7_11. [DOI] [PubMed] [Google Scholar]
- 45.Parvari R., Hershkovitz E., Grossman N., Gorodischer R., Loeys B., Zecic A., Mortier G., Gregory S., Sharony R., Kambouris M., HRD/Autosomal Recessive Kenny-Caffey Syndrome Consortium Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat. Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 46.Martin N., Jaubert J., Gounon P., Salido E., Haase G., Szatanik M., Guénet J.L. A missense mutation in Tbce causes progressive motor neuronopathy in mice. Nat. Genet. 2002;32:443–447. doi: 10.1038/ng1016. [DOI] [PubMed] [Google Scholar]
- 47.Zanni G., Scotton C., Passarelli C., Fang M., Barresi S., Dallapiccola B., Wu B., Gualandi F., Ferlini A., Bertini E., Wei W. Exome sequencing in a family with intellectual disability, early onset spasticity, and cerebellar atrophy detects a novel mutation in EXOSC3. Neurogenetics. 2013;14:247–250. doi: 10.1007/s10048-013-0371-z. [DOI] [PubMed] [Google Scholar]
- 48.Eggens V.R., Barth P.G., Niermeijer J.M., Berg J.N., Darin N., Dixit A., Fluss J., Foulds N., Fowler D., Hortobágyi T. EXOSC3 mutations in pontocerebellar hypoplasia type 1: novel mutations and genotype-phenotype correlations. Orphanet J. Rare Dis. 2014;9:23. doi: 10.1186/1750-1172-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Namavar Y., Barth P.G., Kasher P.R., van Ruissen F., Brockmann K., Bernert G., Writzl K., Ventura K., Cheng E.Y., Ferriero D.M., PCH Consortium Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain. 2011;134:143–156. doi: 10.1093/brain/awq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wibom R., Lasorsa F.M., Töhönen V., Barbaro M., Sterky F.H., Kucinski T., Naess K., Jonsson M., Pierri C.L., Palmieri F., Wedell A. AGC1 deficiency associated with global cerebral hypomyelination. N. Engl. J. Med. 2009;361:489–495. doi: 10.1056/NEJMoa0900591. [DOI] [PubMed] [Google Scholar]
- 51.Wolf N.I., van der Knaap M.S. AGC1 deficiency and cerebral hypomyelination. N. Engl. J. Med. 2009;361:1997–1998. doi: 10.1056/NEJMc091723. author reply 1998. [DOI] [PubMed] [Google Scholar]
- 52.Mochel F., Boildieu N., Barritault J., Sarret C., Eymard-Pierre E., Seguin F., Schiffmann R., Boespflug-Tanguy O. Elevated CSF N-acetylaspartylglutamate suggests specific molecular diagnostic abnormalities in patients with white matter diseases. Biochim. Biophys. Acta. 2010;1802:1112–1117. doi: 10.1016/j.bbadis.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bizzi A., Castelli G., Bugiani M., Barker P.B., Herskovits E.H., Danesi U., Erbetta A., Moroni I., Farina L., Uziel G. Classification of childhood white matter disorders using proton MR spectroscopic imaging. AJNR Am. J. Neuroradiol. 2008;29:1270–1275. doi: 10.3174/ajnr.A1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baird F.J., Bennett C.L. Microtubule defects & neurodegeneration. J. Genet. Syndr. Gene Ther. 2013;4:203. doi: 10.4172/2157-7412.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakraborti S., Natarajan K., Curiel J., Janke C., Liu J. The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton (Hoboken). 2016 doi: 10.1002/cm.21290. Published online March 2, 2016. [DOI] [PubMed] [Google Scholar]
- 56.Poirier K., Saillour Y., Bahi-Buisson N., Jaglin X.H., Fallet-Bianco C., Nabbout R., Castelnau-Ptakhine L., Roubertie A., Attie-Bitach T., Desguerre I. Mutations in the neuronal β-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet. 2010;19:4462–4473. doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iqbal K., Alonso Adel.C., Chen S., Chohan M.O., El-Akkad E., Gong C.X., Khatoon S., Li B., Liu F., Rahman A. Tau pathology in Alzheimer disease and other tauopathies. Biochim. Biophys. Acta. 2005;1739:198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Ren Y., Jiang H., Hu Z., Fan K., Wang J., Janoschka S., Wang X., Ge S., Feng J. Parkin mutations reduce the complexity of neuronal processes in iPSC-derived human neurons. Stem Cells. 2015;33:68–78. doi: 10.1002/stem.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cappelletti G., Surrey T., Maci R. The parkinsonism producing neurotoxin MPP+ affects microtubule dynamics by acting as a destabilising factor. FEBS Lett. 2005;579:4781–4786. doi: 10.1016/j.febslet.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 60.Patel V.P., Chu C.T. Decreased SIRT2 activity leads to altered microtubule dynamics in oxidatively-stressed neuronal cells: implications for Parkinson’s disease. Exp. Neurol. 2014;257:170–181. doi: 10.1016/j.expneurol.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephan R., Goellner B., Moreno E., Frank C.A., Hugenschmidt T., Genoud C., Aberle H., Pielage J. Hierarchical microtubule organization controls axon caliber and transport and determines synaptic structure and stability. Dev. Cell. 2015;33:5–21. doi: 10.1016/j.devcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C.S., Cruaud C., Dürr A., Wincker P. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 63.Tarrade A., Fassier C., Courageot S., Charvin D., Vitte J., Peris L., Thorel A., Mouisel E., Fonknechten N., Roblot N. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum. Mol. Genet. 2006;15:3544–3558. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
- 64.Fassier C., Tarrade A., Peris L., Courageot S., Mailly P., Dalard C., Delga S., Roblot N., Lefèvre J., Job D. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knockout mice. Dis. Model. Mech. 2013;6:72–83. doi: 10.1242/dmm.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., Lauring B.P. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sferra A., Baillat G., Rizza T., Barresi S., Flex E., Tasca G., D'Amico A., Bellacchio E., Ciolfi A., Caputo V. TBCE mutations cause early-onset progressive encephalopathy with distal spinal muscular atrophy. Immunity. 2016;45 doi: 10.1016/j.ajhg.2016.08.006. Published online September 22, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.