Abstract
Massively parallel whole-genome sequencing (WGS) data have ushered in a new era in human genetics. These data are now being used to understand the role of rare variants in complex traits and to advance the goals of precision medicine. The technological and computing advances that have enabled us to generate WGS data on thousands of individuals have also outpaced our ability to perform analyses in scientifically and statistically rigorous and thoughtful ways. The past several years have witnessed the application of whole-exome sequencing (WES) to complex traits and diseases. From our analysis of NHLBI Exome Sequencing Project (ESP) data, not only have a number of important disease and complex trait association findings emerged, but our collective experience offers some valuable lessons for WGS initiatives. These include caveats associated with generating automated pipelines for quality control and analysis of rare variants; the importance of studying minority populations; sample size requirements and efficient study designs for identifying rare-variant associations; and the significance of incidental findings in population-based genetic research. With the ESP as an example, we offer guidance and a framework on how to conduct a large-scale association study in the era of WGS.
Introduction
Early in 2015, President Obama used his State of the Union address to champion the pursuit of “precision medicine,” i.e., utilizing genetic and molecular techniques to individually tailor treatments and preventive measures for chronic diseases. The head of the US NIH and National Cancer Institute swiftly followed suit, describing the over-arching goals and plans for the Precision Medicine Initiative (PMI),1 which may eventually include whole-genome sequencing of a longitudinal cohort of 1 million or more Americans. To support the PMI at the National Heart, Lung, and Blood Institute (NHLBI), the Trans-Omics for Precision Medicine Program (TOPMed) will use WGS data along with molecular, environmental, and clinical data to investigate the etiology of heart, lung, blood, and sleep disorders. As the PMI and TOPMed programs are launched, many tens of thousands of whole-genome sequences will be generated, providing researchers with access to genetic data on a scale of unprecedented size and complexity.
Our ability to thoughtfully analyze and interpret large-scale WGS data will be a significant scientific and computational challenge. However, there are a number of recently completed, large-scale, population-based sequencing studies that offer empirical guidance. One such study, the NHLBI Exome Sequencing Project (ESP), was launched in 2009. Funded through the American Reinvestment and Recovery Act (ARRA), the ESP was conceived to identify rare, putatively functional, protein-coding variants associated with heart-, lung-, and blood-related diseases and traits. The ESP generated high read depth data on both European Americans (EAs) and African Americans (AAs) and was used to study genetic associations with more than 70 traits. Many of the analytic and logistical challenges we encountered in ESP provide a useful starting point for thinking about WGS studies. Here we describe the major findings and methodological advances from the ESP and the implications they have for the design and analysis of future large-scale sequencing projects.
Material and Methods
Study Design
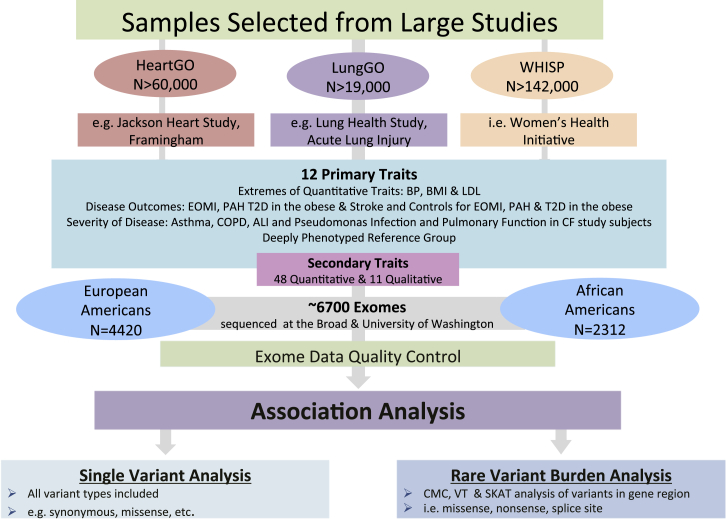
The original design of the ESP was focused on several phenotypes of high public health significance, as defined by the NHLBI Strategic Plan. Given the cost of deep sequencing at that time, only modest samples sizes were affordable. To enhance statistical power and enrich for variants with strong effects, the ESP employed two selection strategies for many of the subsets: sampling the extremes of quantitative traits and selection of individuals with early age at onset of disease. Several large population-based cohort and case-control studies (comprising >220,000 individuals) with detailed phenotype information and available DNA were used to select the 7,034 individuals (4,405 EAs, 2,954 AAs, and 35 of other ancestry) in the ESP (Figure 1).
Figure 1.
Schematic of the Work Flow for Sample Selection and Data Analysis in ESP
Primary traits were selected from large, population-based studies with widely available data on secondary traits. Both European and African American samples were selected for sequencing. Association analyses were conducted using both genes and single variants as units of analysis.
Primary clinical disease-related phenotypes included acute lung injury (ALI), asthma (MIM: 600807), chronic obstructive pulmonary disease (COPD [MIM: 606963]), early-onset myocardial infarction (EOMI [MIM: 608446]), ischemic stroke (MIM: 601367), type 2 diabetes (T2D [MIM: 125853]) with obesity as a co-morbidity, and pulmonary arterial hypertension-systemic sclerosis (PAH-ssa [MIM: 178600]). Several quantitative cardiovascular risk factors were studied in ESP, including blood pressure (BP), body mass index (BMI), and low-density lipoprotein (LDL), by selecting individuals with either extremely high or low trait values. In addition to the 12 primary traits, many of the ESP participants had data on up to 59 secondary phenotypes, including 48 quantitative biochemical, anthropometric, and subclinical measures of cardiovascular, blood, lung, and kidney disease/function. Detailed descriptions of the sample selection criteria, phenotype definitions, and contributing studies can be found in the Supplemental Data. All participants provided informed consent and the study was approved by the Institutional Review Board of each participating study.
Data Generation and Quality Control
The ESP generated exome-sequence data on 7,034 individuals that had been previously recruited through several large, NHLBI-funded cohort studies and deeply phenotyped on traits of public health importance. After rigorous quality control (QC), data were available on 4,392 EAs and 2,307 AAs. Exome sequencing of the DNA samples was performed at the Broad Institute of Harvard and MIT (n = 3,199) and the University of Washington (n = 3,893). Sequencing was performed to an average read depth of ∼90×. Reads were aligned to the human reference sequence (hg19) using the Burrows-Wheeler Alignment tool (BWA2) and the resulting binary alignment map (BAM)3 files were used to call single-nucleotide variants (SNVs) across all samples, i.e., multi-sample calling.
Although we report a 90× mean read-depth, coverage is very unbalanced across the exome and our goal was to obtain at least a 20× read depth for 80% of the exome. As illustrated in Figure S2, there are many regions with low read depth (e.g., <10). Variant sites in these regions would not be accurately genotyped without multi-sample calling. Additionally, multi-sample calling has clear advantages over single-sample calling for variant filtering and for creating a “squared off” call-set where genotypes are called at the same variant sites across all individuals. For these reasons, future uses of the ESP data that seek to combine with other datasets for association testing should consider re-analyzing the BAM files with multi-sample calling.
To identify potentially false-positive variant sites, a support vector machine classifier was used to separate likely true-positive from false-positive variant sites.4 Sites deemed false positive were excluded from further analyses.
Multidimensional scaling (MDS) was performed in order to validate self-reported EA and AA ancestry.5 Exomes were screened for cryptic relatedness and sample duplicates using KING software.6 Both cryptic and intentionally related and duplicate samples were uncovered; duplicate samples (n = 52) were included as part of the QC process. We found that including intentionally duplicated samples significantly helped us calibrate our QC procedures, although intentional duplicates represent a minimum marginal additional cost (0.74%).
QC that is too stringent can lead to a loss of power if causal variants are removed. On the other hand, inclusion of an excess of false-positive variant sites or incorrect genotypes can increase type I errors as well as reduce power. With this in mind, we sought to maximize the concordance rates of known duplicate samples and transition-transversion ratios while minimizing the amount of data removed during QC. As with the generation and analysis of genotype array data,7 implementation of standardized protocols for the generation and QC of sequence data will be important for future studies.
A complete description of the exome-sequencing and variant calling protocols has been described in detail in the Supplementary Methods of Fu et al.8 Details of the variant and sample-level quality control that were implemented in ESP are comprehensively described in Crosby et al.9 The final, cleaned dataset that was used for association analysis is referred to as the ESP6800.
Phenotype QC
We removed duplicate pairs and first- to third-degree relative pairs by retaining only the sample with the higher call rate. For each phenotype, we removed gross outliers by visual inspection and implausible values (e.g., BMI > 90). We also winsorised trait values to the 0.05% and 99.5%, i.e., trait values greater than the 99.5 quantile or less than the 0.005 quantile were truncated to the 99.5 and 0.005 quantiles, respectively. If necessary, quantitative traits were log-transformed for normality without winsorization. On the final set of samples that were used in the association analysis, we ran principal-component analyses, stratified by genetic ancestry. This was done using the MDS option in PLINK.5
Covariates
For each trait we used a model selection procedure to select covariates to be included in the association tests. All regressions included the first two ancestry-specific principal components. Other possible covariates were selected from the following list: age, age2, sex, BMI, smoking, and an indicator variable representing the capture-array and primary phenotype group for each sample.
Variant-Level Association Testing
We ran per-variant analyses to assess whether any individual variants were associated with an increase or decrease in the quantitative trait (or an increase or decrease in the odds for qualitative traits). Within each genetic ancestry group and for each di-allelic variant with at least 10 observed minor alleles in at least 30 samples, we tested for association between genotype and phenotype with a linear regression model. p values were obtained empirically with an adaptive permutation procedure. For computational efficiency, we also ran linear regression for the qualitative traits. Because our p values were obtained empirically, the tests were still statistically valid.10 In the autosome, we assumed an additive genetic model as described above. On the X chromosome, we assumed a dominance model in order to have consistent results across both males and females.
Results were meta-analyzed across genetic ancestries if there were at least 10 minor alleles and at least 30 observations present in both ancestry groups. For quantitative traits, meta-analysis was performed using the inverse-variance weighted technique; for qualitative traits, meta-analysis was performed using the sample-size weighted technique.11
Gene-Level Association Testing
We ran three different types of gene-level tests: Combined Multivariate Collapsing (CMC),12 Variable Threshold (VT),10 and Sequence Kernel Association Test (SKAT).13 Only missense, nonsense, and splice-variants were considered for inclusion in the gene-level tests. We annotated the variants using the SeattleSeq annotation server v.134, with the hg19 build of the human reference genome and the NCBI full genes (NM, XM) gene model option.
We noticed that missing genotype data can cause aggregate tests to have increased type I and type II errors. We therefore removed those variant sites missing greater than 10% of their data and imputed missing genotypes to the mean value.14
In general, aggregate rare variant association tests suffer from a loss of power when non-causal variants are included in the unit of analysis. For this reason, we a priori excluded synonymous variants, even though a fraction of them may be causal.15 Although there are a number of tools available for predicting the impact of nonsynonymous variants (e.g., PolyPhen-216 and CADD17), there are no gold standard approaches; thus, we chose not to include these predictions in our rare variant association testing pipeline.
For the CMC tests, we considered only variants with a within ancestry minor allele frequency ≤ 0.01 that was calculated from the entire ESP6800 call-set. Furthermore, we considered only genes for which the cumulative MAF ≥ 0.005. p values were obtained empirically with an adaptive permutation procedure. p values were meta-analyzed across ancestries if the cumulative MAF ≥ 0.005 in both genetic ancestry groups. Meta-analysis was performed using the sample-size weighted technique.
For the VT tests, we considered only variants with a within ancestry minor allele frequency ≤ 0.05 that was calculated from the entire ESP6800 call-set. For the VT, we considered only genes for which the cumulative MAF ≥ 0.005 at the MAF cutoff that attained the maximum test statistic. p values were meta-analyzed across ancestries if the cumulative MAF ≥ 0.005 in both genetic ancestry groups. Meta-analysis was performed using the sample-size weighted technique.
For the SKAT tests, we considered only variants with a within ancestry minor allele frequency ≤ 0.05 that was calculated from the entire ESP6800 call-set. p values were meta-analyzed across ancestries if the cumulative MAF ≥ 0.005 in both genetic ancestry groups. Meta-analysis was performed using Fisher’s Product Method. All association testing was conducted using the Variant Association Tools software.18
Imputation
In addition to the direct analyses of the exome-sequence data, ESP investigators utilized imputation in additional samples that were derived from some of the same parent NHLBI cohorts, who were genotyped (but not sequenced). Genotype imputation (in silico genotyping) is a statistical technique for predicting genotypes at variants that are not directly measured.19 Genotype imputation utilizes a set of reference samples that have been densely genotyped to identify segments of haplotypes that are shared with the study or “target” population. Prior to ESP, 1,692 AAs and 471 EAs from ESP had been genotyped on the Affymetrix 6.0 array. Using these 4,336 haplotypes as a reference panel, we imputed coding variants from the ESP into ∼13,000 AA samples with Affymetrix 6.0 GWAS data. The imputation was performed in several stages throughout the course of the project; details of the imputation at each stage have been previously reported.20, 21, 22, 23
Power Simulations
Using a simplistic model assigning every variant in the exome the same effect size, we estimated the sample sizes necessary to detect an association at exome-wide significance. We simulated EA samples using parameters adapted from recently published demographic models24 as input to the forward-time simulator Variant Simulation Tools.25 DNA sequences of ∼1.2 million haplotypes were simulated for all coding regions of CCDS genes on hg19 in the presence of purifying selection.26 A binary disease with a prevalence of 1% was simulated, assuming all rare variants in the gene have an odds ratio of 1.5. Quantitative traits were simulated similarly, with an effect size of 0.5σ, where σ is the standard deviation of the trait. We evaluated sample size requirements for the CMC and burden of rare variants (BRV)14 fixed effect tests, as well as the random effects SKAT method at a significance level of 2.5 × 10−6 assuming all rare variants in a gene are causal. A binary search method was used to obtain empirical sample size estimates for 80% power to detect associations.27
Results
Most Coding Variation Is Rare and Population Specific
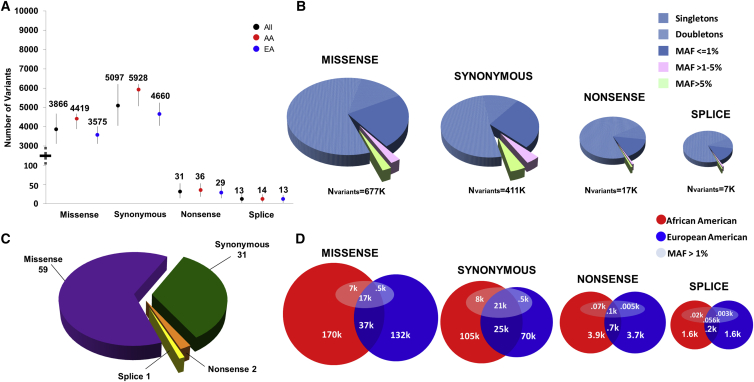
A total of 1,788,563 variant sites were observed in ESP, classified as missense (677,277), synonymous (410,554), nonsense (16,538), and splice (7,049) coding variant sites (Figure 2A). Rare (MAF < 1%) variants comprised the majority within all variant classes: 95.28% missense, 91.11% synonymous, 98.28% nonsense, and 98.25% splice. The majority of coding variants were singletons: 59% missense, 51% synonymous, 71% nonsense, and 72% splice (Figure 2B). Even with a preponderance of singletons, the average number of unique coding variants per individual is <100 (Figure 2C). For all classes of coding variant sites, AAs had, on average, a greater number of variant sites than EAs (Figure 2D). Although a different allelic architecture was observed for EAs and AAs, there was overlap of variant sites: synonymous 20.1% (95% CI 19.95%–20.28%), missense 14.8% (95% CI 14.67%–14.90%), nonsense 9.7% (CI 9.04%–10.31%), and splice 8.2% (CI 7.33%–9.18%). For variant sites that were exclusive to one population with a MAF > 1%, a larger proportion was unique to AAs (missense [p < 2.2 × 10−16], synonymous [p < 2.2 × 10−16], nonsense [p = 7.3 × 10−16], and splice [p = 1.5 × 10−5]) compared to those distinct to EAs (Figure 2D). For variants outside of coding regions, both the UK10K and 1000 Genomes projects report that most common genetic variants are shared across the world and that most rare variants are specific to closely related populations.28, 29
Figure 2.
Coding Variants Observed in the NHLBI-ESP
(A) The average number of missense, synonymous, nonsense, and splice site variants per study subject for 2,307 African Americans and 4,392 European Americans and all study subjects (n = 6,699) for the intersect of all four targets. The vertical lines display the smallest and largest number of variants of each type observed per person.
(B) The number of missense, synonymous, nonsense, and splice sites observed for NHLBI-ESP (n = 6,699) study subjects. Represented in each pie chart is the number of singletons, doubletons, and variant sites with an MAF of ≤1%, >1%–5%, and >5%.
(C) The average number of unique missense, synonymous, nonsense, and splice site variants per individual. The variants are not only exclusive to the NHLB-ESP but also are not observed in either dbSNP or 1000 Genomes.
(D) Comparison of the number of coding variant sites observed in AAs and EAs. The number of missense, synonymous, nonsense, and splice site variants that are unique to each population are observed in both populations and have a MAF of ≥1%. The numbers displayed are exclusive to one category. In order to fairly compare the number of variant sites in African Americans and European Americans, equal numbers of African Americans (n = 2,312) and European Americans (n = 2,312) were studied.
Non-synonymous coding variants showed evidence of evolutionary constraint, consistent with purifying selection of deleterious alleles.30 A modified Out-of-Africa demographic model with accelerated population growth beginning approximately 5,000 years ago demonstrated that the observed excess of rare variation is attributable largely to explosive population growth,30 with 73% of protein-coding variants in the ESP estimated to have arisen in the past 5,000–10,000 years.8 This increased mutational load has led to increased allelic and genetic heterogeneity of traits.8 For disease gene mapping, these results suggest that the complexity we observe in many traits is due, in part, to recent explosive population growth. The implications for association testing are clear: most variants are very rare and testing them individually for association will be under-powered.
Rare-Variant Associations, Imputation, and Replication
We did not observe any systematic inflation of significance from association testing (Figure S1). Ancestry-specific results were examined individually, as well as meta-analyzed for both single variant and aggregate rare variant analyses using a sample-size weighted approach.11 We noticed that many of our association results were not concordant between EAs and AAs. Though this may be due to false positives within each ancestry group, it may also be due to population-specific allelic architectures where the same rare variants have different effect sizes or simply do not underlie complex trait etiology across major ancestry groups.
The design of ESP was structured around identifying rare variants with large effects. A trade off of this design was that ESP was under-powered to detect common variant associations of modest effects. Indeed, for most traits, we did not identify novel associations for common variants, but rather replicated many known hits. For instance, coding variants in APOE (MIM: 107741) and PCSK9 (MIM: 607786) were associated with LDL cholesterol levels.31 However, we did report a common missense variant in PDE4DIP (MIM: 608117) that was not in linkage disequilibrium with any tag SNPs on any commercial GWAS array and was associated with risk for ischemic stroke.32
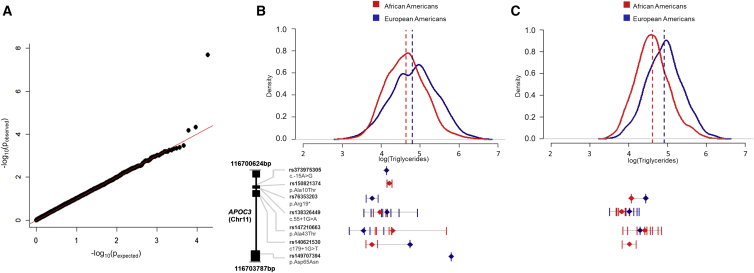
We identified several trait associations where a burden of rare variants within a gene accounted for the association.9, 31, 33, 34 Of note, we identified multiple nonsynonymous variants in APOC3 (MIM: 107720) associated with lower triglyceride levels in both EAs and AAs (Figure 3).9 A burden of multiple, rare nonsynonymous variants were also found in DCTN4 (MIM: 614758) to be associated with time to first psuedomonas infection in cystic fibrosis (MIM: 219700).34 The extremes of LDL cholesterol were combined with other non-LDL extremes to identify a burden of rare and low-frequency variants in PNPLA5 (MIM: 611589) associated with higher LDL cholesterol levels;31 rare, nonsynonymous variants in LDLR (MIM: 606945) and APOA5 (MIM: 606368) were shown to be associated with risk of early-onset myocardial infarction.33 However, very few of the ∼70 traits analyzed in ESP resulted in exome-wide significant associations (Figure S1).
Figure 3.
Triglyceride Rare Variant Association Analysis and Association of Rare Variants in APOC3
(A) QQ plot of the meta-analysis for African Americans and European Americans of rare variant burden analysis of triglyceride levels. Base 10 –log values of the observed p values are displayed versus their expected values. Rare variant association analysis was performed separately for African Americans (n = 1,654) and European Americans (n = 2,074) using the CMC analyzing those variant sites with a MAF ≤ 0.01.
(B) Distribution of triglyceride levels for NHLBI-ESP study subjects and triglyceride levels for individuals with an APOC3 variant. The quantitative trait distribution of triglycerides after natural log transformation for African Americans and European Americans who are study subjects in the NHLBI-ESP. For the 27 individuals (8 African American and 19 European American) who are heterozygous for one of the 7 coding variants (3 splice, 1 stop-gain, and 3 missense), a tick represents their triglyceride levels after natural log transformation. For each variant site a diamond (red for African Americans and blue for European Americans) represents the average triglyceride levels for carriers of that variant.
(C) Distribution of triglyceride levels for study subjects from the Women’s Health Initiative (WHI) and triglyceride levels for individuals with an APOC3 variant. The quantitative trait distribution of triglycerides after natural log transformation for African Americans (n = 1,820) and European Americans (n = 1,643) who are study subjects from the WHI. The DNA samples from the study subjects were genotyped on the exome chip. Of the seven variants that were observed in NHLBI-ESP, four were represented on the exome chip.
In order to replicate significant findings and to follow up in larger populations those associations that did not attain exome-wide significance, the ESP contributed to the development of the Exomechip, a custom genotyping array of rare variant content. Specifically, loss-of-function mutations in APOC3 and NPC1L1 (MIM: 608010) were included on the Exomechip and facilitated discovery and replication of associations between these variants and coronary heart disease.9, 35
In addition to the Exomechip, we utilized genotype imputation to increase sample sizes and enhance our power to detect associations. We created a custom imputation reference panel using samples with both ESP and GWAS data. This reference panel out-performed imputation using 1000 Genomes data as a reference panel22 and led to the discovery of multiple associations with hematologic and anthropometric traits.20, 21, 23, 36 Of note, none of these associations were exome-wide significant in the ESP data alone. Only after augmenting our samples size through genotype imputation were we able to identify these associations.
Power to Detect Associations
For the analysis of individual variants, the power to detect an association is affected by disease prevalence, allele frequency, effect size, sample size, and significance (alpha) level.37 For example, for a disease with a 1% prevalence, a sample size of 40,000 case subjects and 40,000 control subjects is required to have 80% power to detect an association with α = 5 × 10−8 for a variant with MAF = 0.5% and an odds ratio (OR) = 1.5. In addition to these parameters, when aggregating rare variants into a larger unit of analysis (e.g., a gene), association tests are affected by the allelic architecture within a gene (the number of variant sites, their cumulative MAF, the direction and size of their effects, and the proportion of non-causal variants). Thus, for any given trait, the power to detect an aggregate rare variant association varies across genes.
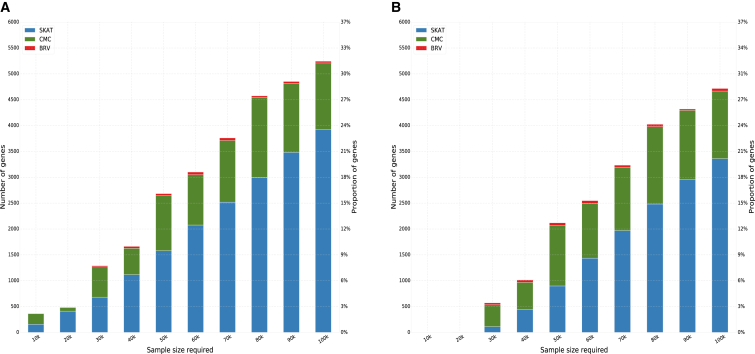
The power to detect aggregate rare-variant associations varies considerably across the exome, with most genes requiring >100,000 samples in order to robustly allow detection of an association (Figure 4). We detect associations with 30% of genes using a sample size of 100,000. Of particular relevance to ESP is that under these idealized conditions, only 1.25% of genes have sufficient power to allow detection of association with 10,000 samples.
Figure 4.
An Analysis of Statistical Power to Detect Associations across the Exome
(A) Sample sizes necessary to detect associations for a binary trait across the exome.
(B) Sample sizes for a quantitative trait.
Results from the SKAT, CMC, and BRV rare-variant association tests are shown in blue, green, and red, respectively.
Our simulations suggest that even when implementing aggregate rare-variant association methods, even larger sample sizes than those used in large-scale common-variant GWASs will be required to detect associations of modest effects using rare-variant association methods. Due to the differences in allelic architecture between AAs and EAs, sample size calculations for gene-level associations will need to be distinct between these two major ethnic groups as well.
In addition to our simulations with aggregate rare-variant association tests, we sought to quantifiably estimate the additional power obtained from the extreme trait design compared to random ascertainment. If 10,000 samples were selected from the extremes of the quantitative trait values for a cohort of 220,000 individuals, which is the equivalent size of ESP, an association could be detected at an exome-wide significance of 2.5 × 10−6 using the aggregate rare-variant association test for ∼80% of the genes in the exome where the effect size is 0.35 σ per each missense, nonsense, and splice site rare variant, with MAF < 1%. This represents a significant increase in power compared to analyzing 10,000 randomly ascertained samples, where an association could be detected for only < 1.0% of genes. We also found that the increase in power is most significant as the QT threshold goes from 1% to 5%, and for quantitative trait thresholds greater than 5%, although the sample sizes are larger, the power gains are marginal (Figure S3). Thus, if the underlying cohort is large enough to permit extreme sampling beyond the 5th and 95th percentiles in sufficient numbers, and the focus of the study is on a single quantitative trait, we recommend an extreme trait sampling design to boost power for detecting associations.
Discussion
Based on the experiences of the ESP and several similar recent WES projects, data generation will not represent a major technical hurdle for future sequencing-based studies of rare-variant associations. Nonetheless, as throughput continues to increase with decreasing sequencing costs, the data-management, variant-calling, QC, and analysis of these data will continue to pose challenges to the scientific community. Through the efforts of hundreds of investigators, the ESP helped establish best practices to turn terabytes of raw sequence data into genetic discoveries for complex traits and diseases.
There were a number of issues involved in having two separate sequencing facilities process and sequence the DNA for this project. The main advantages were competition and innovation: both sequencing centers were actively involved in optimizing their capture and sequencing protocols that led to improvements in coverage and data quality. However, differences between centers due to capture re-agents and analysis strategies created batch effects that we had to control for in the downstream analyses. Although the use of joint-calling over all samples mitigated some of these effects, in retrospect, the analysis pipeline would have been benefitted from uniform alignment and processing strategies, e.g., use of the same capture array at the two centers. Our experience highlights the importance of good experimental design; for instance, balancing case and control subjects across sequencing centers. In order to control type I and II errors, our genetic association analysis incorporated the study design, by including dummy variables to represent different sequencing centers, capture re-agents, and the source and ascertainment of samples.
With improvements in capture re-agents and consistency in coverage across the exome, we anticipate that future WES projects may be able to successfully sequence at lower depth. Specifically, an average depth per variant of about 25× appears to be a sweet spot where variant sites are covered at about the same depth as invariant sites (see Figure S2). This is slightly less than the recommended read depth of 30×, which is used for whole-genome sequencing and which produces substantially more even coverage than exome sequencing. Indeed, one or our rationale for choosing 90× read depth was to provide 80% coverage of the target bases with at least 20× read depth.
As more sequence data continues to be generated, studies will inevitably encounter a situation similar to ESP, where most observed variants are rare and population specific. In order to detect phenotypic associations from these data, the ESP pioneered several approaches for increasing statistical power. Samples were drawn from extremes of continuous traits and early-onset cases of complex diseases, special statistical methods were used to leverage the extreme trait data, rare variants were aggregated into larger units of analysis (i.e., genes), and through imputation and genotyping, large sample sizes were utilized for both discovery and replication.
Importantly, the ESP data represent the largest single collection of AA exomes to date. The AA exomes in the ESP generated multiple discoveries that would have been impossible to detect in US populations of European ancestry.20, 23 Indeed, AAs had on average a greater number of variant sites than EAs and a larger proportion of rare variants were exclusive to AAs compared to EAs. Although AAs are traditionally an under-studied population in human genetics, the ESP showed that the genetic diversity in AA genomes can be harnessed for uncovering rare-variant associations. In particular, variants in APOC3 were identified in both EAs and AAs and were associated with lower triglyceride levels in both ethnicities. Our work with imputation of the ESP AA sequence data also identified several variants that were monomorphic in EAs but reached exome-wide significance in AAs.
The sample selection of ESP presented distinct challenges: (1) heterogeneity between cohorts introduced noise that could not be entirely mitigated through phenotype harmonization; and (2) a simpler design focused on fewer traits with larger sample sizes may have yielded more discoveries. Nonetheless, our findings from AAs point to a major advantage of a heterogeneous sample of multiple ancestry groups, focused on a number of different traits. By including data from large, deeply phenotyped, US cohort studies, we were able to scan 71 different traits for genetic associations in two major ancestry groups. And though it complicated our analyses of both primary and secondary traits, we were able to sample from the extremes of multiple large cohorts, providing us with much more extreme trait values than if we had sampled from a single cohort. Consequently when results are based on an extreme sampling design, caution should be used in generalizing the results to the larger population, because they may be different from the extremes in systematic and unanticipated ways.
Incidental Findings
Prior to the ESP, no large-scale study had generated sufficient quantities of protein-coding sequence variants to enable the estimation of the number of medically actionable genetic variants per individual. Both an initial (based on 500 EA and AA participants) and a final (based on 6,503 participants) analysis of the ESP data provided robust estimates of the carrier frequency of adults with high-penetrance actionable or likely pathogenic variants.38, 39 The ESP data also provided estimates of carrier burden for complex traits such as age-related macular degeneration and drug response.40 These studies from ESP demonstrated the many challenges in variant classification and association with heterogeneous human disease burden. Future sequencing studies will need to grapple with these challenges as more populations are studied and the data expand outside of protein-coding regions.
Statistical Considerations with Association Testing
Due to its unprecedented scale and unique study design, the ESP prompted development of statistical methods and software to handle both extreme trait sampling and rare variant association testing. Quantitative trait data that have been generated by an extreme trait design (as in the ESP) are not normally distributed and should not be analyzed with standard methods. Modifications to likelihood-based approaches (such as conditional likelihood) can overcome the inherent bias in such a design.41 Secondary traits (traits that were not used as the basis of sample selection) from an extreme trait design likewise require special consideration.42
Using the ESP Data
The ESP data are available to investigators through the NIH database of genotypes and phenotypes (dbGaP). As a cautionary note, the ESP variant data should not be used as convenience controls for rare variant association testing. Doing so may significantly inflate association signals. If the ESP data or any other data from sequence-based studies are to be combined with sequence data from other projects, we recommend recalling all genotypes from the underlying BAM files using multi-sample calling, in order to avoid batch effects.
In our analytic pipeline for analyzing the ESP data, we removed related individuals in order to satisfy the assumption that the observations are independent in our regression framework. This required removal of only a few individuals and at the time, family-based methods for rare-variant association analysis using linear mixed models were not available. The advantages of mixed models are that all individuals can be retained in the analysis, and type I inflations due to either relatedness or cryptic relatedness are avoided.
Implications for Future Studies
As new resources emerge (e.g., data from the Encyclopedia of DNA Elements [ENCODE] and Roadmap projects) for interpreting DNA variation outside of coding regions, and as sequencing costs continue to decline,43 sequencing-based studies will not be limited to the exome. However, the exome offers a natural unit of analysis (i.e., a gene) for aggregate rare-variant association methods. It is unclear how best to aggregate association signals outside of coding regions. Will the genetic effects in enhancers, promoters, and other elements related to gene regulation be detectable by the same methods that were used for the exome? For future WGS studies to uncover aggregate signals outside of coding regions, methods development in this area will be crucial. In addition, many of the known loci that underlie complex diseases are located in regulatory elements outside of coding regions.44 Our experiences in the ESP confirm that for many traits and diseases, collaboration with other sequencing consortia (e.g., CHARGE, T2DGENES) will be necessary to accumulate the tens of thousands of samples required to detect associations with low-frequency and rare variants.26, 45 Imputation will also play an important role in future studies. The advantages of using a study-specific ESP reference panel for imputing rare variants appear to generalize to the whole genome.46, 47 With larger reference panels for imputation such as from the Haplotype Reference Consortium (HRC),48 the ability to impute rare variants across the genome with higher accuracy will continue to improve; current estimates suggest the HRC panel can impute variants to 0.1% MAF.
Just as in GWASs of common variants, the human genetics community is coalescing around the notion that the path to discovering insights into the biological mechanisms that underlie complex diseases is through data sharing and large-scale consortia. With the assistance of the NCBI’s database of genotypes and phenotypes (dbGaP), the ESP was a pioneer in data sharing and rapid analysis of large-scale sequence data. As new data continue to be generated for the study of complex trait genetics, this model of large-scale collaboration and data sharing should be emulated.
Consortia
The NHLBI GO Exome Sequencing Project consists of the following individuals. From BroadGO: Stacey B. Gabriel, David M. Altshuler, Gonçalo R. Abecasis, Hooman Allayee, Sharon Cresci, Mark J. Daly, Paul I. W. de Bakker, Mark A. DePristo, Ron Do, Peter Donnelly, Deborah N. Farlow, Tim Fennell, Kiran Garimella, Stanley L. Hazen, Youna Hu, Daniel M. Jordan, Goo Jun, Sekar Kathiresan, Hyun Min Kang, Adam Kiezun, Guillaume Lettre, Bingshan Li, Mingyao Li, Christopher H. Newton-Cheh, Sandosh Padmanabhan, Gina Peloso, Sara Pulit, Daniel J. Rader, David Reich, Muredach P. Reilly, Manuel A. Rivas, Steve Schwartz, Laura Scott, David S. Siscovick, John A. Spertus, Nathan O. Stitziel, Nina Stoletzki, Shamil R. Sunyaev, Benjamin F. Voight, and Cristen J. Willer. From HeartGO: Stephen S. Rich, Ermeg Akylbekova, Larry D. Atwood, Christie M. Ballantyne, Maja Barbalic, R. Graham Barr, Emelia J. Benjamin, Joshua Bis, Eric Boerwinkle, Donald W. Bowden, Jennifer Brody, Matthew Budoff, Greg Burke, Sarah Buxbaum, Jeff Carr, Donna T. Chen, Ida Y. Chen, Wei-Min Chen, Pat Concannon, Jacy Crosby, L. Adrienne Cupples, Ralph D’Agostino, Anita L. DeStefano, Albert Dreisbach, Josée Dupuis, J. Peter Durda, Jaclyn Ellis, Aaron R. Folsom, Myriam Fornage, Caroline S. Fox, Ervin Fox, Vincent Funari, Santhi K. Ganesh, Julius Gardin, David Goff, Ora Gordon, Wayne Grody, Myron Gross, Xiuqing Guo, Ira M. Hall, Nancy L. Heard-Costa, Susan R. Heckbert, Nicholas Heintz, David M. Herrington, DeMarc Hickson, Jie Huang, Shih-Jen Hwang, David R. Jacobs, Nancy S. Jenny, Andrew D. Johnson, Craig W. Johnson, Steven Kawut, Richard Kronmal, Raluca Kurz, Ethan M. Lange, Leslie A. Lange, Martin G. Larson, Mark Lawson, Cora E. Lewis, Daniel Levy, Dalin Li, Honghuang Lin, Chunyu Liu, Jiankang Liu, Kiang Liu, Xiaoming Liu, Yongmei Liu, William T. Longstreth, Cay Loria, Thomas Lumley, Kathryn Lunetta, Aaron J. Mackey, Rachel Mackey, Ani Manichaikul, Taylor Maxwell, Barbara McKnight, James B. Meigs, Alanna C. Morrison, Solomon K. Musani, Josyf C. Mychaleckyj, Jennifer A. Nettleton, Kari North, Christopher J. O’Donnell, Daniel O’Leary, Frank S. Ong, Walter Palmas, James S. Pankow, Nathan D. Pankratz, Shom Paul, Marco Perez, Sharina D. Person, Joseph Polak, Wendy S. Post, Bruce M. Psaty, Aaron R. Quinlan, Leslie J. Raffel, Vasan S. Ramachandran, Alexander P. Reiner, Kenneth Rice, Jerome I. Rotter, Jill P. Sanders, Pamela Schreiner, Sudha Seshadri, Steve Shea, Stephen Sidney, Kevin Silverstein, David S. Siscovick, Nicholas L. Smith, Nona Sotoodehnia, Asoke Srinivasan, Herman A. Taylor, Kent Taylor, Fridtjof Thomas, Russell P. Tracy, Michael Y. Tsai, Kelly A. Volcik, Chrstina L Wassel, Karol Watson, Gina Wei, Wendy White, Kerri L. Wiggins, Jemma B. Wilk, O. Dale Williams, Gregory Wilson, James G. Wilson, Phillip Wolf, and Neil A. Zakai. From ISGS and SWISS: John Hardy, James F. Meschia, Michael Nalls, Stephen S. Rich, Andrew Singleton, and Brad Worrall. From LungGO: Michael J. Bamshad, Kathleen C. Barnes, Ibrahim Abdulhamid, Frank Accurso, Ran Anbar, Terri Beaty, Abigail Bigham, Phillip Black, Eugene Bleecker, Kati Buckingham, Anne Marie Cairns, Wei-Min Chen, Daniel Caplan, Barbara Chatfield, Aaron Chidekel, Michael Cho, David C. Christiani, James D. Crapo, Julia Crouch, Denise Daley, Anthony Dang, Hong Dang, Alicia De Paula, Joan DeCelie-Germana, Allen Dozor, Mitch Drumm, Maynard Dyson, Julia Emerson, Mary J. Emond, Thomas Ferkol, Robert Fink, Cassandra Foster, Deborah Froh, Li Gao, William Gershan, Ronald L. Gibson, Elizabeth Godwin, Magdalen Gondor, Hector Gutierrez, Nadia N. Hansel, Paul M. Hassoun, Peter Hiatt, John E. Hokanson, Michelle Howenstine, Laura K. Hummer, Seema M. Jamal, Jamshed Kanga, Yoonhee Kim, Michael R. Knowles, Michael Konstan, Thomas Lahiri, Nan Laird, Christoph Lange, Lin Lin, Xihong Lin, Tin L. Louie, David Lynch, Barry Make, Thomas R. Martin, Steve C. Mathai, Rasika A. Mathias, John McNamara, Sharon McNamara, Deborah Meyers, Susan Millard, Peter Mogayzel, Richard Moss, Tanda Murray, Dennis Nielson, Blakeslee Noyes, Wanda O’Neal, David Orenstein, Brian O’Sullivan, Rhonda Pace, Peter Pare, H. Worth Parker, Mary Ann Passero, Elizabeth Perkett, Adrienne Prestridge, Nicholas M. Rafaels, Bonnie Ramsey, Elizabeth Regan, Clement Ren, George Retsch-Bogart, Michael Rock, Antony Rosen, Margaret Rosenfeld, Ingo Ruczinski, Andrew Sanford, David Schaeffer, Cindy Sell, Daniel Sheehan, Edwin K. Silverman, Don Sin, Terry Spencer, Jackie Stonebraker, Holly K. Tabor, Laurie Varlotta, Candelaria I. Vergara, Robert Weiss, Fred Wigley, Robert A. Wise, Fred A. Wright, Mark M. Wurfel, Robert Zanni, and Fei Zou. From SeattleGO: Deborah A. Nickerson, Mark J. Rieder, Phil Green, Jay Shendure, Joshua M. Akey, Michael J. Bamshad, Kristine L. Bucasas, Carlos D. Bustamante, David R. Crosslin, Evan E. Eichler, P. Keolu Fox, Wenqing Fu, Adam Gordon, Simon Gravel, Gail P. Jarvik, Jill M. Johnsen, Mengyuan Kan, Eimear E. Kenny, Jeffrey M. Kidd, Fremiet Lara-Garduno, Suzanne M. Leal, Dajiang J. Liu, Sean McGee, Timothy D. O’Connor, Bryan Paeper, Peggy D. Robertson, Joshua D. Smith, Jeffrey C. Staples, Jacob A. Tennessen, Emily H. Turner, Gao Wang, and Qian Yi. From WHISP: Rebecca Jackson, Kari North, Ulrike Peters, Christopher S. Carlson, Garnet Anderson, Hoda Anton-Culver, Themistocles L. Assimes, Paul L. Auer, Shirley Beresford, Chris Bizon, Henry Black, Robert Brunner, Robert Brzyski, Dale Burwen, Bette Caan, Cara L. Carty, Rowan Chlebowski, Steven Cummings, J. David Curb, Charles B. Eaton, Leslie Ford, Nora Franceschini, Stephanie M. Fullerton, Margery Gass, Nancy Geller, Gerardo Heiss, Barbara V. Howard, Li Hsu, Carolyn M. Hutter, John Ioannidis, Shuo Jiao, Karen C. Johnson, Charles Kooperberg, Lewis Kuller, Andrea LaCroix, Kamakshi Lakshminarayan, Dorothy Lane, Ethan M. Lange, Leslie A. Lange, Norman Lasser, Erin LeBlanc, Cora E. Lewis, Kuo-Ping Li, Marian Limacher, Dan-Yu Lin, Benjamin A. Logsdon, Shari Ludlam, JoAnn E. Manson, Karen Margolis, Lisa Martin, Joan McGowan, Keri L. Monda, Jane Morley Kotchen, Lauren Nathan, Judith Ockene, Mary Jo O’Sullivan, Lawrence S. Phillips, Ross L. Prentice, Alexander P. Reiner, John Robbins, Jennifer G. Robinson, Jacques E. Rossouw, Haleh Sangi-Haghpeykar, Gloria E. Sarto, Sally Shumaker, Michael S. Simon, Marcia L. Stefanick, Evan Stein, Hua Tang, Kira C. Taylor, Cynthia A. Thomson, Timothy A. Thornton, Linda Van Horn, Mara Vitolins, Jean Wactawski-Wende, Robert Wallace, Sylvia Wassertheil-Smoller, and Donglin Zeng. From NHLBI GO ESP Project Team: Deborah Applebaum-Bowden, Michael Feolo, Weiniu Gan, Dina N. Paltoo, Jacques E. Rossouw, Phyliss Sholinsky, and Anne Sturcke.
Acknowledgments
We acknowledge the support of the National Heart, Lung and Blood Institute (NHLBI), the contributions of the research institutions that participated in this study, and the study investigators, field staff, and study participants who created this resource for biomedical research. Funding for the GO (Grand Opportunity) Exome Sequencing Project was provided by NHLBI grants RC2 HL-103010 (Heart GO), RC2 HL-102923 (Lung GO), and RC2 HL-102924 (WHISP). The exome sequencing was supported by NHLBI grants RC2 HL-102925 (Broad GO) and RC2 HL-102926 (Seattle GO).
Published: September 22, 2016
Footnotes
Supplemental Data include Supplemental Notes containing descriptions of the sample selection criteria, phenotype definitions, and contributing studies, three figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.08.012.
Web Resources
Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE), http://www.chargeconsortium.com
ENCODE, https://www.encodeproject.org/
Exome Chip Design, http://genome.sph.umich.edu/wiki/Exome_Chip_Design
NHLBI Exome Sequencing Project, https://esp.gs.washington.edu/drupal/
OMIM, http://www.omim.org/
Precision Medicine Initiative (PMI), NIH, https://www.nih.gov/precision-medicine-initiative-cohort-program
Roadmap, http://www.roadmapepigenomics.org/
SeattleSeq Annotation Server, http://snp.gs.washington.edu/SeattleSeqAnnotation134/
T2D-GENES Consortium, https://t2d-genes.sph.umich.edu
The Haplotype Reference Consortium, http://www.haplotype-reference-consortium.org/home
Trans-Omics for Precision Medicine (TOPMed) Program, https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed
Supplemental Data
References
- 1.Collins F.S., Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jun G., Wing M.K., Abecasis G.R., Kang H.M. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosby J., Peloso G.M., Auer P.L., Crosslin D.R., Stitziel N.O., Lange L.A., Lu Y., Tang Z.Z., Zhang H., Hindy G., TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auer P.L., Wang G., Leal S.M. Testing for rare variant associations in the presence of missing data. Genet. Epidemiol. 2013;37:529–538. doi: 10.1002/gepi.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheidecker S., Etard C., Haren L., Stoetzel C., Hull S., Arno G., Plagnol V., Drunat S., Passemard S., Toutain A. Mutations in TUBGCP4 alter microtubule organization via the γ-tubulin ring complex in autosomal-recessive microcephaly with chorioretinopathy. Am. J. Hum. Genet. 2015;96:666–674. doi: 10.1016/j.ajhg.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G.T., Peng B., Leal S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 20.Auer P.L., Johnsen J.M., Johnson A.D., Logsdon B.A., Lange L.A., Nalls M.A., Zhang G., Franceschini N., Fox K., Lange E.M. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du M., Auer P.L., Jiao S., Haessler J., Altshuler D., Boerwinkle E., Carlson C.S., Carty C.L., Chen Y.D., Curtis K., National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project Whole-exome imputation of sequence variants identified two novel alleles associated with adult body height in African Americans. Hum. Mol. Genet. 2014;23:6607–6615. doi: 10.1093/hmg/ddu361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan Q., Liu E.Y., Auer P.L., Zhang G., Lange E.M., Jun G., Bizon C., Jiao S., Buyske S., Franceschini N. Imputation of coding variants in African Americans: better performance using data from the exome sequencing project. Bioinformatics. 2013;29:2744–2749. doi: 10.1093/bioinformatics/btt477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnsen J.M., Auer P.L., Morrison A.C., Jiao S., Wei P., Haessler J., Fox K., McGee S.R., Smith J.D., Carlson C.S., NHLBI Exome Sequencing Project Common and rare von Willebrand factor (VWF) coding variants, VWF levels, and factor VIII levels in African Americans: the NHLBI Exome Sequencing Project. Blood. 2013;122:590–597. doi: 10.1182/blood-2013-02-485094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazave E., Ma L., Chang D., Coventry A., Gao F., Muzny D., Boerwinkle E., Gibbs R.A., Sing C.F., Clark A.G., Keinan A. Neutral genomic regions refine models of recent rapid human population growth. Proc. Natl. Acad. Sci. USA. 2014;111:757–762. doi: 10.1073/pnas.1310398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng B. Reproducible simulations of realistic samples for next-generation sequencing studies using Variant Simulation Tools. Genet. Epidemiol. 2015;39:45–52. doi: 10.1002/gepi.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kryukov G.V., Shpunt A., Stamatoyannopoulos J.A., Sunyaev S.R. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl. Acad. Sci. USA. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G.T., Li B., Santos-Cortez R.P., Peng B., Leal S.M. Power analysis and sample size estimation for sequence-based association studies. Bioinformatics. 2014;30:2377–2378. doi: 10.1093/bioinformatics/btu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange L.A., Hu Y., Zhang H., Xue C., Schmidt E.M., Tang Z.Z., Bizon C., Lange E.M., Smith J.D., Turner E.H., NHLBI Grand Opportunity Exome Sequencing Project Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auer P.L., Nalls M., Meschia J.F., Worrall B.B., Longstreth W.T., Jr., Seshadri S., Kooperberg C., Burger K.M., Carlson C.S., Carty C.L., National Heart, Lung, and Blood Institute Exome Sequencing Project Rare and coding region genetic variants associated with risk of ischemic stroke: The NHLBI Exome Sequence Project. JAMA Neurol. 2015;72:781–788. doi: 10.1001/jamaneurol.2015.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Do R., Stitziel N.O., Won H.H., Jørgensen A.B., Duga S., Angelica Merlini P., Kiezun A., Farrall M., Goel A., Zuk O., NHLBI Exome Sequencing Project Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emond M.J., Louie T., Emerson J., Zhao W., Mathias R.A., Knowles M.R., Wright F.A., Rieder M.J., Tabor H.K., Nickerson D.A., National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project. Lung GO Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat. Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stitziel N.O., Won H.H., Morrison A.C., Peloso G.M., Do R., Lange L.A., Fontanillas P., Gupta N., Duga S., Goel A., Myocardial Infarction Genetics Consortium Investigators Inactivating mutations in NPC1L1 and protection from coronary heart disease. N. Engl. J. Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naik R.P., Derebail V.K., Grams M.E., Franceschini N., Auer P.L., Peloso G.M., Young B.A., Lettre G., Peralta C.A., Katz R. Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. JAMA. 2014;312:2115–2125. doi: 10.1001/jama.2014.15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 38.Amendola L.M., Dorschner M.O., Robertson P.D., Salama J.S., Hart R., Shirts B.H., Murray M.L., Tokita M.J., Gallego C.J., Kim D.S. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor H.K., Auer P.L., Jamal S.M., Chong J.X., Yu J.H., Gordon A.S., Graubert T.A., O’Donnell C.J., Rich S.S., Nickerson D.A., Bamshad M.J., NHLBI Exome Sequencing Project Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am. J. Hum. Genet. 2014;95:183–193. doi: 10.1016/j.ajhg.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin D.Y., Zeng D., Tang Z.Z. Quantitative trait analysis in sequencing studies under trait-dependent sampling. Proc. Natl. Acad. Sci. USA. 2013;110:12247–12252. doi: 10.1073/pnas.1221713110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu D.J., Leal S.M. A unified method for detecting secondary trait associations with rare variants: application to sequence data. PLoS Genet. 2012;8:e1003075. doi: 10.1371/journal.pgen.1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayden E.C. Technology: The $1,000 genome. Nature. 2014;507:294–295. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 44.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiezun A., Garimella K., Do R., Stitziel N.O., Neale B.M., McLaren P.J., Gupta N., Sklar P., Sullivan P.F., Moran J.L. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deelen P., Menelaou A., van Leeuwen E.M., Kanterakis A., van Dijk F., Medina-Gomez C., Francioli L.C., Hottenga J.J., Karssen L.C., Estrada K., Genome of Netherlands Consortium Improved imputation quality of low-frequency and rare variants in European samples using the ‘Genome of The Netherlands’. Eur. J. Hum. Genet. 2014;22:1321–1326. doi: 10.1038/ejhg.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pistis G., Porcu E., Vrieze S.I., Sidore C., Steri M., Danjou F., Busonero F., Mulas A., Zoledziewska M., Maschio A. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 2015;23:975–983. doi: 10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016 doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.