Abstract
Fusion transcripts (i.e. chimeric RNAs) resulting from gene fusions have been used successfully for cancer diagnosis, prognosis, and therapeutic applications. In addition, many fusion transcripts are found in normal human cell lines and tissues, with some data supporting their role in normal physiology. Besides chromosomal rearrangement, intergenic splicing can generate them. Global identification of fusion transcripts becomes possible with the help of Next Generation Sequencing (NGS) technology like RNA-Seq. In the past decade, major advancements have been made for chimeric RNA discovery due to the development of advanced sequencing platform and software packages. However, current software tools behave differently in terms of specificity, sensitivity, time, and computational memory usage. Recent benchmarking studies showed that none of the tools are inclusive. The development of high performance (accurate and fast), and user-friendly fusion detection tool/pipeline is still an open quest. In this article, we review the existing software packages for fusion detection. We explain the methods of the tools, and discuss various factors that affect fusion detection. We summarize conclusions drawn from several comparative studies, and then discuss some of the pitfalls of these studies. We also describe the limitations of current tools, and suggest directions for future development.
INTRODUCTION
Fusion transcript describes a phenomenon in which a hybrid RNA is composed of transcripts of two separate genes. Traditionally, such chimeric RNAs were known to be the result of gene fusions and are associated with cancer. BCR-ABL1 resulted from t(9;22) in chronic myelogenous leukemia (CML) is the classic example [1]. Gene fusions like BCR-ABL are found in many tumors, including hematological cancers [2,3] and in solid tumors such as prostate cancer [4], lung cancer [5], glioblastoma [6], fibrolamellar hepatocellular carcinoma [7], breast cancer [8], skin cancer [9], lymphoid cancers [10], and sarcoma samples [11]. These fusions are good targets for cancer diagnosis, prognosis, and therapeutic applications [2,12]. Several classic fusions, such as: BCR-ABL, PML-RAR, and EML4-ALK, are the poster children for successful targeted cancer therapy [13,14].
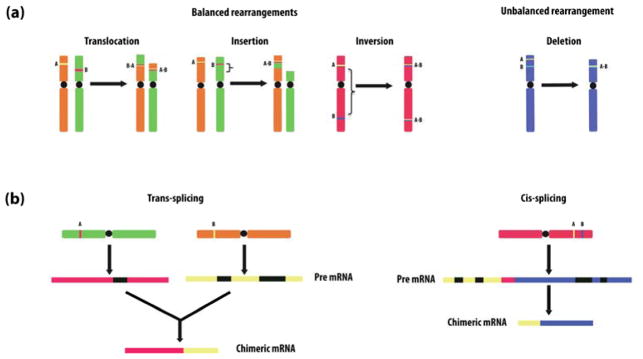
Even though chimeric RNAs are mostly known to be the products of chromosomal rearrangement at the DNA level (Figure 1(a)), fusion transcripts can also be produced by trans-splicing (Figure 1(b)) [15,16] and cis-splicing between neighboring genes (Figure 1(b)) [17,18]. Such “intergenically” produced fusion transcripts can be detected in normal human cell lines [18] and tissues [19–22]. Some of them were shown to have functional relevance [16].
Figure 1. Mechanism of fusion formation.
(a) Fusions formed at DNA level (i.e. chromosomal rearrangements). At DNA level, gene fusion may originate through ‘balanced’ and ‘unbalanced’ chromosome rearrangements. ‘Balanced’ changes comprise translocations, insertion and inversion, whereas ‘unbalanced’ change that leads to fusion genes can be a deletion of an interstitial chromosomal segment. (b) RNA level fusions may occur through trans-splicing or cis-splicing between neighboring genes. Black blocks represent introns. In the “cis-splicing” mechanism, the red block represents intergenic region. The sizes of exon, intron and intergenic regions are not drawn to scale.
With the advancement of Next Generation Sequencing (NGS), fusions can be detected either in genomic DNA sequencing datasets [23,24], or transcriptome sequencing (RNA-Seq) datasets [9,25]. They can be detected from both single-end reads [26] and paired-end reads [27] of various lengths. The main advantage of paired-end reads is that the connectivity information between the sequenced ends is available. In 2009, two studies shows the initial efforts in detecting fusion transcripts from paired RNA-Seq data [27,28]. In these studies, no specific fusion detection tool was used. In 2010, first dedicated tool for the fusion detection (i.e. FusionSeq) was published [29]. Since 2010, around 33 computational tools have been developed for detecting fusion transcripts using RNA-Seq data (see a comprehensive list of 33 methods in Table 1). However, there are various challenges associated with these tools. These tools have been trained and tested on different types of datasets. Their behavior changes with datasets. A large amount of time and computational memory is often required. In addition to missing true fusion events, these tools can also produce false positives [22]. Practical knowledge of these tools in terms of time consumption, computational memory usage, sensitivity, and specificity is urgently needed. This review discusses the common features, methodology, requirements, and other important issues regarding all of the fusion detection tools available to the scientific community. A summery of the benchmarking studies of fusion detection tools, and several pitfalls associated with such studies are listed in this review. Several limitations of current tools and some future directions are provided here.
Table 1.
Summery of computational tools for fusion transcripts detection.
FUSION DETECTION TOOLS
Most of the fusion detection tools have been developed in the last six years. Table 1 includes most if not all of the popular ones. We can classify the fusion detection tools on the basis of the input reads format, type of input reads, and the reference used to align the reads (Table 2). Other features of fusion detection tools, and default detection and filtering parameters are listed In Table 3.
Table 2.
Features of fusion transcripts detection tools.
| Tools | Reads format | Input data | Reference | |||
|---|---|---|---|---|---|---|
| Single-end | Paired-end | WGS | RNA-Seq | Genome | Transcriptome | |
| Bellerophontes | ✔ | ✔ | ✔ | |||
| BreakFusion | ✔ | ✔ | ✔ | ✔ | ||
| ChimeraScan | ✔ | ✔ | ✔ | ✔ | ||
| Comrad | ✔ | ✔ | ✔ | ✔ | ✔ | |
| CRAC | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Dissect | ✔ | ✔ | ✔ | ✔ | ||
| deFuse | ✔ | ✔ | ✔ | ✔ | ||
| EricScript | ✔ | ✔ | ✔ | |||
| FusionAnalyser | ✔ | ✔ | ✔ | ✔ | ||
| FusionCatcher | ✔ | ✔ | ✔ | ✔ | ✔ | |
| FusionFinder | ✔ | ✔ | ✔ | ✔ | ||
| FusionHunter | ✔ | ✔ | ✔ | ✔ | ||
| FusionMap | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| FusionMatcher | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| FusionQ | ✔ | ✔ | ✔ | ✔ | ||
| FusionSeq | ✔ | ✔ | ✔ | ✔ | ||
| IDP-fusion | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| INTEGRATE | ✔ | ✔ | ✔ | ✔ | ||
| JAFFA | ✔ | ✔ | ✔ | ✔ | ||
| MapSplice | ✔ | ✔ | ✔ | ✔ | ||
| NCLscan | ✔ | ✔ | ✔ | ✔ | ||
| nFuse | ✔ | ✔ | ✔ | ✔ | ✔ | |
| PRADA | ✔ | ✔ | ✔ | ✔ | ✔ | |
| ShortFuse | ✔ | ✔ | ✔ | ✔ | ||
| SnowShoes-FTD | ✔ | ✔ | ✔ | ✔ | ||
| SOAPFuse | ✔ | ✔ | ✔ | ✔ | ||
| SOAPfusion | ✔ | ✔ | ✔ | |||
| TopHat-Fusion | ✔ | ✔ | ✔ | ✔ | ✔ | |
| TRUP | ✔ | ✔ | ✔ | |||
Table 3.
Other features of fusion detection tools, and default detection and filtering parameters.
| Tools | Anchor Length Filter | Read Through transcript filter | Supported reads filter | PCR artifact filter | Homology based filter | Alignment tool |
|---|---|---|---|---|---|---|
| Bellerophontes | N | Y | Y | Y | Y | TopHat |
| BreakFusion | N | N | N | N | N | BWA/BLAT |
| ChimeraScan | 10 | Y | 4 | N | N | Bowtie/BWA |
| EricScript | N | Y | 3/1 | Y | Y | BWA/BLAT |
| FusionAnalyser | Y | Y | Y | N | Y | BWA |
| FusionCatcher | 10 | Y | 3/1 | N | Y | Bowtie/STAR/BLAT/Bowtie2 |
| FusionFinder | N | Y | N | N | Y | Bowtie |
| FusionHunter | 10 | Y | 3/1 | Y | Y | Bowtie |
| FusionMap | Y | Y | Y | Y | Y | GSNAP |
| FusionQ | 10 | N | 3/1 | N | Y | Bowtie |
| FusionSeq | N | Y | Y | Y | Y | ELAND |
| JAFFA | N | Y | 3/1 | N | Y | Bowtie/BLAT |
| MapSplice | N | N | N | N | N | Bowtie |
| deFuse | 10 | Y | 3/1 | N | Y | Bowtie/BLAT |
| SOAPFuse | 10 | N | 3/1 | N | N | Soap2/BWA/BLAT |
| TopHat-Fusion | 10 | Y | 3/1 | N | Y | Bowtie |
| PRADA | N | N | N | N | N | BWA/BLAST |
| ShortFuse | N | N | Y | N | N | Bowtie |
| SnowShoes-FTD | N | Y | 2/N | Y | Y | Bowtie/BWA |
FUSION FORMATION MECHANISM
As discussed above in the introduction section, chimeric RNAs can be generated at both DNA level (through chromosomal rearrangements) (Figure 1(a)) and RNA level (through cis- or trans-splicing) (Figure 1(b)). Mechanism of generating identical fusion transcripts may differ in cancer and normal cases. The fusions JAZF1-JJAZ1 and PAX3-FOXO1 are the two such examples of fusion transcripts generated by chromosomal translocation (i.e. DNA level) in cancer cells but by trans-splicing (i.e. RNA level) in normal cells [30,31]. Although all the fusion detection tools provide the information about the fusion type (i.e. ‘interchromosomal’ and ‘intrachromosomal’ etc.), the prediction of detail mechanism of the fusions (translocation, deletion, trans-splicing, cis-spicing between neighboring genes, etc.) is impossible with RNA-Seq data alone. Therefore, popular fusion detection tools such as SOAPfuse, EricScript, deFuse, Chimerascan, which use only RNA-Seq reads will not predict fusion-generating mechanism reliably. Some tools such as Comrad, CRAC, FusionMap, IPD-fusion and nFuse use both RNA-Seq reads and WGS reads to predict the mechanism of fusions. Those tools can be used to differentiate fusion events that occur at DNA level or only at RNA level.
FACTORS AFFECTING FUSION DETECTION
The detection of fusions from NGS data relies on the type of sequencing data and the computational strategies utilized to process that data.
WGS and RNA-Seq for the fusion detection
Although Whole Genome Sequencing (WGS) has been used to detect several important gene fusion events [32–34], this technique requires a great amount of sequencing, and exhaustive computational analysis. The cost of WGS of human samples is generally higher when compared to RNA-Seq. In addition, WGS will only detect fusion events that occur at the DNA level. This is a limitation, as WGS will miss all of the fusion events that occur at the RNA splicing level. However, this feature could be desirable in cases where researchers only want to find subset of fusions that are generated by chromosomal rearrangement.
RNA-Seq sequencing only sequences a small part (~2%) of the genome that is transcribed and spliced into mature mRNA [29]. In addition to the traditional gene fusions, RNA-Seq will detect ‘intergenically’ spliced fusions that only occur at the RNA level. RNA-Seq also allows for the detection of multiple alternative splice variants resulting from fusions. Low cost and quick turnaround time make RNA-Seq very popular in fusion transcript studies. However, the limitations of RNA-Seq are: 1) it can not detect fusion events involving non-transcribed events [35]; 2) detecting fusion events not occurring at the DNA level is a double-edged sword; 3) tissue-specificity and the broad dynamic range of expression in the human transcriptome are two factors that complicate RNA-Seq data analysis [36].
Even with these limitations, RNA-Seq is often the method of choice for fusion detection due to its previously mentioned advantages over WGS, and investigators’ interest in transcribed genes. As shown in the Table 2, all these tools can utilize paired-end RNA-Seq reads as input. In addition to the RNA-Seq reads, some of the tools, such as: Comrad [37], CRAC [38], FusionMap [39], IDP-fusion [40], and nFuse [41], also consider Whole Genome Sequencing (WGS) reads as input. IDP-fusion and JAFFA [42] are the recently developed tools, which can also consider the long input reads (i.e. >1000 bp) generated by Single molecule real time sequencing (SMRT) sequencing technologies.
Effect of Reference sequences
Most of the fusion detection tools require both genome and transcriptome sequences as references (Table 2). Some tools, such as: Bellerophontes [43], CRAC, Dissect [44], MapSplice [45], and TRUP [46], require only the genome sequences as reference, whereas EricScript and JAFFA consider only transcriptome reference sequences. As a general limitation of using any particular reference, a tool may miss fusions involving novel sequences that are not represented in that reference. For instance, fusions with novel exons cannot be detected by using transcriptome as a reference as in the case of EricScript and JAFFA. Some tools include the assembly software packages to construct new reference sequences. FusionQ [47], BreakFusion, FusionCatcher, and JAFFA use Cufflinks [48], TIGRA-SV [49], Velvet [50], and Oases [51] respectively. As a trade off, these tools tend to require more computational time and memory.
Effect of single-end versus paired-end reads
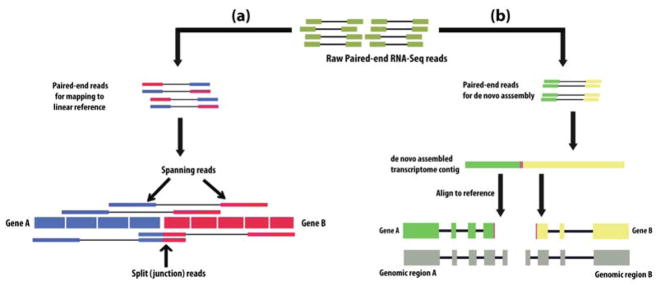
WGS and RNA-Seq experiments can be performed in two formats: single-end, or paired-end. Initially, some studies detect fusions by using single-end reads [26]. Sequencing of both ends of a set of longer DNA/cDNA fragments produced paired short reads, i.e. paired-end reads. A read that harbors a fusion junction is called a ‘split read’ or ‘encompassing read’ (Figure 2(a)). These ‘split reads’ do not directly align with the reference sequence. Analyzing the alignment of ‘split reads’ can thus identify fusion events. If the reads from the ends match two different reference entities, but the fusion junction is not included in either read (Figure 2(a)), these two reads are called ‘spanning reads’, or a ‘spanning pair’. The discordant mappings are a characteristic of fusion events. Using both ‘spanning reads’ and ‘split reads’, Maher et al. [27] and Ha et al. [52], achieved improved sensitivity of fusion detection with paired-end data. However, if only ‘split reads’ are used to identify fusions, data shows that the single-end reads have the ability to detect fusions as well [35,39].
Figure 2. Two approaches for the initial steps of fusion detection.
(a) ‘Mapping first’ approach of fusion detection. Paired-end RNA-Seq reads are first aligned to detect the fusion breakpoint through the alignment of ‘spanning reads’ and ‘split reads’ to the reference sequences. (b) ‘Assembly first’ approach of fusion detection. In this approach, paired-end RNA-Seq reads are first assembled into the contigs (i.e. de novo transcriptome contigs) and then the contigs, having fusion junction, are aligned to reference sequences.
Assembly and mapping of reads
Most of the fusion detection tools first align the reads to reference DNA/RNA sequence, and then find fusion breakpoints from the resulting alignment patterns [11]. This approach is the ‘mapping first’ approach (Figure 2(a)). Another approach, the ‘assembly first’, involves assembling the input reads, and then aligning the assembled contigs to reference DNA/RNA (Figure 2(b)). This approach was used for discovery of several important fusions transcripts in case of acute myeloid leukemia (AML) [53]. Of the above two approaches, the ‘mapping first’ approach is faster and dominant in the field of NGS-based detection of fusions. The main disadvantage of the ‘assembly first’ approach is that the assembly of short reads is time consuming and error prone. BreakFusion [54] and JAFFA [42] use a combination of both approaches.
Other important issues
Other important issues regarding fusion detection include: sequencing coverage, insert length, read length, and quality of reads. Carrara et al. noticed a positive correlation between read length and false positive rates with FusionMap and defuse [22]. They also found a correlation between quality score and false fusion detection with MapSplice. In 2015, Liu et al. discussed the effects of coverage, insert length, read length, and quality of reads on fusion detection [55]. An increase in sequencing coverage led to an increase in the sensitivity of fusion detection of nearly all of the tools they reviewed in their study.
Recently, our research group also discussed the effects of quality and length of RNA-Seq reads. We found that both read length and the quality of RNA-Seq reads affect the false positive fusion predictions [56].
FUSION DETECTION METHODOLOGY
In this section, we focus on to the methodology used by the fusion detection tools. The flow of fusion detection can be divided into three steps: (1) reads mapping and filtering, (2) fusion junction detection, and (3) fusion assembly and selection.
Reads mapping and filtering
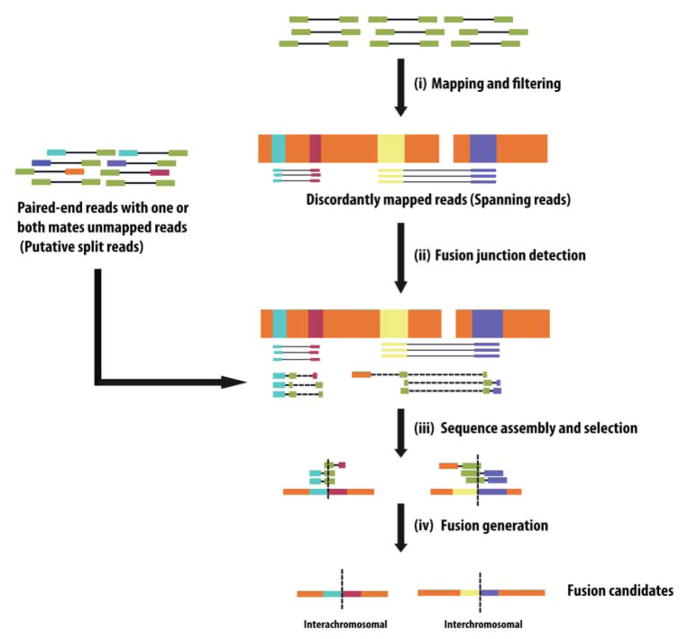
Most of the fusion detection tools utilize mapping as their initial step in analysis (Figure 3(i)). After mapping, the alignment of each read (pair) is evaluated, and the reads that are irrelevant to fusions are removed. Some tools based primarily on ‘split reads’, such as FusionMap and TopHat-Fusion, to filter out all of the mapped reads. Methods, which use ‘spanning reads’, such as SnowShoes-FTD, preserve all discordantly mapped pairs of reads. In addition to this, these methods also keep unmapped reads (potential ‘split reads’) in order to assist in the selection of fusion candidates [29,57–59] (Figure 3(ii)). Most of the methods have the filtering techniques to further discard the reads that are less likely to harbor fusions (Table 3). For example, FusionSeq [29], has more than ten filters to eliminate spurious fusions. One important filter is related to the conformation of intrachromosomal fusions. Two neighboring genes on the same chromosome can produce a read-through transcript. Developers of some tools have decided to eliminate this type of fusion, using a threshold value of distance between the fusion partners, as in the case of FusionMap, FusionHunter [60], ShortFuse [59], SnowShoes-FTD, and TopHat-Fusion.
Figure 3. Fusion detection-using split reads and spanning reads.
(i) Paired-end reads are mapped to reference sequences and discordantly mapped reads (spanning reads) are directly aligned to the target fusion genes. (ii) For paired-end reads with one or both ends unaligned (potential split reads), the unmapped mate is cut into several pieces to be aligned to estimated fusion boundaries. Here, the two pieces of same read are connected by dashed line. (iii) Fusion candidate sequences are assembled. (iv) Assembled fusion sequences with highest probability to be selected as real fusion. Vertical dotted line represents the fusion junction.
Fusion junction detection
The identification of fusion junctions through ‘split read’ mapping is the second step of the procedure [11,35,39,60]. The unmapped reads from the previous step are broken into several pieces, and the first and last segment of each ‘split read’ are then independently aligned to the reference sequences (Figure 3(ii)). Once the alignment pattern is found, adjusting the boundaries of the original fragments, and performing realignment can accurately find the location of the fusion junction (Figure 3(ii)). The length of the partitioned segments influences the ‘split read’ mapping. Short length segments increase the sensitivity for fusion candidates, but also increase the false positive prediction rate. To balance the sensitivity and false positive prediction rates, these methods either break the read into two segments [60,61], or use a fixed length of end segments.
Another approach to identify fusion junctions is to infer fusion breakpoints from ‘spanning reads’, and then extract the candidates that are confirmed by ‘split reads’ [29,57–59]. Discordant alignments are first collected into clusters, each having a maximal set of reads that share the same pair of breakpoints. After this, the boundary region of each candidate fusion junction is identified from its cluster. Next, fusion junction loci are assumed, and putative fusion transcripts are predicted. In the final step, unmapped reads are aligned to the predicted fusion transcripts. The predictions, to which the highest numbers of unmapped reads are aligned, are proposed as candidate fusions.
Fusion assembly and selection
After the identification of fusion junctions, joining the two partner genes together can generate the fusion sequences (Figure 3(iii)). Previously unmapped reads are then aligned to the candidate fusions. Reads mapped in this step (supporting reads) provide an additional layer of confidence to the fusion candidates. Like ‘split reads’, ‘spanning reads’ can also provide supporting evidence, as they comprise fusion junctions in their insert sequences. More supporting reads can eliminate a large number of false candidates; however, in doing this, the risk of discarding true fusions expressed at low transcription levels increases. To overcome this problem, some tools have scoring functions to rank fusion candidates [11,29,35,39,59]. These scoring functions are mainly based on features including read depth, mapping quality, and number of supporting reads. The scores are either generated analytically and empirically (e.g. FusionSeq [29]), or learned from known data using machine learning techniques (e.g. deFuse [11]).
COMPARATIVE ASSESSMENT OF FUSIONS DETECTION TOOLS
To process RNA-Seq data, fusion transcript detection tools require large amounts of time and computational memory, (i.e. RAM). The behavior of these tools also changes with the datasets. As mentioned in the introduction, software tools can not only miss the true fusions events, but also produce false fusions [22]. Therefore, there is a need for practical knowledge of these software packages, or pipelines, in terms of computational time and memory usage, sensitivity, and specificity. In 2013, Carrara et al. compared the performance of six fusion detection tools: FusionHunter, FusionMap, FusionFinder, MapSplice, deFuse, and TopHat-Fusion in terms of sensitivity (on positive dataset) and false fusion detection (on negative dataset) [22]. In this study, they prepared the negative datasets of different read lengths and quality scores, which allows for detecting dependency of the tools on both of these features. Computation time and memory of fusion detection tools were not done in this study. On the basis of their analysis, Carrara et al. [22] concluded that; 1) almost all tools (except FusionHunter) were error prone, with high variability among the tools, identifying some fusions not present in the synthetic dataset, 2) FusionMap has the best balance between specificity and sensitivity, and 3) the sensitivity of the tools does not seem to be sufficient in providing consistent results.
Recently, our research group evaluated the performance of 12 of the best tools available for fusion detection [56]. We evaluated the sensitivity, false discovery rate, computing time, and memory usage of these tools with four different datasets (positive, negative, mixed, and test), and ranked these tools on the basis of TOPSIS analysis. On the basis of this study, we conclude that, 1) EricScript had a high positive predictive value (PPV) (100% on the mix dataset (positive + negative dataset)), and reasonable sensitivity (78%, on the positive dataset), 2) it also requires the least amount of time and memory utilization, 3) JAFFA and SOAPfuse have features that appear to give them the advantage over the older tools, but these tools consumed more time and computational memory on all of the datasets, and 4) read length, quality score of reads, and coverage affect fusion detection.
While our manuscript was undergoing revision review, Liu et al. reported the comparative evaluation of 15 fusion transcript detection tools on the synthetic data sets of different coverage, read length, and background noises [55]. They also checked the performance of these tools on three real data sets with experimental validations [55]. In this study, fusion detection tools were also compared in terms of the amount of time consumed to analyze different datasets. Based on this study, they conclude that, 1) no tool performed dominantly best with all synthetic and real data sets, 2) the performance of SOAPfuse was consistently better for both synthetic and real data sets, followed by FusionCatcher, JAFFA, and PRADA, 3) EricScript and ChimeraScan performed well on synthetic data, but poorly on the three real data sets, and 4) the performance of each tool appeared to be data-dependent, and not always consistent between synthetic and real data. In terms of time, SOAPfuse was one of the most costly software packages.
These comparative studies are useful as they provide guidance for end users to choose proper tools based on their needs. When performing the benchmark study and comparing our analyses with others, we realize some common limitations and biases that influenced some conclusions. They are listed here for future researches to consider.
Firstly, the number of reads in the simulated positive datasets is low (i.e. < 1 million) in all three studies, including ours. So, these datasets do not mimic real RNA-Seq data analyses, which have usually above 50 million reads, for fusion detection. We, as well as Liu et al. [55] tried to address this issue by mixing the positive dataset with a simulated negative dataset, generating a dataset of 70 million reads. Ideally, a bigger simulated positive dataset should be used for the positive dataset.
Secondly, some studies included “negative” datasets with normal human tissue RNA-Seq runs with the assumption that the fusion transcripts should be only present in tumor samples. However, this assumption is unjustified, as numerous studies have shows that the fusion transcripts are also present in normal cell lines and tissues samples [16,18,19,22,62]. So, by using the normal tissues or cell lines as negative datasets, some fusion detection tools were punished for detecting fusions, and deemed to have low specificity. In this regard, we believe that a simulated negative dataset is still ideal to compare false fusion discovery rates.
Lastly, previously published experimental datasets were often included to demonstrate that the fusion detection tools can pick up known fusions. This is important. However, in some comparative studies, fusions detected in addition to the known fusions were considered false positives [55]. We have to keep in mind that known fusions identified through real experimental studies were found using the particular software tool chosen in that particular study. Using these datasets to evaluate precision and recall rates is not justified, because it is based on the assumption that only the known fusions in these datasets are real fusions. Again, some tools were deemed to have high false positive rates based on this assumption. We believe that real experimental datasets can be used as an additional test for sensitivity, but should not be used to compare specificity, as no one knows all the fusions that truly exist in a real experimental dataset.
LIMITATIONS OF CURRENT SEQUENCING TECHNOLOGY
Small overlaps between different tools
In the past, researchers found a very little overlap among the results of different fusion detection algorithms [43,63]. In the recent studies of Liu et al. [55], and our own [56], fusions detected by different software tools have small overlaps. We both concluded that none of the tools are inclusive. This could be due, partially, to the different reference sequences these different tools use (some use only transcriptome, whereas some use both transcriptome and genome), as well as the false positives and false negatives different tools detect.
Read-through fusions
Read-through fusions are considered differently than other fusions. In some software, such fusions are filtered off, as in the situations of TopHat-Fusion and ChimeraScan. In other software tools, they are listed as a separate group, such as in EricScript (read-through) and SOAPfuse (INTRACHR-SS-OGO-0GAP). How does one prove that a read-through is a fusion between two genes, or an alternative splicing of the same gene? The situation becomes more complicated when dealing with overlapping genes. We have noted that some of the fusion detection tools discard overlapping genes. ChimeraScan nominates the chimeras on the basis of the alignment of the discordant reads to the reference genome/transcriptome. Reads aligned to overlapping transcripts are not considered as discordant. FusionCatcher removes the overlapping genes on the same strand according to publically known databases, such as Ensembl, UCSC, or RefSeq. SOAPfuse filters out reads from overlapping gene regions. However, two genes overlapping, does not nullify the possibility of a fusion transcript. Ultimately, we need experimental evidence to prove that the overlapping genes are truly two separate genes, and not a multi-exonic gene that is historically mistaken as two genes. In addition, different gene annotation databases can give different results in terms of overlapping genes. For instance, RTFDC1 and FAM209A are overlapping genes transcribing on the same strand according to Ensembl, but not overlapping according to RefSeq.
Highly similar sequences
Most of the tools use filters to remove highly similar sequences, such as paralogous genes, in order to eliminate false positives due to template switching during library preparation. For instance, EricScript filters out discordant alignments between paralogous genes, and does not consider them for the downstream analysis. It is a reasonable strategy to improve specificity. However, true positives might also be eliminated. Trans-splicing could produce such fusions, as the transcripts from paralogous genes may be processed via the same transcription machinery. To prove whether or not a fusion exists between two genes with highly similar sequences, experimental evidence is ultimately needed.
Conclusion
Though a total of ~33 tools for fusion detection are available for the scientific community, the computational study of fusion genes/transcripts is at preliminary stage. Based on the limitations and performances of current software tools, a better tool with high sensitivity, high specificity, and efficient time and computational memory consumption is needed. Future tools should keep the needs of end users in mind. They should incorporate user-friendly features, such as fusion flanking sequences to guide RT-PCR primer design, fusion junction relative to the exon position, and potential effects on the protein-coding frame. Ideal pipelines should also cross check existing fusion database, such as the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, and ChiTaRS. For users who wish to select the subset of fusions that occur at DNA level, or desire more predictions of fusion mechanism, such pipelines should be able to compare results from WGS and RNA-Seq. For users who wish to identify pathological fusions, the tools should also be able to extract fusions that are not present under normal physiology. This can be done through filtering off fusions detected in normal matched samples, as well as cross checking with existing databases composed of known fusions in normal tissues and cells. These features will help to prioritize fusions, and guide functional study. In addition, single cell sequencing will be advantageous to study the heterogeneity feature of both physiologically normal tissue and tumor samples. Future tools should be compatible with the long input reads (i.e. >1000 bp) generated by Single Molecule Real Time (SMRT) sequencing technologies. Deeper reads and longer read lengths will also certainly help. Other technologies such as PacBio, which can sequence through a transcript, may also make fusion detection more reliable.
As of now, our recommendations to detect fusion events are to: 1) use paired-end format; 2) perform RNA-Seq if interested in fusion transcripts not limited to chromosomal rearrangement; 3) achieve reasonable reads length (>70bp); 4) achieve reasonable reads depth (>50 million); and 5) select appropriate software based on needs.
Acknowledgments
We thank Dr. Daniel Nicorici for helpful discussion. We thank Loryn Facemire for her help on editing the manuscript.
Funding
This work is supported by National Cancer Institute, grant [CA190713]. HL is a Research Scholar Grant [126405-RSG-14-065-01-RMC] from the American Cancer Society, and a St. Baldrick’s V Scholar.
Footnotes
Further Reading
In this review, we focused on the fusion transcripts detection tools, mechanism, factors alter fusion detection, methodology, several efforts made by the researchers to compare the tools and pitfalls of these studied. In addition to this, readers can also go through the comparative studies to gain more insight on the performance of individual tools [22,55,56,75] and other latest review covering discovery and understanding of oncogenic gene fusions [76].
References
- 1.Tkachuk DC, Westbrook CA, Andreeff M, et al. Detection of bcr-abl fusion in chronic myelogeneous leukemia by in situ hybridization. Science. 1990;250:559–62. doi: 10.1126/science.2237408. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 3.Morgan GJ, Wiedemann LM. Molecular biology of the Philadelphia positive leukaemias. Recenti Prog Med. 1989;80:508–19. [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–9. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honeyman JN, Simon EP, Robine N, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–4. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guffanti A, Iacono M, Pelucchi P, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–27. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steidl C, Shah SP, Woolcock BW, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–81. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X, Zhan P, Gavine PR, et al. FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br J Cancer. 2014;110:967–975. doi: 10.1038/bjc.2013.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dancey JE, Bedard PL, Onetto N, et al. The genetic basis for cancer treatment decisions. Cell. 2012;148:409–20. doi: 10.1016/j.cell.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Wolyniec K, Carney DA, Haupt S, et al. New Strategies to Direct Therapeutic Targeting of PML to Treat Cancers. Front Oncol. 2013;3:124. doi: 10.3389/fonc.2013.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–11. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Wang J, Ma X, et al. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle. 2009;8:218–22. doi: 10.4161/cc.8.2.7358. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Gong M, Yuan H, et al. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 18.Qin F, Song Z, Babiceanu M, et al. Discovery of CTCF-sensitive Cis-spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babiceanu M, Qin F, Xie Z, et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magrangeas F, Pitiot G, Dubois S, et al. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor alpha-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J Biol Chem. 1998;273:16005–10. doi: 10.1074/jbc.273.26.16005. [DOI] [PubMed] [Google Scholar]
- 21.Frenkel-Morgenstern M, Lacroix V, Ezkurdia I, et al. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22:1231–42. doi: 10.1101/gr.130062.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrara M, Beccuti M, Cavallo F, et al. State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinformatics. 2013;14(Suppl 7):S2. doi: 10.1186/1471-2105-14-S7-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell PJ, Stephens PJ, Pleasance ED, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampton OA, Den Hollander P, Miller CA, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–77. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin JZ, Berger MF, Adiconis X, et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maher CA, Kumar-Sinha C, Cao X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maher CA, Palanisamy N, Brenner JC, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgren H, Murumagi A, Kangaspeska S, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sboner A, Habegger L, Pflueger D, et al. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010 doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Wang J, Mor G, et al. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–61. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 31.Yuan H, Qin F, Movassagh M, et al. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 32.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Totoki Y, Tatsuno K, Yamamoto S, et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet. 2011;43:464–9. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 34.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–76. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor BS, Ladanyi M. Clinical cancer genomics: how soon is now? J Pathol. 2011;223:318–26. doi: 10.1002/path.2794. [DOI] [PubMed] [Google Scholar]
- 37.McPherson A, Wu C, Hajirasouliha I, et al. Comrad: detection of expressed rearrangements by integrated analysis of RNA-Seq and low coverage genome sequence data. Bioinformatics. 2011;27:1481–8. doi: 10.1093/bioinformatics/btr184. [DOI] [PubMed] [Google Scholar]
- 38.Philippe N, Salson M, Commes T, et al. CRAC: an integrated approach to the analysis of RNA-seq reads. Genome Biol. 2013;14:R30. doi: 10.1186/gb-2013-14-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge H, Liu K, Juan T, et al. FusionMap: Detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- 40.Weirather JL, Afshar PT, Clark TA, et al. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 2015;43:e116. doi: 10.1093/nar/gkv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPherson A, Wu C, Wyatt AW, et al. nFuse: discovery of complex genomic rearrangements in cancer using high-throughput sequencing. Genome Res. 2012;22:2250–61. doi: 10.1101/gr.136572.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson NM, Majewski IJ, Oshlack A. JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7:43. doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abate F, Acquaviva A, Paciello G, et al. Bellerophontes: an RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts334. [DOI] [PubMed] [Google Scholar]
- 44.Yorukoglu D, Hach F, Swanson L, et al. Dissect: detection and characterization of novel structural alterations in transcribed sequences. Bioinformatics. 2012;28:i179–87. doi: 10.1093/bioinformatics/bts214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Singh D, Zeng Z, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Cuesta L, Sun R, Menon R, et al. Identification of novel fusion genes in lung cancer using breakpoint assembly of transcriptome sequencing data. Genome Biol. 2015;16:7. doi: 10.1186/s13059-014-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Ma J, Chang CJ, et al. FusionQ: a novel approach for gene fusion detection and quantification from paired-end RNA-Seq. BMC Bioinformatics. 2013;14:193. doi: 10.1186/1471-2105-14-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz MH, Zerbino DR, Vingron M, et al. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–92. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ha KCH, Lalonde E, Li L, et al. Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genomics. 2011;4:75. doi: 10.1186/1755-8794-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen K, Wallis JW, Kandoth C, et al. BreakFusion: targeted assembly-based identification of gene fusions in whole transcriptome paired-end sequencing data. Bioinformatics. 2012;28:1923–4. doi: 10.1093/bioinformatics/bts272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Tsai W-H, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Vo AD, Qin F, et al. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asmann YW, Hossain A, Necela BM, et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinsella M, Harismendy O, Nakano M, et al. Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics. 2011;27:1068–75. doi: 10.1093/bioinformatics/btr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Chien J, Smith DI, et al. FusionHunter: identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
- 61.Piazza R, Pirola A, Spinelli R, et al. FusionAnalyser: a new graphical, event-driven tool for fusion rearrangements discovery. Nucleic Acids Res. 2012;40:e123. doi: 10.1093/nar/gks394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo J-H, Liu S, Zuo Z-H, et al. Discovery and Classification of Fusion Transcripts in Prostate Cancer and Normal Prostate Tissue. Am J Pathol. 2015;185:1834–45. doi: 10.1016/j.ajpath.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Wu N, Liu J, et al. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol. 2015;10:131. doi: 10.1186/s13000-015-0310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, White NM, Schmidt HK, et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2016;26:108–18. doi: 10.1101/gr.186114.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoogstrate Y, Böttcher R, Hiltemann S, et al. FuMa: reporting overlap in RNA-seq detected fusion genes. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv721. [DOI] [PubMed] [Google Scholar]
- 66.Chuang T-J, Wu C-S, Chen C-Y, et al. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qadir MA, Zhan SH, Kwok B, et al. ChildSeq-RNA: A next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16:361–70. doi: 10.1016/j.jmoldx.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 68.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. 2014 bioRxiv. [Google Scholar]
- 69.Abate F, Zairis S, Ficarra E, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torres-Garcia W, Zheng S, Sivachenko A, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–2226. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia W, Qiu K, He M, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14:R12. doi: 10.1186/gb-2013-14-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Supper J, Gugenmus C, Wollnik J, et al. Detecting and visualizing gene fusions. Methods. 2013;59:S24–8. doi: 10.1016/j.ymeth.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 73.Benelli M, Pescucci C, Marseglia G, et al. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28:3232–9. doi: 10.1093/bioinformatics/bts617. [DOI] [PubMed] [Google Scholar]
- 74.Francis RW, Thompson-Wicking K, Carter KW, et al. FusionFinder: a software tool to identify expressed gene fusion candidates from RNA-Seq data. PLoS One. 2012 doi: 10.1371/journal.pone.0039987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carrara M, Beccuti M, Lazzarato F, et al. State-of-the-art fusion-finder algorithms sensitivity and specificity. Biomed Res Int. 2013;2013:340620. doi: 10.1155/2013/340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw282. [DOI] [PMC free article] [PubMed] [Google Scholar]