Summary
The RNA-guided endonuclease Cas9 from Streptococcus pyogenes (SpCas9) forms the core of a powerful genome editing technology. DNA cleavage by SpCas9 is dependent on the presence of a 5’-NGG-3’ protospacer adjacent motif (PAM) in the target DNA, restricting the choice of targetable sequences. To address this limitation, artificial SpCas9 variants with altered PAM specificities have recently been developed. Here we report crystal structures of the VQR, EQR, and VRER SpCas9 variants bound to target DNAs containing their preferred PAM sequences. The structures reveal that the non-canonical PAMs are recognized by an induced fit mechanism. Besides mediating sequence-specific base recognition, the amino acid substitutions introduced in the SpCas9 variants facilitate conformational remodeling of the PAM region of the bound DNA. Guided by the structural data, we developed a SpCas9 variant that specifically recognizes NAAG PAMs. Taken together, these studies inform further development of Cas9-based genome editing tools.
Introduction
The RNA-guided endonuclease Cas9 is the defining component of prokaryotic type II CRISPR systems, in which it provides sequence-specific targeting of DNA viruses and mobile genetic elements (Barrangou, 2014; Sorek et al., 2013). The enzyme associates with an RNA guide structure consisting of a mature CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA), forming a ribonucleoprotein complex that binds DNA sequences complementary to a 5’-terminal 20-nt guide sequence in the crRNA (Jinek et al., 2012; Karvelis et al., 2013). Upon RNA-DNA hybridization and formation of an R-loop structure, the Cas9 RuvC and HNH endonuclease domains catalyze site-specific cleavage of the two strands in the target DNA, generating a double-strand break (DSB) (Gasiunas et al., 2012; Jinek et al., 2012). DNA cleavage is strictly dependent on the presence of a protospacer adjacent motif (PAM) immediately downstream of the target site and requires a near-perfect match between the target DNA and a “seed” region in the crRNA guide sequence (Gasiunas et al., 2012; Jinek et al., 2012).
Structural studies employing X-ray crystallography and electron microscopy have revealed the molecular mechanism of Cas9 (Anders et al., 2014; Jiang et al.; 2016; 2015; Jinek et al., 2014; Nishimasu et al., 2014). Guide RNA binding to apo-Cas9 results in a dramatic structural transition from an open state to a closed conformation in which the enzyme is primed to engage its DNA target (Jiang et al., 2015; Jinek et al., 2014). PAM recognition is thought to be the initial step in target DNA binding, with the PAM acting as a licensing element to trigger strand separation in the target DNA and formation of the crRNA-target DNA heteroduplex (Sternberg et al., 2014). Crystal structures of Cas9 enzymes bound to PAM-containing target DNAs have revealed the mechanism of PAM recognition. In the Streptococcus pyogenes Cas9 (SpCas9), the canonical 5’-NGG-3’ PAM is read out by a pair of arginine residues inserted into the major groove of the PAM DNA duplex (Anders et al., 2014). Staphylococcus aureus Cas9 also employs a major-groove recognition mechanism involving direct and water-mediated hydrogen bonding interactions with the 5’-NNGRRT-3’ PAM (Nishimasu et al., 2015). Upon target binding and R-loop formation, the Cas9 enzyme undergoes a further conformational rearrangement that positions the HNH nuclease domain for cleavage of the target DNA strand (Jiang et al., 2016; Sternberg et al., 2015).
In recent years, Cas9 nucleases have been repurposed to develop a transformative genome editing technology (Doudna and Charpentier, 2014). Programmed by dual crRNA-tracrRNA guides or chimeric single-molecule guide RNAs (sgRNAs), Cas9 can efficiently generate DSBs at specific genomic loci in eukaryotic cells (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013b). These in turn induce DNA repair mechanisms that can be exploited to introduce genetic modifications in the vicinity of the Cas9-generated DSB. Cas9-mediated genome editing has been demonstrated in numerous organisms and cell types and holds great promise for clinical and industrial applications (Barrangou and May, 2015; Hsu et al., 2014). Catalytically inactive variants of Cas9 have also been repurposed for RNA-guided gene expression control and for cellular imaging applications (Chen et al., 2013; Gilbert et al., 2013; Konermann et al., 2015; Mali et al., 2013a). The SpCas9 endonuclease is currently the most widely used Cas9 enzyme. However, its utility is restricted to target sites juxtaposed to the canonical 5’-NGG-3’ motif. To address this inherent limitation and expand the spectrum of targetable genomic sites, engineered variants of SpCas9 have recently been developed by utilizing a bacterial selection-based directed evolution to recognize alternative PAMs (Kleinstiver et al., 2015b). The VQR (D1135V/R1335Q/T1337R) and EQR (D1135E/R1335Q/T1337R) variants primarily recognize 5’-NGAN-3’ and 5’-NGNG-3’ PAMs while the VRER variant (D1135V/G1218R/R1335E/T1337R) is specific for the 5’-NGCG-3’ motif.
To obtain structural insights into the molecular mechanisms underlying PAM recognition in engineered SpCas9 proteins, we determined the crystal structures of the VQR, EQR, and VRER Cas9 variants in complexes with sgRNA guides and target DNAs containing their preferred PAMs. The structures reveal that besides providing chemical moieties for sequence-specific interactions with the PAM bases, the amino acid substitutions introduced in these variants simultaneously induce structural rearrangements in the bound DNA that accommodate those interactions. We subsequently used these structures to guide the engineering of a SpCas9 variant specific for 5’-NAAG-3’ PAMs. Together, these results provide a structural framework for the continued development of Cas9-based genome editing technologies.
Results
Crystal Structures of VQR, EQR, and VRER Cas9 Variants
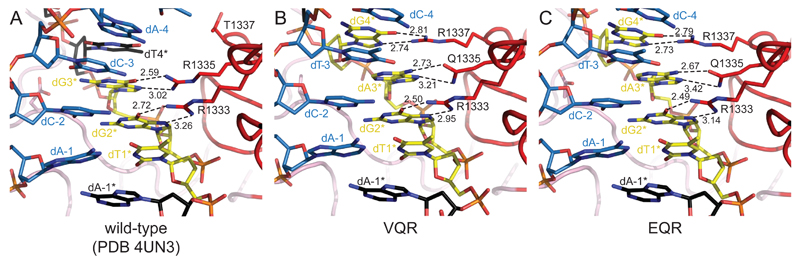
The molecular genome editing toolkit has recently been expanded by the engineering of three variants of the SpCas9 enzyme - VQR and EQR, which recognize NGAN and NGNG PAMs, and VRER, which recognizes the NGCG PAM (Kleinstiver et al., 2015b). Our previous structural work revealed the structural basis for the recognition of the canonical NGG PAM by wild-type (WT) SpCas9, which occurs in the major groove of the PAM and relies on sequence-specific contacts between arginine residues Arg1333 and Arg1335 and the first and second guanine base of the PAM, respectively (Anders et al., 2014). The two arginines project from the PAM-interacting motif ([PIM], residues 1,328-1,338), which forms a beta-strand within the C-terminal PAM-interacting domain of SpCas9 (residues 1,099-1,368). The common feature of the VQR, EQR, and VRER variants is the T1337R substitution. In WT SpCas9, Thr1337 is positioned adjacent to Arg1335 within the PIM and faces the major groove of the PAM DNA (Figure 1A). This suggests that the T1337R substitution could mediate sequence-specific recognition of an additional guanine base, which is supported by the observations that all three engineered variants display preference for a 4-nt PAM containing a guanine in the fourth position (Kleinstiver et al., 2015b). To elucidate the structural basis of PAM recognition by the artificial Cas9 variants, we sought to determine the atomic structures of their DNA-bound complexes by X-ray crystallography. To this end, we took advantage of a crystal form previously obtained for the WT SpCas9-sgRNA-target DNA complex in which neither the PAM motif itself nor the PAM-interacting domain of SpCas9 participate in crystal-packing interactions and can therefore accommodate a variety of DNA sequences and protein amino acid substitutions (Anders et al., 2014). To optimize DNA binding, we designed target DNAs containing a TGAG PAM sequence for VQR and EQR variants and TGCG PAM for the VRER variant (Figure S1). We determined the structure of the VRER variant complex at a resolution of 2.7 Å and an Rfree of 24.7 %, while the VQR and EQR variant complexes were solved at 2.50 Å and 2.68 Å resolution and with Rfree factor values of 25.4 % and 23.6 %, respectively (Table 1). The conformations of all three SpCas9 variants are almost identical to that of WT SpCas9. The VQR, EQR, and VRER variants superimpose with the structure of WT SpCas9 with root-mean-square deviations (rmsd) of 0.90 Å, 0.28 Å, and 0.24 Å, respectively. Within the C-terminal PAM-interacting domain, the superpositions yield respective rmsd values of 0.30 Å, 0.27 Å, and 0.26 Å for the VQR, EQR, and VRER variants, indicating that the amino acid substitutions introduced in these variants do not perturb the conformation of the PAM-interacting domain (Figure S2A and S2B). This implies that in these variants the PAM-interacting domains behave as rigid structural scaffolds that accommodate the binding of their cognate PAM sequences by induced fit.
Figure 1. TGAG PAM Recognition by VQR and EQR SpCas9 Variants.
(A) Zoom-in view of the major groove of the TGG PAM region of the DNA in the structure of the WT SpCas9-sgRNA-target DNA complex. PAM nucleotides are colored yellow. Target DNA strand is colored blue. Non-PAM nucleotides of the non-target DNA strand are colored black. Arginine residues making sequence-specific contacts with the PAM are shown in stick format. Numbers indicate interatomic distances in Å.
(B) Zoomed-in view of the TGAG PAM in the VQR variant complex.
(C) Zoomed-in view of the TGAG PAM in the EQR variant complex. Both (B) and (C) are shown in the same orientation as the WT SpCas9 complex in (A). Full sequences of the RNA guide and target DNA are provided in Figure S1.
Table 1. Crystallographic Data Collection and Refinement Statistics for Cas9-sgRNA-PAM DNA Complexes.
| Dataset (Cas9) | VQR | EQR | VRER |
|---|---|---|---|
| PAM sequence | NGAG | NGAG | NGCG |
| X-ray source | SLS PXIII | SLS PXIII | SLS PXIII |
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 176.6, 67.0, 187.2 | 177.7, 68.2, 188.2 | 177.5, 67.8, 187.7 |
| α, β, γ (°) | 90.0, 111.1, 90.0 | 90.0, 110.9, 90.0 | 90.0, 111.1, 90.0 |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 |
| Resolution (Å)* | 47.62–2.50 (2.65–2.50) | 48.25–2.68 (2.75–2.68) | 47.98–2.70 (2.77–2.70) |
| Rsym (%)* | 9.6 (77.0) | 12.3 (77.4) | 12.7 (89.5) |
| I/σI* | 11.9 (1.7) | 13.2 (2.2) | 13.2 (1.9) |
| Completeness (%)* | 99.4 (98.2) | 99.3 (91.7) | 99.9 (100.0) |
| Redundancy* | 3.5 (3.6) | 7.0 (6.5) | 7.0 (6.6) |
| Refinement | |||
| Resolution (Å) | 47.62–2.50 | 48.25–2.68 | 47.98–2.70 |
| No. reflections | 71,146 | 59,668 | 57,665 |
| Rwork / Rfree | 0.218 / 0.254 | 0.205 / 0.236 | 0.218 / 0.247 |
| No. atoms | |||
| Macromolecules | 13,198 | 13,289 | 13,349 |
| Ion | 15 | 15 | 15 |
| Water | 264 | 130 | 85 |
| B-factors | |||
| mean | 52.5 | 47.6 | 55.7 |
| Macromolecules | 52.7 | 47.7 | 55.7 |
| Ion | 64.2 | 70.7 | 74.7 |
| Water | 44.8 | 41.8 | 47.4 |
| Rmsds | |||
| Bond lengths (Å) | 0.014 | 0.004 | 0.004 |
| Bond angles (°) | 0.82 | 0.79 | 0.88 |
| Ramachandran plot | |||
| % favored | 98.2 | 97.2 | 95.1 |
| % allowed | 1.8 | 2.8 | 4.9 |
| % outliers | 0.0 | 0.1 | 0.0 |
| Molprobity | |||
| Clashscore | 10.4 | 9.5 | 11.2 |
Values in parentheses denote highest resolution shell
Structural Basis of Sequence-Specific NGAG Recognition by the VQR and EQR Variants
The structures of the VQR and EQR variant complexes show that their PAM binding modes are virtually identical. In both complexes, the side chains of Arg1333, Gln1335, and Arg1337 are inserted into the major groove of the PAM-containing portion of the bound DNA and form a triple stack that recognizes the NGAG PAM by sequence-specific hydrogen bonding interactions (Figures 1B and 1C). As in WT SpCas9, the side chain of Arg1333 forms bidentate hydrogen bonds with N7 and O6 of the guanine base in the second PAM position (dG2*). The amide group of Gln1335 contacts N7 and N6 of dA3* at the third PAM position, as expected. The Gln1335 side chain is further stabilized by a hydrogen bonding interaction with the neighboring Glu1219 (Figure S3). Finally, Arg1337 makes canonical bidentate hydrogen bonds with the major groove edge of dG4*. Base-specific recognition of a guanine base in the fourth PAM position is consistent with both VQR and EQR variants having highest preference for the NGAG PAM and displaying partial activity with other NGNG PAMs in PAM depletion and gene disruption assays in vivo (Kleinstiver et al., 2015b). Being located at the top of the stack, Arg1337 is partly solvent-exposed and not constrained by side chain packing. It is therefore possible for the side chain to adopt other conformations, allowing it to form hydrogen bonding interactions with other bases present in the fourth position. This could explain the previously observed promiscuity of the VQR and EQR variants toward other NGAN PAMs (Kleinstiver et al., 2015b). In vitro, both variants displayed highest activity with TGAG and TGCG PAMs and were also partially active with TGGT, TGTT and TAAG PAMs (Figure S4A).
NGAG PAM Binding Is Achieved by Distortion of the PAM-Containing DNA Strand
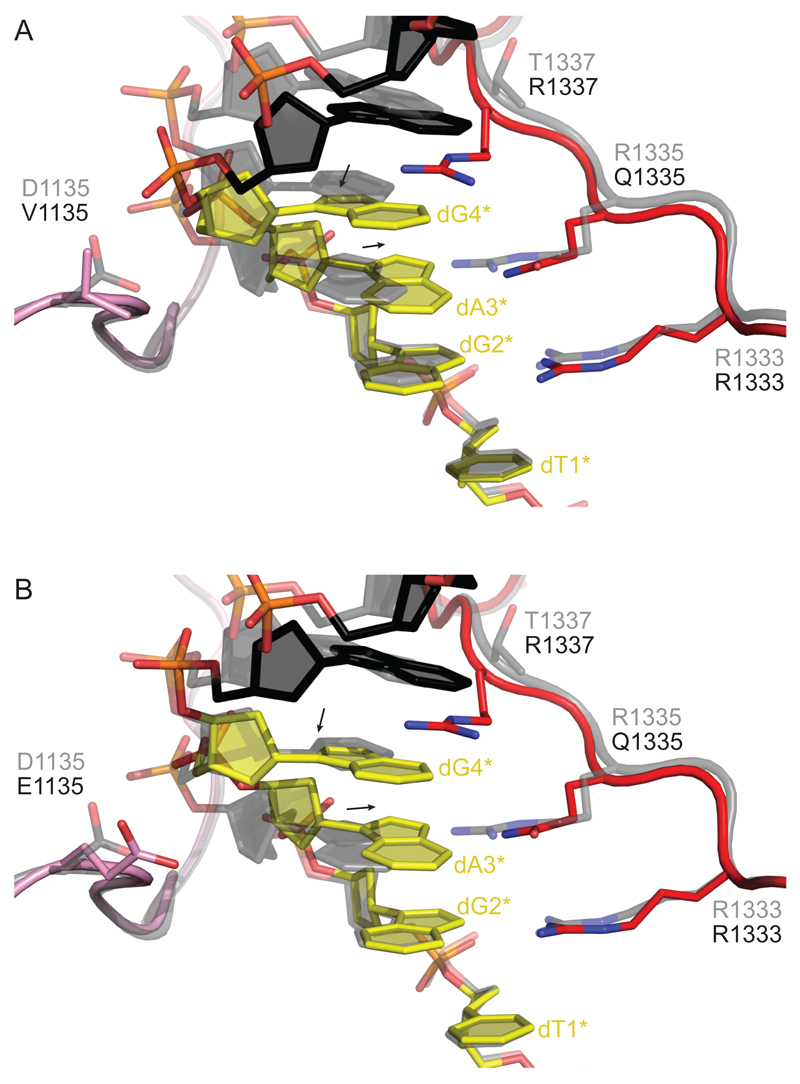
The substitution of Arg1335 with glutamine in SpCas9 was used as the starting point in the directed evolution experiments that led to the VQR and EQR variants, since the substitution alone is not sufficient to confer specificity for a NGA PAM (Kleinstiver et al., 2015b). A plausible explanation for this is that the Gln1335 side chain, being ~1.5 Å shorter than arginine, is not able to reach the major groove edge of the adenine base in the absence of further structural rearrangements in the Cas9-sgRNA-DNA complex. Superimposing the polypeptide backbones of the VQR and EQR variants with the structure of WT SpCas9 complex reveals only minor conformational differences between the polypeptide chains in those structures. This is true even when only considering the regions in immediate contact with the PAM (residues 1,104-1,138 and 1,321-1,338) (Figure S2A and S2B). These observations suggest that the binding of the non-canonical PAMs is achieved by structural remodeling of the PAM. Indeed, closer inspection of the VQR and EQR structures reveals that rather than causing major structural rearrangements in the PAM-interacting domain of SpCas9, the amino acid substitutions in these variants facilitate the recognition of the NGAG PAM by inducing and accommodating structural changes in the bound DNA. In both complexes, the PAM-containing non-target strand of the bound DNA is distorted such that the base and deoxyribose moieties of dA3* are shifted by approximately 1.3 Å toward the PAM-interacting motif, thereby bringing the major groove edge of dA3* within hydrogen bonding distance of Gln1335 (Figure 2). This is compensated by concomitant rearrangements in the deoxyribose-phosphate backbone of nucleotides dG4* and dA5* that displace the two nucleotides ~1.0 Å away from the PIM. This is in part necessary to accommodate the side chain of Arg1337 in the major groove of the DNA as it interacts with dG4*. The PAM distortion thus appears to be largely driven by sequence-specific hydrogen bonding interactions with Gln1335 and Arg1337. The function of the D1135V and D1135E substitutions in the VQR and EQR variants are less clear but presumably contribute to the accommodation of the PAM duplex in a distorted conformation. In the VQR variant, the deoxyribose moiety of dG4* is able to pack against the side chain of Val1135 via van der Waals contacts. Additionally, the lack of negative charge at the 1135 position might improve the energetics of DNA binding by reducing electrostatic repulsion. In the EQR variant, the glutamate side chain is able to adopt a conformation that avoids a steric clash with the DNA backbone in part due to a salt bridge interaction with the side chain of Arg114. However, the sterics and energetics of DNA binding might not be optimal, thus accounting for the lower DNA cleavage activity of the EQR variant observed in gene disruption assays in vivo (Kleinstiver et al., 2015b). Despite structural rearrangements in the PAM-containing non-target DNA strand, base pairing with the target (i.e., complementary) DNA strand is still maintained in part because the target DNA strand is not contacted by Cas9 within the PAM region. However, analysis of base pair parameters using the 3DNA server (Lu and Olson, 2008) reveals that the distortion of the deoxyribose-phosphate backbone results in increased buckling and opening of the dT(-3)-dA3* base pair (Table S1). Together, the structures of the VQR and EQR Cas9 variants reveal that NGAG PAM recognition is achieved by a combination of sequence-specific recognition of the PAM sequence coupled with an induced distortion of the DNA backbone in the PAM-containing DNA strand. The residues involved in sequence-specific interactions (i.e., Gln1335 and Arg1337) appear to be the major drivers of the structural rearrangement, while the Val1135 (in VQR) and Glu1135 (EQR) play auxiliary, accommodating roles. This is consistent with the observation that the R1335Q/T1337R variant is almost as active as the VQR or EQR variants in a cell-based EGFP gene disruption assay (Kleinstiver et al., 2015b).
Figure 2. The VQR and EQR Variants Induce a Structural Distortion in the PAM DNA.
(A) Close-up view of the superimposed structures of the VQR variant with WT SpCas9. The PAM-interacting regions of the VQR Cas9 variant (colored pink and red) are not perturbed (Figure S2). The non-target DNA strand is colored black with the PAM highlighted in yellow. The superimposed WT SpCas9 structure is shown in gray. Arrows indicate shifts of the base and deoxyribose moieties of dA3* and dG4* in the VQR variant relative to the WT SpCas9 structure.
(B) Close-up view of the EQR variant superimposed with WT SpCas9. Color coding is as in (A)
NGCG PAM Recognition by the VRER Variant
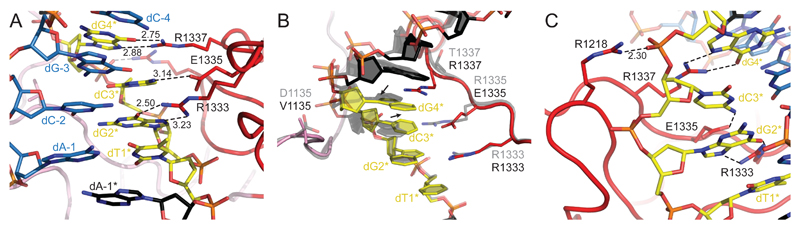
The structure of the VRER SpCas9 variant bound to a TGCG PAM-containing DNA reveals that this variant also relies on inducing a distortion of the non-target DNA strand to achieve sequence-specific recognition of the PAM bases. As in the VQR and EQR variants, the polypeptide backbone of the PAM-interacting domain in the VRER variant remains largely unperturbed by the amino acid substitutions (Figure S2C). The TGCG PAM is contacted in the major groove by the triple stack of Arg1333, Glu1335, and Arg1337 side chains, with Arg1333 and Arg1337 contacting dG2* and dG4*, respectively, by canonical sequence-specific hydrogen bonding interactions while the carboxyl group of Glu1335 forms a single hydrogen bond with the N4 of dC3* (Figure 3A). To enable these interactions to take place, the deoxyribose-phosphate backbone of the PAM-containing DNA strand undergoes a distortion that results in dC3* shifting toward and dG4* away from the PAM-interacting motif harboring the Arg1333-Glu1335-Arg1337 triple stack (Figure 3B), similar to the distortions observed in the VQR and EQR variants. This is accompanied by prominent buckling of the dG(-3)-dC3* base pair and increased twist of the bases (Table S1). As in the VQR variant, the D1135V substitution in the VRER Cas9 might contribute to steric accommodation of the structural distortions in the DNA. The introduction of an arginine residue at position 1,218 in the VRER variant in turn results in the formation of a salt bridge contact to the backbone phosphate group of dG4* (Figure 3C). The recognition of cytosine in the third PAM position by a single hydrogen bonding interaction to Glu1335, together with the juxtaposition of the carboxyl groups of Glu1335 and Glu1219 results in an energetically suboptimal arrangement, which likely relies on energetic compensation from the additional hydrogen bonding and electrostatic interactions mediated by Arg1337 and Arg1218. Consistent with this, the VRER variant has strict requirement for the presence of a guanine in the fourth position of the PAM (Kleinstiver et al., 2015b), while it is partially active with a TGAG PAM in vitro (Figure S4A).
Figure 3. NGCG Recognition and Induced Fit by the VRER SpCas9 Variant.
(A) Zoom-in view of the major groove of the TGCG PAM region of the DNA in the structure of the VRER variant complex. PAM nucleotides are colored yellow. The structure is depicted as the WT, VQR, and EQR SpCas9 complexes in Figure 1.
(B) Cartoon diagram of the TGCG PAM and the PAM-interacting regions of the VRER SpCas9, overlaid with the structure of the WT SpCas9 complex (gray). The bound DNA is depicted in the same manner as in Figure 2.
(C) Close-up view of the salt bridge interaction between Arg1218 and the phosphate group of dG4*.
Toward Structure-Guided Engineering of an NAAG-Specific SpCas9 Variant
Previous attempts to change the PAM specificity of SpCas9 to an NAA PAM by simply introducing arginine to glutamine substitutions at positions 1,333 and 1,335 were unsuccessful (Anders et al., 2014; Kleinstiver et al., 2015b), suggesting that additional amino acid substitutions would be required to induce conformational rearrangements in the bound DNA or in the PAM-interacting domain. Guided by our structural analysis of the VQR, EQR, and VRER SpCas9 variants, we attempted to expand the targeting potential of SpCas9 by developing a variant specific for NAAG PAMs. We reasoned that recognition of adenine and guanine bases in the third and fourth PAM positions, respectively, could be achieved by retaining the substitutions R1335Q, T1337R, and G1218R, while the R1333Q substitution would be required for sequence-specific recognition of an adenine at the second PAM positions. A SpCas9 protein harboring these four substitutions displayed very low or no activity on targets containing NAAN as well as NGNN PAMs (Figures S4B and S4C). We therefore sought to introduce additional substitutions that would facilitate hydrogen bonding contacts between Gln1333 and the major groove edge of the second PAM nucleotide by locally restructuring the polypeptide backbone of the PAM-interacting motif. Upstream of position 1,333 within the PIM, the side chain of Ile1331 packs against the hydrophobic interior of the PAM-interacting domain, while the peptide carbonyl group of Asp1332 is held in place by a hydrogen bonding interaction with the amide side chain group of Asn1286 (Figure 4A). To reposition the backbone of the beta strand, effectively pushing it deeper into the major groove of the PAM DNA, we introduced a bulky phenylalanine residue at position 1,331 and further substituted Asn1286 with a glutamine to maintain the hydrogen bonding interaction with the peptide backbone (Figure 4B). To improve the electrostatics of the interactions with the NAAG PAM DNA, we additionally substituted the aspartate residue at position 1,332 with a lysine. We generated the septuple mutant G1218R/N1286Q/I1331F/D1332K/R1333Q/R1335Q/T1337R (hereafter referred to as the QQR1 variant) and tested its endonuclease activity against linearized target plasmid DNAs containing either TGGT or TAAG PAMs. Whereas WT SpCas9 efficiently cleaved the TGGT-containing target and displayed no activity on the TAAG PAM, the opposite was observed for the QQR1 variant (Figure 4C). Further analysis confirmed that the QQR1 variant is highly specific for NAAG, as no other NAAN PAM supported efficient target DNA cleavage (Figures 4D, S4B, and S4C). The cleavage rate of the QQR1 variant was substantially slower than that of WT SpCas9 (Figure 4C), suggesting that further optimization by directed evolution will likely be needed to improve the catalytic activity of QQR1 SpCas9. Nonetheless, these results demonstrate the feasibility of structure-guided engineering of PAM specificities in Cas9 enzymes.
Figure 4. Structure-Guided Engineering of an NAAG-Specific QQR1 Variant.
(A) Close-up view of the PIM of the VQR variant bound to the non-target DNA strand. PAM nucleotides are highlighted in yellow. Hydrogen bonding interactions are indicated with dashed lines.
(B) Schematic model of amino acid substitutions introduced in the QQR1 variant in order to induce repositioning of the polypeptide backbone of the PAM-interacting motif and facilitate hydrogen bonding contacts between Gln1335 and an adenine base in the second PAM position.
(C) Nuclease activity assays of WT and QQR1 SpCas9 enzymes. WT and QQR1 SpCas9 were programmed using an identical sgRNA (sequence listed in Table S2) and incubated with linearized plasmids containing a target sequence adjacent to either TGGT or TAAG PAMs. Samples were taken at indicated time points and analyzed by agarose gel electrophoresis.
(D) PAM specificity of the QQR1 variant. WT and QQR1 SpCas9 proteins were incubated with a sgRNA and linearized plasmid DNAs harboring targets flanked by the indicated PAMs.
Discussion
By virtue of their programmability using short RNA molecules, Cas9 endonucleases have revolutionized genome editing. As an obligatory step in the DNA binding and cleavage mechanism of Cas9, PAM recognition ensures the fidelity of DNA targeting. This however constrains the endonuclease activity of Cas9 to genomic sites adjacent to cognate PAMs. Single-nucleotide resolution is highly desirable for a number of genome editing approaches; for example, precise insertions by homology-directed repair, targeted disruption of short genetic elements such as transcription factor or microRNA binding sites by non-homologous end joining, or allele-specific gene targeting. SpCas9, currently the most widely used Cas9 enzyme, is highly specific for NGG PAMs. One possibility to overcome the inherent limitation of its PAM specificity is to exploit other Cas9 orthologs with different PAM specificities, as has been done with Cas9 enzymes from Staphylococcus aureus (Kleinstiver et al., 2015b; Ran et al., 2015), Streptococcus thermophilus (Glemzaite et al., 2015; Müller et al., 2015), Neisseria meningitidis (Hou et al., 2013) or Brevibacillus laterosporus (Karvelis et al., 2015), or the class II crRNA-guided endonuclease Cpf1 (Zetsche et al., 2015). However, distinct Cas9 and Cpf1 orthologs invariably use distinct guide RNA structures. The other option is to utilize selection-based directed evolution strategies to engineer artificial SpCas9 variants with altered PAM specificities that would use the same guide RNA structures as the naturally occurring enzyme. The recent developments of the VQR, EQR, and VRER SpCas9 variants as well as the KKH variant of Staphylococcus aureus Cas9 represent the first steps in this direction (Kleinstiver et al., 2015a, 2015b).
In this study, we conducted structural analysis of the PAM recognition modes of engineered SpCas9 variants. We reveal that the PIMs in all three variants are nearly identical in their conformations to WT SpCas9 and function as rigid structural scaffolds. Sequence-specific PAM recognition occurs through induced fit achieved by structural distortions in the bound DNA. The specific amino acid substitutions introduced in each SpCas9 variant contribute to the induced-fit PAM recognition mechanism in three complementary ways: (i) by providing base-specific contacts to the PAM, (ii) by permitting steric accommodation of the induced distortions in the PAM DNA, and (iii) by improving the thermodynamics of DNA binding to compensate for the energetic cost of distorting the bound DNA and to offset sub-optimal interactions. Notably, by playing all three roles, the T1337R substitution effectively extends the PAM by one nucleotide and confers a preference (in VQR and EQR variants) or a strict requirement (in the VRER variant) for a guanine base in the fourth position of the PAM. The D1135V substitution in the VQR and VRER variants most likely contributes by enabling steric accommodation of the bound DNA and removing a potentially repulsive electrostatic interaction.
Collectively, our structural observations demonstrate that SpCas9, and specifically its PAM-interacting domain, provides a robust DNA binding platform that enables structural plasticity of PAM recognition, which could be exploited in the development of additional SpCas9 variants by combining both structure-guided computational design and selection-based directed evolution approaches. It is conceivable, however, that certain PAM sequences can only be recognized by mechanisms that also involve remodeling of the PAM-interacting domain of Cas9. By combining amino acid substitutions designed to effect restructuring of the PIM in SpCas9 with those mediating sequence-specific base recognition and inducing structural distortions in the bound DNA, we engineered a SpCas9 variant (QQR1) that displays preference for NAAG PAMs. We anticipate that further refinement of this variant will be needed to improve the kinetics of DNA cleavage for efficient genome editing. Collectively, our studies set the stage for ongoing structure-guided engineering of the Cas9 platform and will thus contribute toward the development of new genome editing tools and technologies.
Experimental Procedures
Guide RNA Generation
Guide RNAs were prepared by in vitro transcription using double-stranded DNA templates and T7 RNA polymerase as described previously (Anders and Jinek, 2014). Transcription templates were generated by PCR. All RNA and DNA oligonucleotide sequences are provided in Table S2. Transcribed guide RNAs were purified by gel electrophoresis on an 8 % denaturing (7 M urea) polyacrylamide gel followed by ethanol precipitation.
Mutagenesis
Point mutations in Streptococcus pyogenes Cas9 were introduced into pMJ806 or pMJ841 plasmids (Addgene #39312 and #39318, www.addgene.org) by inverse PCR and mutations were verified by DNA sequencing.
Protein Expression and Purification
WT and mutant SpCas9 proteins were expressed and purified as described for WT SpCas9 previously (Anders and Jinek, 2014; Anders et al., 2014). Cells were lysed in 20 mM Tris (pH 8.0), 500 mM NaCl, 5 mM imidazole (pH 8.0) by sonication. Proteins were purified by nickel-affinity and cation exchange chromatographic steps. For use in nuclease activity assays, proteins were further purified by size exclusion chromatography using a Superdex200 16/600 column (GE Healthcare) in 20 mM HEPES (pH 7.5) and 250 mM KCl. Purified proteins were concentrated to 2-4 mg/ml, flash frozen in liquid nitrogen, and stored at -80°C. For crystallization, Cas9-sgRNA-target DNA complexes were reconstituted by mixing Cas9 protein with sgRNA and pre-hybridized target DNA strands in a 1:1:2 ratio. Ribonucleoprotein complexes were purified in 20 mM HEPES (pH 7.5), 500 mM KCl, and 2 mM MgCl2 by size exclusion chromatography using a Superdex200 16/600 column. Purified complexes were concentrated to 4-8 mg/ml, flash frozen in liquid nitrogen, and stored at -80°C.
Crystallization and Structure Determination
Purified complexes were diluted to 1-2 mg/ml in 20 mM HEPES (pH 7.5), 250 mM KCl, and 2 mM MgCl2. Crystals were obtained using the hanging drop vapor diffusion method by mixing 1 µl complex solution with 1 µl reservoir solution (100 mM Tris-acetate [pH 8.5], 300–400 mM KSCN, 15%–21% PEG 3350). Crystals were cryoprotected by transfer into 100 mM Tris-acetate (pH 8.5), 200 mM KSCN, 30% PEG 3350, 10% ethylene glycol, and flash cooled in liquid nitrogen. X-ray diffraction data were collected at beam line X06DA (PXIII) of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) and processed using XDS (Kabsch, 2010). Data collection statistics are shown in Table 1. Structures were determined by molecular replacement using the Phaser module of the Phenix package (McCoy et al., 2007; Zwart et al., 2008). The atomic structures of Cas9 (PDB 4UN3, chain b) and the guide and target nucleic acids (PDB 4UN3, chain a, chain c residues 9-28, and chain d residues 2-4) were used as separate search models. The PAM-interacting side chains and PAM bases were omitted from the models. Model building and refinement were carried out using COOT (Emsley and Cowtan, 2004) and Phenix.refine (Afonine et al., 2012).
Endonuclease Activity Assay
All target plasmids were prepared by inserting a 20-nt target site and adjacent PAM sequence into pUC19 using the BamHI and EcoRI restriction sites and verified by DNA sequencing. Target sequences are listed in Table S2. Target plasmids were linearized using SspI. For plasmid cleavage assays, equimolar amounts (60 nM final concentration) of Cas9 and sgRNA were incubated in CutSmart Buffer (New England BioLabs) at room temperature for 5 min. Following addition of 550 ng (6 nM final concentration) of linearized target plasmid, the reactions (total volume 55 µl) were incubated at 37°C for 2 hr. 10 µl samples were taken at indicated time points, quenched by addition of EDTA (final concentration 50 mM) and flash-frozen in liquid nitrogen. Samples were treated with 30 µg Proteinase K for 30 min at room temperature. Cleavage products were resolved by gel electrophoresis on a 1% agarose gel stained with GelRed dye (Biotium) and visualized using a Typhoon FLA9500 scanner (GE Healthcare).
Supplemental Information
Supplemental information includes four figures, and two tables, and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.02.020.
Acknowledgements
We thank Vincent Olieric, Meitian Wang, and Takashi Tomizaki at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for assistance with X-ray diffraction experiments. We are grateful to Alexa Burger, Christian Mosimann, Andrew May, and members of the Jinek group for discussion and critical reading of the manuscript. This work was supported by the University of Zurich, the European Research Council (ERC) Starting Grant ANTIVIRNA (Grant no. 337284), and the Bert L & N Kuggie Vallee Foundation Young Investigator Award.
Footnotes
Accession Numbers
The atomic coordinates of the Streptococcus pyogenes Cas9 variant structures have been deposited in the Protein Data Bank at PDB: 5FW1 (VQR), 5FW2 (EQR), and 5FW3 (VRER).
Author Contributions
C.A. and M.J. designed experiments. C.A. prepared and crystallized Cas9 complexes, collected X-ray data, determined crystals structures, and carried out enzymatic activity assays. K.B. assisted with sample preparation. M.J. assisted with X-ray structure determination and refinement and supervised the project. C.A. and M.J. wrote the manuscript.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Jinek M. In vitro enzymology of Cas9. Meth Enzymol. 2014;546:1–20. doi: 10.1016/B978-0-12-801185-0.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Jinek M. In Vitro Reconstitution and Crystallization of Cas9 Endonuclease Bound to a Guide RNA and a DNA Target. Meth Enzymol. 2015;558:515–537. doi: 10.1016/bs.mie.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. RNA events. Cas9 targeting and the CRISPR revolution. Science. 2014;344:707–708. doi: 10.1126/science.1252964. [DOI] [PubMed] [Google Scholar]
- Barrangou R, May AP. Unraveling the potential of CRISPR-Cas9 for gene therapy. Expert Opin Biol Ther. 2015;15:311–314. doi: 10.1517/14712598.2015.994501. [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci US A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glemzaite M, Balciunaite E, Karvelis T, Gasiunas G, Grusyte MM, Alzbutas G, Jurcyte A, Anderson EM, Maksimova E, Smith AJ, et al. Targeted gene editing by transfection of in vitro reconstituted Streptococcus thermophilus Cas9 nuclease complex. RNA Biology. 2015;12:1–4. doi: 10.1080/15476286.2015.1017209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu L-F, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci US A. 2013 doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016 doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biology. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, Siksnys V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015a;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015b;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther. 2015 doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015:1–14. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.