Abstract
Acute lymphoblastic leukemia (ALL) has many features in common with normal B-cell progenitors, including their ability to respond to diverse signals from the bone marrow microenvironment (BMM) resulting in regulation of cell cycle progression and survival. Bone marrow derived cues influence many elements of both steady state hematopoiesis and hematopoietic tumor cell phenotypes through modulation of gene expression. MicroRNAs (miRNAs) are one regulatory class of small non-coding RNAs that have been shown to be increasingly important in diverse settings of malignancy. In the current study, miRNA profiles were globally altered in ALL cells following exposure to primary human bone marrow niche cells including bone marrow stromal cells (BMSC) and primary human osteoblasts (HOB). Specifically, mature miR-221 and miR-222 transcripts were decreased in ALL cells co-cultured with BMSC or HOB, coincident with increased p27 (CDKN1B), a previously validated target. Increased p27 protein in ALL cells exposed to BMSC or HOB is consistent with accumulation of tumor cells in the G0-phase of the cell cycle and resistance to chemotherapy induced death. Overexpression of miR-221 in ALL cells during BMSC or HOB co-culture prompted cell cycle progression and sensitization of ALL cells to cytotoxic agents, blunting the protective influence of the BMM. These novel observations indicate that BMM regulation of miR-221/222 contributes to marrow niche supported tumor cell quiescence and survival of residual cells.
Implications
Niche influenced miR-221/222 may define a novel therapeutic target in ALL to be combined with existing cytotoxic agents to more effectively eradicate refractory disease that contributes to relapse.
Keywords: Microenvironment, MicroRNA, Acute Lymphoblastic Leukemia, Quiescence, Chemosensitivity
Introduction
Acute lymphoblastic leukemia (ALL) is a disease that initiates, progresses and frequently relapses in the bone marrow. Within the marrow space tumor cells occupy the same niche that supports healthy hematopoiesis and have the capacity to respond to cues in that niche which regulate diverse processes including hematopoietic cell quiescence (1,2). As such, it is not surprising that the bone marrow is the site of initiation, progression, and often relapse of aggressive hematopoietic malignancies (3). Bone marrow derived signals influence many elements of both steady state hematopoietic cell development and tumor cell biology through modulation of gene expression. Cues from human bone marrow stromal cells (BMSC) and osteoblasts (HOB), elements of the bone marrow niche that influence leukemic cell biology, have been shown to influence crucial signaling associated with resistant ALL survival and progression (4,5). We have previously reported microenvironment influence on survival pathways (6–9) and the maintenance of a “tumor stem cell” phenotype (10) driven by alteration of tumor cell gene expression profiles and regulation of protein expression or activity. In the current study we investigated tumor cell microRNA (miRNA) regulation influenced by stromal cells or osteoblasts as a novel mechanism by which the bone marrow niche may influence leukemic cells.
MiRNAs define one non-coding RNA regulatory class that has been shown to be increasingly important in hematopoietic cell biology (11). MiRNAs are small single-stranded non-coding RNA molecules around 22-nucleotides (nt) long that regulate target genes post-transcriptionally. Binding of miRNAs to complementary sequences in the 3′-UTR of target mRNA prevents their translation through either translation termination, mRNA cleavage, or mRNA destabilization. This regulation is mediated through the RNA induced silencing complex (RISC). MiRNAs are essential in normal cell development and maintenance, and their dysfunction has been shown to induce tumorigenesis in a variety of settings (12).
Regulation of miRNAs can occur transcriptionally, as well as during their processing and maturation (miRNA biogenesis) into active mature miRNAs. The canonical miRNA biogenesis is based on a stepwise processing machinery. MiRNAs are transcribed by RNA polymerase II to produce a 5′-cap and 3′ polyadenylated primary miRNA (pri-miRNA) with one or more imperfect loop structures that are recognized by DGCR8 and cleaved by the enzyme Drosha in the nucleus. Cleavage of the pri-miRNA stem loop structure generates a precursor miRNA (pre-miRNA) that is recognized and transported to the cytoplasm by the Exportin-5 protein. The pre-miRNA is cleaved by the enzyme Dicer (mature miRNA), and loaded into the RNA-induced silencing complex (RISC) containing Argonaute (13,14). Deficiencies in miRNA biogenesis have been shown to contribute to promotion and progression of disease (15) and patient outcome (16) in various malignancies.
Our lab has previously shown ALL cells associated with the bone marrow microenvironment (BMM) have altered cell cycle kinetics coincident with protection from several conventional chemotherapeutic agents (17). Two miRNAs described to regulate leukemic cell proliferation are miR-221 and miR-222 (18). They are clustered miRNAs that are located on the small arm of the X chromosome where they are transcribed on a single pri-miRNA by RNA polymerase II and regulate a nearly identical set of genes due to conservation of their seed sequence. A well validated miR-221/222 target gene in numerous cancers (18–20), including ALL (21), is the cell cycle inhibitor cyclin-dependent kinase inhibitor 1B (CDKN1B; p27). The expression of p27 has been shown to blunt cell cycle progression at the G0/G1 transition with levels of p27 being higher in quiescent cells compared to those that are actively cycling (22). Manipulation of either miR-221 or miR-222 has the potential to result in cell cycle progression of previously quiescent cells, increasing the efficacy of S phase specific chemotherapeutic agents in inducing apoptosis.
The current study demonstrates that exposure of tumor cells to signals from either human primary BMSC or HOB reduce ALL levels of mature miR-221/222 without overt changes in miRNA stability. Notable changes in the expression of miRNA biogenesis proteins, which may contribute to a blunting in mature miR-221/222, were observed in ALL cells following co-culture with either BMSC or HOB. Constitutive overexpression of miR-221/222 resulted in sustained proliferation and chemosensitivity of ALL cells, even during BMSC or HOB co-culture. These data suggest potential niche regulation of miRNAs may define a novel upstream signaling pathway that promotes tumor cell quiescence and coincident resistance to chemotherapy. The ability to interrupt BMM regulation of miR-221/222 in ALL cells provides a possible novel point of therapeutic intervention to eradicate quiescent, chemoresistant ALL cells that contribute to relapse of disease following cessation of treatment.
Materials and Methods
Cell lines and culture conditions
In the following experiments, Philadelphia chromosome positive (Ph+) lymphoblastic cell lines SUP-B15 (ATCC #CRL-1926; date obtained: Aug. 2011) and Nalm-27 (Fujisaki Cancer Center; date obtained: July 2009) and Ph- REH (ATCC#CRL-8286; date obtained: Nov. 2014) were utilized. Cell line authentication is routinely monitored every 6 months by B-cell surface immunostaining and fluorescence in situ hybridization (FISH) analysis for Philadelphia gene status (date tested- Oct. 2015). In addition, primary human leukemic cells were acquired from the West Virginia University Health Sciences Center and West Virginia University Cancer Institute tissue bank. Primary patient sample 1 (P1) is a 43 year old female patient with ALL at diagnosis and primary patient sample 2 (P2) is a 61 year old male patient with CML in blast crisis (blasts considered active lymphoid disease). For primary patient samples, a pathology report accompanying the corresponding tissue of origin confirmed the identity of the samples. Representative elements of the microenvironment are modeled through use of BMSC and HOB. BMSC are isolated from patients who have not received chemotherapy and have no evidence of marrow disease. HOBs (PromoCell) are isolated from femoral trabecular bone tissue from the knee or hip joint region. In tumor-BMSC/HOB co-culture, ALL cells are seeded at 0.5–1.0 x 106 cells/ml on ~85% confluent stromal layer and fed every 4 days at which time leukemic cells are collected for inclusion in experiments with remaining leukemic cells moved to new primary BMSC or HOB adherent layers consistently every 12 days. Cultures are maintained in 5% O2 to model normal bone marrow oxygen tension, reported to range from 1–7% (23). The tumor population used in this study comprise of ALL cells which physically interact with the stromal adherent layer as opposed to the ALL cells in media suspension. The adherent tumor cell subpopulation, which we previously described to be the most chemotherapy resistant (referred to as the phase dim population), were separated from the stromal layers by size exclusion with G10 Sephadex (Sigma) (24) and used in experiments described below.
Chemotherapeutic reagents
Cytarabine (Ara-C; Selleckchem, Cat # S1648) and Vincristine (VCR; Selleckchem, Cat # S1241) were stored per manufacturer recommendations and were diluted in base media immediately prior to use. Experimental concentrations of Ara-C [1μM] or VCR [25 μM] were used to approximate clinically relevant doses reported as serum levels in ALL patients (25,26).
Evaluation of leukemic cell viability
ALL cells were cultured in media alone or co-cultured with BMSC or HOB for 4 days to establish tumor-adherent cell interactions. At day 4, cultures were provided fresh media and exposed to Ara-C or VCR for 48 hours. Viability was evaluated by trypan blue exclusion in triplicate samples.
Antibodies and western blot analysis
Rabbit polyclonal anti-p27 (Cat # 3686), Drosha (Cat # 3364), Dicer (Cat # 5362), and Ago1 (Cat # 5053) were purchased from Cell Signaling Technology and used at a 1:1000 dilution. Mouse polyclonal anti-GAPDH was purchased from Research Diagnostics Inc. RDI. Protein was isolated by lysing cells and concentration was determined using the bicinchoninic acid (BCA) protein assay (Pierce). Proteins were resolved on SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked in TBS 5%/nonfat dry milk 0.05% Tween-20 and probed with the indicated primary antibodies. After incubation with horseradish peroxidase–conjugated secondary antibodies, signal was visualized using enhanced chemiluminescence reagents (Amersham). Densitometry was performed by scanning the developed X-ray film (BioExcell) and quantified using ImageJ.
RNA isolation and quantitative real-time PCR (qRT-PCR)
RNA was isolated from leukemic cells using the MirVana RNA isolation kit with TURBO DNase I digestion (Life Technologies). One-step qRT-PCRs for primary miRNA transcripts were performed in triplicate using 50 ng of RNA per well, with the QuiantiTect SYBR Green RT-PCR kit (Qiagen) and mature miRNA levels were performed in triplicate using 20 ng of RNA per well measured by 2-step qRT-PCR with TaqMan reagents (Life Technologies) on an Applied Biosystems 7500 real-time PCR machine. GUSB and U6 were used as loading controls. The primer sequence for primary miR-221/222 transcript; forward- TGGTAGTAGGTA AGTCCCAGAA and reverse- TCAACACAACTGCCTACTGC. Primer sets designed to amplify mature miR-221/222 were purchased from Life Technologies.
MiRNA microarray profiling and target gene annotation
For initial profiling to identify subsequent targets for more in depth investigation, total RNA was isolated from Sup-B15 ALL cells cultured in media only or in co-culture with BMSC or HOB from triplicate independent cultures. RNA was processed by LC Sciences and miRNA microarray assays completed as previously described (27) (Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4744). Mature miRNAs in SUP-B15 cells co-cultured with BMSC or HOB that had a mean relative intensity of 500 or greater (0 to 30000 intensity scale) and a P value < 0.05 when compared to Sup-B15 cells grown in media without any BMSC or HOB exposure were further analyzed. Heatmap was created using GenePattern software. Using the prediction software TargetScan (28) and DIANA TarBase (29) a list of predicted target genes based on the seed sequence of the miRNA and validated target genes based on previously published work was generated for investigation.
Generation of overexpressing and miRNA sponge leukemic cells
The overexpression of p27 was achieved by lenti-viral transduction of leukemic cells using an open reading frame expression plasmid for p27 validated and purchased from GeneCopoeia. To generate the miR-221 overexpressing lenti-viral vector, a fragment of approximately 1 kilobase corresponding to the precursor miRNA and the surrounding sequences was amplified from normal genomic DNA. A large portion of miRNA surrounding sequence was included in the attempt to allow correct processing of pre-miR-221 to its mature form and to induce overexpression while preserving a physiologic mechanism of miRNA production. MiRNA sponge expressing ALL cells were generated using antisense miRNA binding sites modified from (30). Two binding sites were designed for miR-221 and miR-222 per sponge sequence.
Cell proliferation assay
ALL cells were labeled with cell retention dye CellTrace-FarRed (Life Technologies) as described by manufacturer instructions. Cells were then cultured under normal growth conditions for 4 days in either media only or co-culture with BMSC or HOB. Tumor cell samples were fixed with 3.7% formaldehyde and CellTrace fluorescence intensity was measured by flow cytometry using LSRFortessa (BD Pharmingen). Proliferation indices were calculated using FCS Express4.
Immunofluorescence imaging
Confocal images were acquired using an upright LSM 510 Zeiss microscope and processed using Zen2009 software and Adobe Photoshop with fluorescence intensity held constant for any experiment in which image acquisition would be compared across samples. ALL sub-populations (tumor cells floating freely in co-culture versus those physically attached to or beneath adherent BMSC or HOB) were cytospun on glass slides following G10 Sephadex purification. Cells were fixed with 4% PFA, blocked in 1x PBS 5% FBS 0.3% Triton X-100, washed with 1x PBS, and incubated with rabbit anti-p27 (1:100) followed by anti-rabbit Alexa 647 (1:200). Slides were washed with PBS and mounted to coverslips using Prolong® Gold anti-fade/DAPI overnight (Life Technologies).
Statistical analysis
All data are presented as the mean ± standard error (with the exception for qPCR data which was mean ± standard deviation) and the statistical significances between conditions were determined by the Student t test or 2-way ANOVA test using GraphPad software. All results are representative of at least 3 independent experiments (p < 0.05 indicated by an * to denote statistical significance).
Results
ALL cells have reduced expression of miRNA biogenesis proteins when influenced by BMM
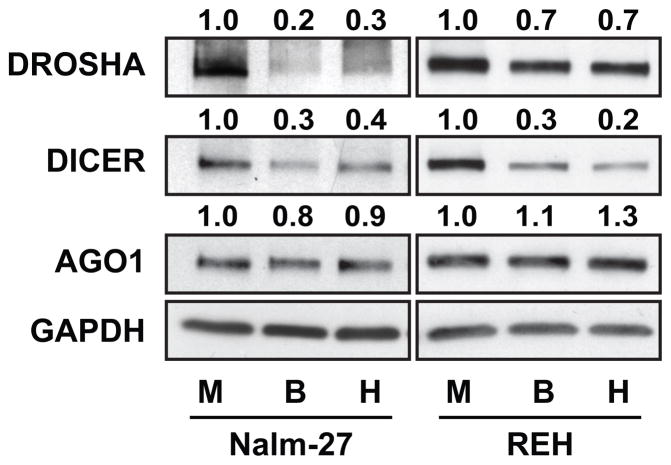
It has been well documented that BMM niches that support hematopoiesis are hypoxic (31). Hypoxia and other factors associated with these niches have been reported by others to regulate miRNA biogenesis (32). To determine whether miRNA biogenesis proteins are regulated in ALL cells exposed to BMSC or HOB, Nalm-27 and REH ALL cell lines were co-cultured with BMSC or HOB and protein abundance measured by western blot. Drosha, a major component of the nuclear microprocessing complex during miRNA biogenesis, was downregulated in ALL cells co-cultured with BMSC or HOB compared to media only control (Fig. 1). A repression of Dicer, a cytoplasmic RNase III that cleaves precursor miRNA into mature miRNA duplexes, was also observed in ALL cells in co-culture, but not the RISC component Ago1 (Fig. 1) or Ago 2 (DNS). These data suggest an alteration in miRNA biogenesis processing in ALL cells influenced by the BMM.
Figure 1.
The BMM alters miRNA biogenesis protein expression in ALL. Drosha, Dicer, and Ago-1 protein abundance was measured in Nalm-27 and REH ALL cells in media only or co-cultured with BMSC or HOB by western blot. GAPDH was used as loading control. The western blots are representative images of 3 independent experiments. M, media; B, BMSC; H, HOB.
The BMM regulates leukemic miRNAs
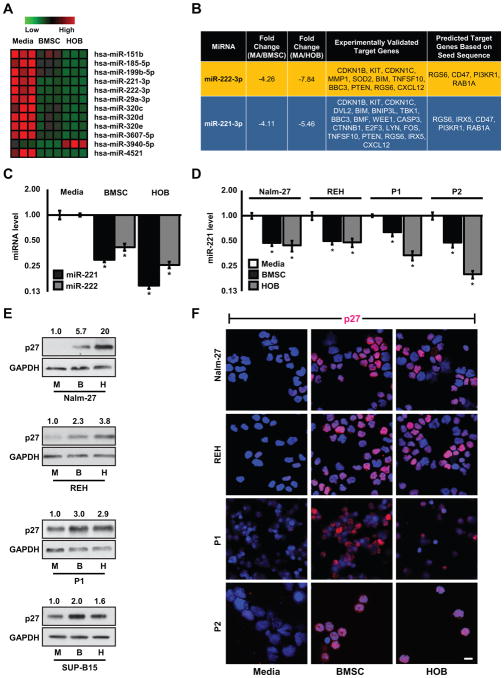
To determine if leukemic cells co-cultured with BMSC or HOB have altered miRNA profiles as a result of dysregulation in the expression of proteins involved in miRNA biogenesis (Fig. 1), we performed a miRNA microarray on SUP-B15 ALL cell line co-cultured with BMSC or HOB or cultured in media only. A subset of miRNAs had altered abundance when influenced by the BMM (Fig. 2A). We focused on miR-221 and miR-222 because they had the highest significant repression in leukemic cells co-cultured with either BMSC or HOB compared to media alone control and they have been previously shown to target cell cycle and survival genes (Fig. 2B) (18). Validation by qRT-PCR of miR-221 and miR-222 levels presented a 2- to 8-fold repression in SUP-B15 cells co-cultured with BMSC or HOB relative to media alone control cells (Fig. 2C). This observation was consistent in Nalm-27 and REH ALL cell lines as well as primary patient samples when co-cultured with BMSC or HOB (Fig. 2D). Cell cycle inhibitor CDKN1B (p27) protein abundance, which is known to be negatively regulated by both miR-221 and miR-222 (18,20), was increased in leukemic cell lines and patient samples when tumor cells were co-cultured with BMSC or HOB as shown by western blot (Fig. 2E) and immunofluorescence based imaging (Fig. 2F) compared to leukemic cells grown in media alone. Consistent with p27 two other validated miR-221/222 target genes which have been previously shown to play a role in cellular quiescence and survival, PTEN and CDKN1C (p57), were assessed in leukemic cells under conditions mentioned above. Both PTEN and p57 protein abundance was increased in leukemic cells co-cultured with BMSC or HOB compared to leukemic cells grown in media alone (Supplementary Fig. S1A,B) further demonstrating the potential miR-221/222 alteration is eliciting.
Figure 2.
ALL cells have miRNA alterations when influenced by the BMM. A, The heat map represents miRNA microarray data comparing SUP-B15 ALL cells co-cultured with BMSC or HOB to those grown in media only. B, The table represents miR-221 and miR-222 relative fold changes from the microarray along with experimentally validated target genes taken from TarBase (29) and predicted target genes taken from TargetScan (28). C, qRT-PCR validating miRNA microarray data measuring fold change of miR-221 and miR-222 in SUP-B15 ALL cells from the same conditions as A. D, qRT-PCR of mature miR-221 fold change in Nalm-27, REH, and 2 patient derived leukemic samples co-cultured with BMSC or HOB compared to those grown in media only. E, Western blots and F, immunofluorescence images of p27 abundance in Nalm-27, REH, and patient derived leukemic samples (due to lack of adequate sample, western blot analysis was unable to be performed for P2). Western blot values depict fold change relative to media only controls and normalized to housekeeping gene GAPDH. Error bars represent standard deviation of the mean with samples performed in triplicate. *, P < 0.05. Scale bars, 10 μm. P1, patient sample 1; P2, patient sample 2.
Increased abundance of p27 in ALL leads to chemoresistance
In order to determine if alterations in p27 protein abundance affect ALL cell phenotype, a constitutively expressed p27 construct was generated in order to increase p27 protein abundance in ALL cells. REH leukemic cells transduced with the p27 overexpression construct had a 35-fold increase in protein abundance compared to empty vector control and were comparable to leukemic cells co-cultured with BMSC or HOB when assessed by western blot analysis (Supplementary Fig. S2A). Coincident with the increase in p27 protein abundance, leukemic cells expressing the p27 construct are more chemoresistant to Ara-C and Vincristine relative to empty vector control cells (Supplementary Fig. S2B).
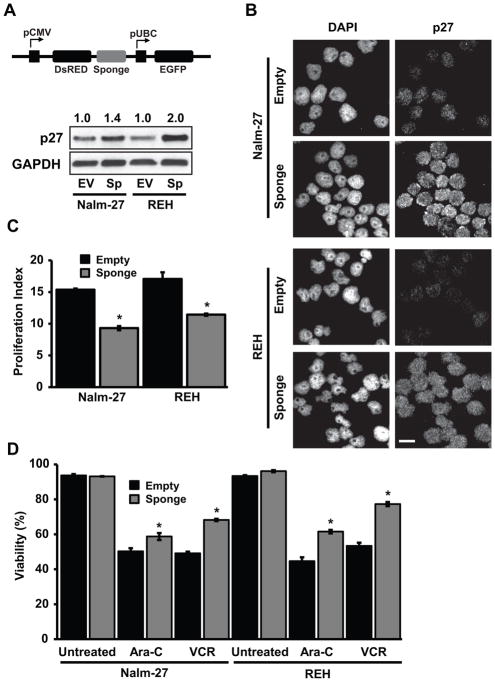
Sequestering miR-221 and miR-222 creates a more quiescent, chemoresistant ALL phenotype
To determine the contribution of miR-221/222 to ALL cell cycle progression and chemotherapy resistance, a constitutively expressed miRNA sponge targeting miR-221/222 was generated to sequester miR-221/222 by serving as a competitive binding partner to the two mature miRNAs and subsequently blunting their ability to bind to endogenous target genes (Fig. 3A). The effectiveness of sequestering the two miRNAs is shown in Figure 3A by measuring intracellular p27 protein abundance by western blot in leukemic cells under media only growth conditions as a validated target. This observation was confirmed by immunofluorescence imaging of p27 (Fig. 3B). Blunting miR-221/222 regulation of p27 was associated with leukemic cells proliferating at a slower rate than empty vector control cells assessed by the proliferation index measured after 4 days using generational tracking (Fig. 3C). Coincident with reduced proliferation, miR-221/222 sponge cells are more chemoresistant to Ara-C and Vincristine relative to empty vector control cells (Fig. 3D).
Figure 3.
MiR-221/222 sponge ALL cells have increased p27, with reduced proliferation and chemosensitivity. A, Schematic of lentiviral miRNA sponge construct along with western blot of p27 abundance alterations within Nalm-27 and REH miR-221/222 sponge cells. Values depict fold change relative to empty vector control and normalized to housekeeping gene GAPDH. B, Immunofluorescent images of p27 in miR-221/222 sponge cells compared to empty vector control. C, Proliferation index was measured 4 days after cell retention dye addition in cells described in A. D, The viability of these cells after 48 hour exposure to Ara-C and VCR were measured by trypan blue exclusion. Error bars represent standard error of the mean with samples performed in triplicate. *, P < 0.05. Western blot, immunofluorescent images and bar graphs are representative data from 3 independent experiments. Scale bars, 10 μm. EV, empty vector; Sp, sponge; Ara-C, cytarabine; VCR, vincristine.
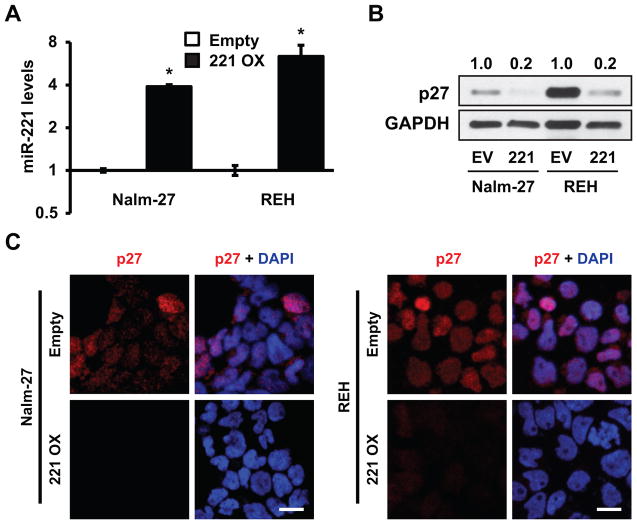
MiR-221 overexpression alters p27 abundance, proliferation rate, and chemosensitivity in ALL influenced by BMM
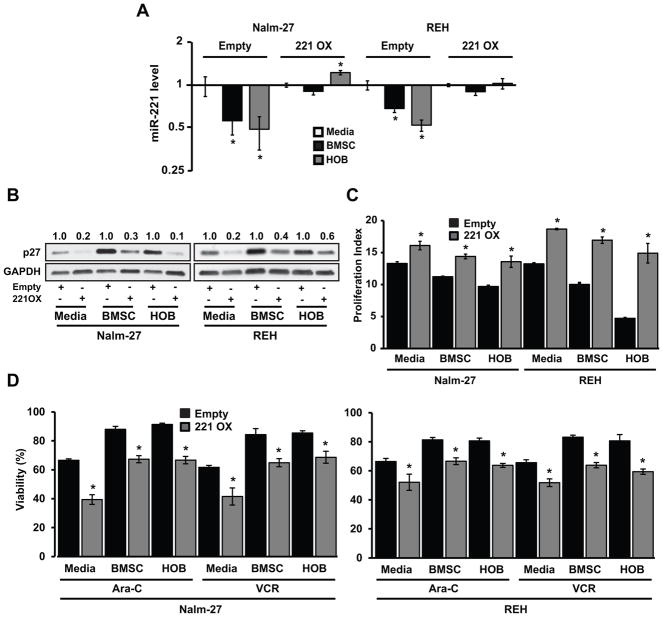
In order to investigate the significance of forced expression of miR-221/222 on ALL cell phenotype, including during BMM co-culture, ALL cell lines constitutively expressing pre-miR-221 were developed. To confirm mature miR-221 overexpression qRT-PCR was completed with a 4- and 6-fold overexpression observed in Nalm-27 and REH ALL cells respectively (Fig. 4A). As demonstrated by western blot in Figure 4B, Nalm-27 and REH ALL cell lines overexpressing miR-221 are characterized by reduced abundance of p27, by approximately five-fold, when compared to empty vector controls. This observation was confirmed by immunofluorescence imaging (Fig. 4C).
Figure 4.
Overexpressing miR-221 reduces p27 abundance in ALL. A, qRT-PCR of miR-221 fold change in Nalm-27 and REH cells overexpressing miR-221 compared to empty vector control. B, Western blot analysis of p27 abundance in Nalm-27 and REH cells overexpressing miR-221 or empty vector control. Values depict fold change relative empty vector control and normalized to housekeeping gene GAPDH. C, Immunofluorescent images of p27 expression under the same conditions as B. Error bars represent standard deviation of the mean with samples performed in triplicate. *, P < 0.05. Western blot, immunofluorescent images and bar graphs are representative data from 3 independent experiments. Scale bars, 10 μm. EV, empty vector; 221/221 OX, miR-221 overexpression.
Based on the observation that miR-221 overexpressing ALL cells had a marked reduction of the downstream target p27 when cultured in media only (Fig. 4), we next determined whether reduction would be maintained in ALL cells influenced by the BMM. Both Nalm-27 and REH empty vector control cells and miR-221 overexpressing cells were co-cultured with BMSC or HOB and the abundance of miR-221 and p27 transcripts and protein was evaluated by qRT-PCR and western blot, respectively. ALL cells overexpressing miR-221 co-cultured with BMSC or HOB had miR-221 abundance comparable to ALL cells grown in media alone (Fig. 5A). When p27 abundance was assessed, miR-221 overexpressing cells had reduced levels of p27 when co-cultured with BMSC or HOB compared to matched empty vector control counterparts (Fig. 5B). To investigate if modulation of miR-221 in ALL cells can overcome the quiescent influence of the BMM, cell proliferation index was measured. ALL cells with empty vector or miR-221 overexpression were co-cultured with BMSC or HOB for 4 days. ALL cells with miR-221 overexpression proliferated significantly more than empty vector control cells when in media only as well as under co-cultured conditions (Fig. 5C). To determine whether ALL cells overexpressing miR-221 could overcome the protective influences of the BMM, the empty vector and miR-221 overexpressing ALL cells were co-cultured with BMSC or HOB and exposed to Ara-C or Vincristine. ALL cells overexpressing miR-221 were significantly more sensitive to chemotherapy exposure than empty vector controls, even during BMSC or HOB co-culture conditions (Fig. 5D).
Figure 5.
MiR-221 overexpression can overcome BMM regulation of ALL proliferation and survival. A, qRT-PCR of miR-221 fold change in Nalm-27 and REH overexpressing miR-221 co-cultured with BMSC or HOB for 4 days compared to media only control. B, p27 abundance was measured in Nalm-27 and REH comparing empty vector control to miR-221 overexpression cells when in media only, co-cultured with BMSC or HOB by western blot. Values depict fold change relative to empty vector control and normalized to housekeeping gene GAPDH. C, Proliferation index was measured 4 days after cell retention dye was added with cells in same conditions as A,B. D, Viability of Nalm-27 and REH cells after 4 day co-culture with BMSC or HOB and exposure to Ara-C and VCR for 48 hours and measured by trypan blue exclusion. Error bars represent standard error of the mean with samples performed in triplicate. *, P < 0.05. Results are representative data from 3 independent experiments. Scale bars, 10 μm. 221 OX, miR-221 overexpression; Ara-C, cytarabine; VCR, vincristine.
BMM influences miR-221/222 biogenesis
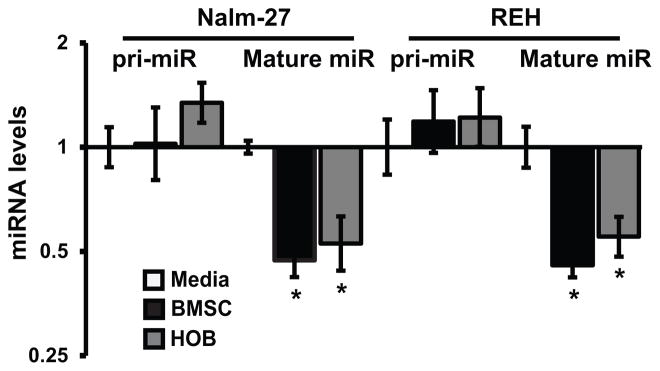
Based on the observation that BMSC or HOB co-culture correlated with repressed miRNA biogenesis proteins (Fig. 1) and reduced mature miR-221/222 transcript abundance (Fig. 2A), investigation of the pri-miRNA levels of miR-221/222 by qRT-PCR was completed. This aspect of the evaluation was aimed to determine if initial expression of the pri-miRNA transcript was potentially upstream of reduced mature transcript levels that were detected. Important to the experimental approach taken is that miR-221 and miR-222 are transcribed from the same pri-miRNA transcript. We observed that miR-221/222 primary transcript levels were unchanged in Nalm-27 and REH cells co-cultured with BMSC or HOB, but mature miR-221 was repressed in ALL cells compared to tumor cells cultured in media only (Fig. 6). Because regulation of miR-221 and miR-222 does not appear to be due to transcriptional alteration, the stability of miR-221 and miR-222 when influenced by the BMM was assessed. No significant change in stability of the miRNAs being investigated was observed when leukemic cells grown in media only or co-cultured with BMSC or HOB were compared after actinomycin D exposure (DNS). This observations suggests that miR-221/222 stability is unaffected by the BMM and not a mechanism by which the BMM affects mature miR-221/222 levels.
Figure 6.
Primary transcript levels of miR-221/222 are unaltered in BMM influenced ALL. Fold change of the pri-miRNA of miR-221/222 and mature miR-221 was measured by qRT-PCR in Nalm-27 and REH cells co-cultured with BMSC or HOB relative to cells cultured with media only. Error bars represent standard deviation of the mean with samples performed in triplicate. *, P < 0.05. Results are representative data from 3 independent experiments. pri-miR, primary miRNA transcript; mature miR, mature miRNA.
Discussion
While significant progress has been made in the treatment of ALL, disease relapse remains a concern that requires identification of mechanism-based innovative treatments. Specifically in children, aggressive treatment strategies required to treat relapsed ALL, with marked potential of late effects and treatment induced secondary malignancies, is a clinical challenge (33). Against this backdrop it is essential to reduce the toxicity of treatment without blunting efficacy. Targeting microenvironment-protected tumor cells which are typically more resistant to chemotherapy treatment (6,7,9) may provide a strategy in the treatment of resistant and refractory ALL. A fundamental understanding of the mechanisms by which the microenvironment is protecting this subset of tumor cells may contribute to the development of novel therapeutic approaches.
Varied perspectives exist regarding whether therapeutic strategies should attempt to maintain cancer cells in a dormant-like state to blunt expansion versus attempting to force these cells through division rendering them responsive to cell cycle specific conventional chemotherapeutic agents. Based on data that correlates the number of tumor cells present in a patient after chemotherapy treatment with the likelihood of relapse (34), it may be ideal to eradicate quiescent residual tumor cells in sites of sanctuary like the bone marrow, when possible. For this approach to become a technical reality, we must better understand the critical elements of microenvironment supported tumor cell quiescence, which we recognize may be disease specific as well as sub-type specific given the heterogeneity within many malignancies. As such, it is not assumed that one model supports broad applicability across all types of lymphoblastic leukemia, but rather insights in a targeted setting may provide insight into signaling pathways that warrant interrogation in other settings in which the marrow microenvironment is well documented to provide a clinically problematic site of sanctuary for tumor cells.
Healthy pro-B cells rely on the bone marrow’s signaling cues for survival, growth and differentiation (35) to sustain one lineage of baseline hematopoiesis. In the setting of ALL these interactions are “hijacked” and provide a sanctuary site for malignant B lineage cells that share many characteristics with their normal counterparts. Earlier work from our lab and many others has characterized altered pro-survival proteins, adhesion molecules, and stem cell characteristics in leukemic cells influenced by the BMM (6–10,36). As our understanding of the complexity of mRNA species has broadened, so have the opportunities to better understand how the marrow niche may be influencing critical signaling pathways with consideration beyond traditional gene expression analysis.
The tumor microenvironment has been shown to play a role in the regulation of miRNA expression in diverse tumor types (37,38), and consistent with previous observations, BMM interaction induced changes in ALL miRNA expression (Fig. 2A). Drosha and Dicer protein expression was repressed in chemotherapy resistant, quiescent BMM influenced ALL cells (Fig. 1). These observations align with recent findings that demonstrate altered miRNA biogenesis correlated to disease progression and overall patient outcome (16,39). Initial interpretation of these results might suggest global repression of mature miRNAs, and in our model we do, in fact, observe that the majority of changes in miRNAs are reduced levels of transcripts being detected in ALL cells co-cultured with BMM derived cells relative to tumor cells grown in media alone (Fig. 2A; Supplementary Table 1,2). However, other studies have demonstrated that miRNAs can be upregulated, even with a reduction in processing, with late stage lung cancer miR-210 providing one example through increased transcription (40). In the current study, two miRNAs down-regulated coincident with BMM interaction are miR-221 and miR-222, previously described as oncomiRs. These oncomiRs have been shown to be dysregulated in various cancer types (41,42) with expression positively correlated with cell proliferation in gastric carcinoma and pancreatic cancer (19,43). The most annotated target gene of miR-221/222 is the quiescence master regulator p27 (18–20). We demonstrate that p27 is abundant in the leukemic cells influenced by the BMM (Fig. 2D) consistent with previous observations of BMM supported ALL quiescence (17).
Targeted reduction of miR-221/222 reduced ALL cell cycle progression and increased chemotherapy resistance in the absence of any BMSC or HOB signal with a trend similar to ALL cells during co-culture of niche cells (Fig. 3). Not surprisingly, given the complexity of the BMM and interaction of multiple signaling cascades, interruption of miR-221/222 in isolation did not completely recapitulate the effects of the niche. However, these data combined with those from ALL cells constitutively overexpressing miR-221/222 suggest miR-221/222 may play a role in the promotion of the quiescent and resistant phenotype observed in ALL cells subsequent to BMM cell interaction.
Signals from BMM adherent BMSC or HOB did not significantly reduce the proliferation of leukemic cells constitutively overexpressing miR-221 (Fig. 5C) and were not able to restore chemoresistance in ALL cells when miR-221 was artificially sustained at or above baseline levels during co-culture (Fig. 5D). This suggests that the ability of the BMM to reduce miR-221 expression in leukemic cells is an important phenotypic change in ALL cells influenced by the BMM associated with increased ALL survival. However, forced miR-221 overexpression did not result in a complete loss of protection when compared to cells expressing the matched empty vector, again consistent with observations from several laboratories that have described complex contributions of the BMM protection of leukemic cells (9,44). It is not suggested that any one pathway will define an absolute point at which niche protection can be eliminated, but rather an increased understanding of critical signaling pathways that may include miRNA regulation may contribute to innovative combination therapy to eradicate subsets of resistant residual cells.
Collectively, these data suggest that the BMM regulates ALL cell quiescence and resistance from chemotherapy exposure by altering signals that converge on reduced levels of miR-221/222. The alteration does not appear to be due to transcriptional regulation of primary miR-221/222 transcript (Fig. 6), but potentially by the microenvironment effecting key components in miRNA biogenesis. This observation is consistent with pathways described in breast cancer (45) and in normal vascular smooth muscle cells (46) influenced by hypoxia or TGF-β, respectively. Anatomical niches that support hematopoietic cells in the BMM have been well characterized as being hypoxic (31) and it was recently shown that hypoxia can mediate the transcriptional repression of both Drosha and Dicer (47) which raises the possibility of hypoxia as a key influence on healthy or malignant hematopoietic cell miRNA profiles resulting in regulation of quiescence. In addition, stromal elements have been reported by several labs to be a source of marrow TGF-β (48,49) and our laboratory has previously described increased activation of TGF-β subsequent to chemotherapy (50). Exposure of hematopoietic tumors to both baseline and increased bioavailable TGF-β in the niche could result in an unintended result of tumor cell quiescence during treatment regulated, in part, by TGF-β regulated miRNA pathways.
Although in this current study miR-221/222’s contribution to the protected phenotype ALL cells influenced by the BMM acquire was focused on exclusively, other miRNAs identified in Figure 2A could impact leukemic cell characteristics as well. For example miR-185 and miR-4521 were shown to be reduced in ALL cells in co-culture with BMSC or HOB. miR-185 was previously shown to regulate the Akt pathway in non-small cell lung carcinomas, acting as a tumor suppressor (51). Therefore, leukemic cells in co-culture with reduced miR-185 abundance may be contributing to the increase in protection of these tumors cells when exposed to chemotherapy agents. Similarly, a reduction in miR-4521 abundance has been shown in chronic lymphocytic leukemia to contribute to the pathogenesis and formation of this disease (52). Although the characterization of these two miRNA in our model is outside the scope of this study, further in-depth investigation will need to be performed to determine their potential role in BMM regulated leukemic cell protection and survival.
The potential for the eradication of the quiescent leukemic cells that persist following traditional treatment makes the miR-221/222-p27 pathway intriguing as a future clinical target in residual disease. Although miRNA therapy is somewhat in its infancy, there have been great strides made not only in the delivery, but also the specificity of miRNA therapy (53). Because novel miRNA based therapies are being developed it is important to understand the mechanisms by which miRNAs are being modulated in specific disease settings, such as therapy resistant ALL, in order to target upstream regulators. Targeted modulation of miRNA expression may provide an innovative approach to achieving the ultimate therapeutic goal of increasing disease-free survival with reduced overall toxicity in ALL. While relevant in general, progress is specifically needed in the pediatric population that comprises the predominant group of patients with the most significant long-term effects associated with aggressive therapy.
Supplementary Material
Acknowledgments
Financial support: Supported by National Institutes of Health (NHLBI) R01 HL056888 (LFG), National Cancer Institute (NCI) RO1 CA134573NIH (LFG), P30 GM103488 (LFG), WV CTR-IDEA NIH 1U54 GM104942, the Alexander B. Osborn Hematopoietic Malignancy and Transplantation Program, and the WV Research Trust Fund.
We are grateful for the support of Dr. Kathy Brundage and the West Virginia University Flow Cytometry Core Facility, supported by NIH S10-OD016165 and the Institutional Development Award (IDeA) from the NIH Institute of General Medical Sciences of the National Institutes of Health (CoBRE P30GM103488 and INBRE P20GM103434) along with Dr. Karen Martin and the West Virginia Microscope Imaging Facility, supported by the Mary Babb Randolph Cancer Center and NIH grants P20 RR016440, P30 GM103488 and P20 GM103434.” We would also like to thank Ian Hare for his assistance with manuscript editing.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Meads MB, Hazlehurst LA, Dalton WS. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:2519–26. doi: 10.1158/1078-0432.CCR-07-2223. [DOI] [PubMed] [Google Scholar]
- 2.Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood. 2013;121:4821–31. doi: 10.1182/blood-2012-12-475483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiarini F, Lonetti A, Evangelisti C, Buontempo F, Orsini E, Evangelisti C, et al. Advances in understanding the acute lymphoblastic leukemia bone marrow microenvironment: From biology to therapeutic targeting. Biochim Biophys Acta BBA - Mol Cell Res. 2016;1863:449–63. doi: 10.1016/j.bbamcr.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Manabe A, Murti KG, Coustan-Smith E, Kumagai M, Behm FG, Raimondi SC, et al. Adhesion-dependent survival of normal and leukemic human B lymphoblasts on bone marrow stromal cells. Blood. 1994;83:758–66. [PubMed] [Google Scholar]
- 5.Shiozawa Y, Pedersen EA, Taichman RS. GAS6/Mer axis regulates the homing and survival of the E2A/PBX1-positive B-cell precursor acute lymphoblastic leukemia in the bone marrow niche. Exp Hematol. 2010;38:132–40. doi: 10.1016/j.exphem.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall BM, Fortney JE, Taylor L, Wood H, Wang L, Adams S, et al. Stromal cells expressing elevated VCAM-1 enhance survival of B lineage tumor cells. Cancer Lett. 2004;207:229–39. doi: 10.1016/j.canlet.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 7.Fortney JE, Zhao W, Wenger SL, Gibson LF. Bone marrow stromal cells regulate caspase 3 activity in leukemic cells during chemotherapy. Leuk Res. 2001;25:901–7. doi: 10.1016/s0145-2126(01)00051-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Fortney JE, Gibson LF. Stromal cell protection of B-lineage acute lymphoblastic leukemic cells during chemotherapy requires active Akt. Leuk Res. 2004;28:733–42. doi: 10.1016/j.leukres.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Mudry RE, Fortney JE, York T, Hall BM, Gibson LF. Stromal cells regulate survival of B-lineage leukemic cells during chemotherapy. Blood. 2000;96:1926–32. [PubMed] [Google Scholar]
- 10.Wang L, O’Leary H, Fortney J, Gibson LF. Ph+/VE-cadherin+ identifies a stem cell like population of acute lymphoblastic leukemia sustained by bone marrow niche cells. Blood. 2007;110:3334–44. doi: 10.1182/blood-2007-01-068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montagner S, Dehó L, Monticelli S. MicroRNAs in hematopoietic development. BMC Immunol. 2014;15:14. doi: 10.1186/1471-2172-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel J-P. MicroRNAs: molecular features and role in cancer. Front Biosci Landmark Ed. 2012;17:2508–40. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen TA, Jo MH, Choi Y-G, Park J, Kwon SC, Hohng S, et al. Functional Anatomy of the Human Microprocessor. Cell. 2015;161:1374–87. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 15.Passon N, Gerometta A, Puppin C, Lavarone E, Puglisi F, Tell G, et al. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012;65:320–6. doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- 16.Lin R-J, Lin Y-C, Chen J, Kuo H-H, Chen Y-Y, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–50. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moses BS, Slone WL, Thomas P, Evans R, Piktel D, Angel PM, et al. Bone marrow microenvironment modulation of acute lymphoblastic leukemia phenotype. Exp Hematol. 2016;44:50–59. e2. doi: 10.1016/j.exphem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfò L, Rossi C, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–59. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27kip1, p57kip2, and PUMA. Am J Cancer Res. 2013;3:465–77. [PMC free article] [PubMed] [Google Scholar]
- 20.Castagnino P, Kothapalli D, Hawthorne EA, Liu S-L, Xu T, Rao S, et al. miR-221/222 compensates for Skp2-mediated p27 degradation and is a primary target of cell cycle regulation by prostacyclin and cAMP. PloS One. 2013;8:e56140. doi: 10.1371/journal.pone.0056140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotani A, Ha D, Hsieh J, Rao PK, Schotte D, den Boer ML, et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–78. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, et al. p57Kip2 and p27Kip1 Cooperate to Maintain Hematopoietic Stem Cell Quiescence through Interactions with Hsc70. Cell Stem Cell. 2011;9:247–61. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Chow DC, Wenning LA, Miller WM, Papoutsakis ET. Modeling pO(2) distributions in the bone marrow hematopoietic compartment. II. Modified Kroghian models. Biophys J. 2001;81:685–96. doi: 10.1016/S0006-3495(01)75733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slone WL, Moses BS, Evans R, Piktel D, Martin KH, Petros W, et al. Modeling Chemotherapy Resistant Leukemia In Vitro. J Vis Exp. 2016;108:e53645. doi: 10.3791/53645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liedtke M, Dunn T, Dinner S, Coutré SE, Berube C, Gotlib J, et al. Salvage therapy with mitoxantrone, etoposide and cytarabine in relapsed or refractory acute lymphoblastic leukemia. Leuk Res. 2014;38:1441–5. doi: 10.1016/j.leukres.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Van den Berg HW, Desai ZR, Wilson R, Kennedy G, Bridges JM, Shanks RG. The pharmacokinetics of vincristine in man: reduced drug clearance associated with raised serum alkaline phosphatase and dose-limited elimination. Cancer Chemother Pharmacol. 1982;8:215–9. doi: 10.1007/BF00255487. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Gong Y, Chao T, Peng X, Yuan J, Ma Z, et al. Identification of differentially expressed microRNAs by microarray: a possible role for microRNAs gene in medulloblastomas. Chin Med J (Engl) 2009;122:2405–11. [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Hsu S-D, Tseng Y-T, Shrestha S, Lin Y-L, Khaleel A, Chou C-H, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kluiver J, Slezak-Prochazka I, Smigielska-Czepiel K, Halsema N, Kroesen B-J, van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–7. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–73. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med. 2013;64:20–30. doi: 10.1016/j.freeradbiomed.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malempati S, Gaynon PS, Sather H, La MK, Stork LC. Outcome After Relapse Among Children With Standard-Risk Acute Lymphoblastic Leukemia: Children’s Oncology Group Study CCG-1952. J Clin Oncol. 2007;25:5800–7. doi: 10.1200/JCO.2007.10.7508. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X-S, Liu Y-R, Zhu H-H, Xu L-P, Liu D-H, Liu K-Y, et al. Monitoring MRD with flow cytometry: an effective method to predict relapse for ALL patients after allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2012;91:183–92. doi: 10.1007/s00277-011-1285-1. [DOI] [PubMed] [Google Scholar]
- 35.Passa O, Tsalavos S, Belyaev NN, Petryk A, Potocnik AJ, Graf D. Compartmentalization of bone morphogenetic proteins and their antagonists in lymphoid progenitors and supporting microenvironments and functional implications. Immunology. 2011;134:349–59. doi: 10.1111/j.1365-2567.2011.03495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Leary H, Akers SM, Piktel D, Walton C, Fortney JE, Martin KH, et al. VE-cadherin Regulates Philadelphia Chromosome Positive Acute Lymphoblastic Leukemia Sensitivity to Apoptosis. Cancer Microenviron Off J Int Cancer Microenviron Soc. 2010;3:67–81. doi: 10.1007/s12307-010-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747–55. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Haag D, Muley T, Warth A, Zapatka M, Toedt G, et al. Tumor-microenvironment interactions studied by zonal transcriptional profiling of squamous cell lung carcinoma. Genes Chromosomes Cancer. 2013;52:250–64. doi: 10.1002/gcc.22025. [DOI] [PubMed] [Google Scholar]
- 39.Jafarnejad SM, Sjoestroem C, Martinka M, Li G. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod Pathol Off J U S Can Acad Pathol Inc. 2013;26:902–10. doi: 10.1038/modpathol.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puisségur M-P, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–78. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–20. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- 43.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B, Li M, McDonald T, Holyoake TL, Moon RT, Campana D, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood. 2013;121:1824–38. doi: 10.1182/blood-2012-02-412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim S-O, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun. 2014;5:5202. doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quante M, Tu SP, Tomita H, Gonda T, Wang SSW, Takashi S, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–72. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nemunaitis J, Tompkins CK, Andrews DF, Singer JW. Transforming growth factor beta expression in human marrow stromal cells. Eur J Haematol. 1991;46:140–5. doi: 10.1111/j.1600-0609.1991.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang L, Clutter S, Benincosa J, Fortney J, Gibson LF. Activation of transforming growth factor-beta1/p38/Smad3 signaling in stromal cells requires reactive oxygen species-mediated MMP-2 activity during bone marrow damage. Stem Cells Dayt Ohio. 2005;23:1122–34. doi: 10.1634/stemcells.2004-0354. [DOI] [PubMed] [Google Scholar]
- 51.Li S, Ma Y, Hou X, Liu Y, Li K, Xu S, et al. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8:11854–62. [PMC free article] [PubMed] [Google Scholar]
- 52.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071–6. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–10. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.