Abstract
Nucleotide excision repair (NER) is a versatile pathway that removes helix-distorting DNA lesions from the genomes of organisms across the evolutionary scale, from bacteria to humans. The serial steps in NER involve recognition of lesions, adducts or structures that disrupt the DNA double helix, removal of a short oligonucleotide containing the offending lesion, synthesis of a repair patch copying the opposite undamaged strand, and ligation, to restore the DNA to its original form. Transcription-coupled repair (TCR) is a subpathway of NER dedicated to the repair of lesions that, by virtue of their location on the transcribed strands of active genes, encumber elongation by RNA polymerases. In this review, I report on recent findings that contribute to the elucidation of TCR mechanisms in the bacterium Escherichia coli, the yeast Saccharomyces cerevisiae and human cells. I review general models for the biochemical pathways and how and when cells might choose to utilize TCR or other pathways for repair or bypass of transcription-blocking DNA alterations.
Keywords: Nucleotide excision repair, transcription-coupled repair, DNA damage, DNA repair
Introduction
The genetic material of all living organisms is protected against the constant threats posed by environmental agents and byproducts of cellular metabolic processes. All organisms studied to date possess mechanisms for the prevention, detection and repair of damage to their DNA. There are pathways dedicated to the repair of small nucleotide modifications such as oxidation or alkylation (base excision repair), the rare but potentially lethal occurrence of chromosomal breaks (double-strand break repair), the binding of the complementary strands of DNA by crosslinking agents (interstrand crosslink repair), the sealing of single-nucleotide loss that may result from various metabolic activities (single-strand break repair), and the ubiquitous nucleotide excision repair (NER) that has evolved to deal with structurally different types of lesions that significantly weaken or distort the structure of DNA (Friedberg et al. 2014; Iyama and Wilson 2013). NER was the first mechanism of DNA excision repair identified in the Setlow and Hanawalt laboratories in the 1960s (Pettijohn and Hanawalt 1964; Setlow and Carrier 1964). In 2015 Aziz Sancar was awarded the Nobel Prize in Chemistry for the identification, purification and characterization of NER factors, and mechanistic studies that have greatly contributed to our current understanding of this important process.
The NER process begins with the recognition of a DNA lesion. Then, dual incisions of the damaged DNA strand, one on either side of the lesion, are produced. The lesion-bearing oligonucleotide is removed, a patch is synthesized using the undamaged strand as a template, and the patch is ligated to the contiguous strand. NER comprises two subpathways that differ only in regards to the first step, recognition of the lesion. Global genomic repair (GGR) detects lesions throughout the genome, whereas transcription-coupled repair (TCR) is initiated when RNA polymerases are blocked by lesions on the template DNA strand. TCR was first described in rodent cells, then in human cells, E. coli, yeast and in other organisms (reviewed in Ganesan et al. 2012; Hanawalt and Spivak 2008; Kamarthapu and Nudler 2015; Mullenders 2015), including halophilic archaea as recently reported (Stantial et al. 2016), and was defined as the recognition and repair of DNA lesions or structures occurring in the transcribed strand of active genes. Interestingly, the first observation of TCR (Mellon et al. 1987) was possible because the model lesions utilized in those initial experiments, cyclobutane pyrimidine dimers (CPDs), are repaired inefficiently in non-transcribed DNA in hamster cells, facilitating detection of the faster repair of transcribed strands by TCR.
Mutations that result in null or reduced functionality of NER proteins cause mild to extreme photosensitivity in humans (reviewed in Spivak and Hanawalt 2015). Xeroderma pigmentosum (XP) was the first disease ascribed to a defect in DNA repair (Cleaver 1968). XP patients have a 1000-4000 higher incidence of cancer of the skin, eyes, and tip of the tongue than unaffected individuals; progressive neurological degeneration can affect 20% of XP patients, and an increased incidence of internal cancers has been reported. XP comprises 7 complementation groups with defective GGR; 5 of these groups exhibit defective TCR, whereas patients in the XPC and XPE complementation groups (with defective XPC or XPE protein respectively) are TCR-proficient. An 8th complementation group, XP variant (XPV), is proficient in NER; the disease is due to mutations in DNA polymerase η, a translesion bypass polymerase that specializes in replicating DNA containing CPDs (Johnson et al. 1999). Cockayne syndrome (CS) is a multi-symptom disease defined by microcephaly, defective growth, and progressive neurologic degeneration due to leucodystrophy, in addition to 3-5 minor criteria such as demyelination of peripheral nerves, pigmentary retinopathy, hearing loss, cachectic dwarfism, and progeria with shortened lifespan. Most CS patients are photosensitive, but their cells are always UV-sensitive. CS cells are completely defective in TCR but they are GGR-competent. CS patients belong to one of 2 complementation groups, CSA or CSB; rare mutations in XPB, XPD, XPG or ERCC1 can result in combined XP/CS phenotypes. Interestingly, no cancers have been reported in CS patients.
Trichothiodystrophy (TTD) patients exhibit intellectual impairment and other symptoms similar to those of CS. The most notable characteristic of TTD is the presence of brittle hair and nails; the tiger-tail patterning of the hair under polarizing light is diagnostic of the disease. There are 6 complementation groups of TTD, but only 3 are photosensitive and are the result of certain mutations in the XPB, XPD or TTDA subunits of the transcription/repair factor TFIIH.
Combined XP/TTD phenotypes have been reported to result from certain mutations in XPD. UV-sensitive syndrome (UVSS) can be caused by mutations in CSA, CSB or UVSSA. Only 8 patients have been reported. These individuals develop normally and are only affected by sun sensitivity and abnormal pigmentation in exposed skin; the relatively mild phenotype suggests that many more patients remain undiagnosed. No cancers have been observed to date in UVSS patients. Additional diseases with defective NER and photosensitivity include De Sanctis-Cacchione syndrome, XFE progeroid syndrome, and cerebro-oculo-facial-skeletal (COFS) syndrome. All the diseases mentioned above are autosomal recessive, and are very rare except in isolated ethnic groups with a high incidence of intermarriage.
Global and transcription-coupled repair in bacteria
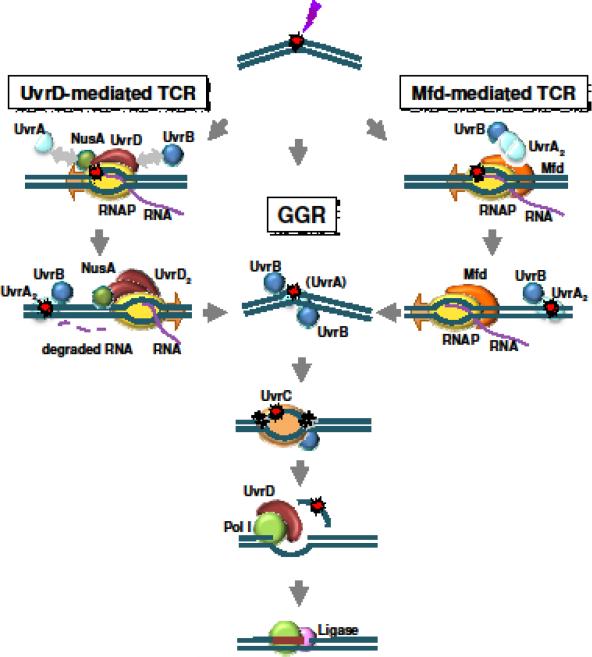
The GGR pathway in E. coli begins when a complex of the UvrA and UvrB proteins recognizes a DNA lesion in a process involving three-dimensional random searching (Kisker et al. 2013). UvrC then associates with UvrB and produces two nicks in the damaged DNA strand, one on either side of the lesion. UvrA dissociates from UvrB, and UvrB then dissociates from the DNA after the dual incisions. DNA helicase I (the UvrD protein) and DNA polymerase I cooperate to remove the oligonucleotide containing the lesion and replace the missing bases using the undamaged complementary strand as a template. Finally, ligase I joins the newly synthesized sequence to the contiguous DNA strand (Kisker et al. 2013; Spivak 2015; Spivak and Ganesan 2014) (Fig. 1 center).
Fig. 1. NER in E. coli.
A DNA lesion that does not block transcription may be detected by the combined action of the UvrAB complex. If transcription arrest occurs, two alternative modes of displacing the RNAP can operate: the Mfd-mediated forward movement that implies transcription termination or bypass of the lesion, and the UvrD/NusA-mediated backtracking that allows resumption of transcription once the lesion is repaired. Both Mfd and UvrD/NusA can recruit the UvrAB complex, which binds to the DNA and recognizes and verifies the damage to be repaired. Repair then proceeds through a common series of steps. UvrA dissociates from the preincision complex leaving 1 or 2 molecules of UvrB bound to the DNA. UvrC interacts with UvrB and catalyzes two nicks in the DNA, one on either side of the lesion. The combined action of UvrD (helicase I) and DNA polymerase I removes the oligonucleotide containing the lesion, as well as UvrB and UvrC, from the DNA and results in the synthesis of a patch using the undamaged complementary strand as a template. DNA ligase I seals the patch to the contiguous DNA strand, thus restoring the DNA to its original form
TCR is triggered upon transcription blockage when the RNAP, rather than the UvrAB complex, recognizes a lesion as it transcribes a gene in a linear mode. The arrested polymerase complex may cover the lesion, necessitating specific activities to remove the RNAP and expose the lesion to the repair complex. The first protein implicated in TCR in E. coli was the mutation frequency decline (Mfd) factor, also known as transcription coupled repair factor (TCRF) (Selby et al. 1991). Mfd is a translocase that binds to the arrested RNAP and promotes its forward movement by pushing it from behind; at the same time, Mfd recruits UvrA through its UvrB homology domain, thus leading to the initiation of GGR (reviewed in Kamarthapu and Nudler 2015; Spivak and Ganesan 2014) (Fig. 1 right).
Nearly 3 decades later, an Mfd-independent mode of TCR has been proposed (Fig. 1 left). In addition to the core RNA polymerase subunits, the elongation complex in vivo contains other proteins, including the regulatory proteins NusA, NusG, and Rho, which modulate the behavior of the complex and have different patterns of assembly and dissociation on different genes (Mooney et al. 2009). NusA associates with RNAP as it leaves the promoter region, and it has termination and antitermination activities; it also functions in translesion DNA synthesis through its interaction with DNA polymerase IV. Mutants defective in NusA are sensitive to DNA damage, including that caused by UV (Cohen et al. 2010). It has been proposed that NusA, which can interact with UvrA (Butland et al. 2005; Cohen et al. 2010), mediates an alternative Mfd-independent pathway of TCR that could account for the fact that the inactivation of mfd has little effect on viability or sensitivity to UV (Cohen et al. 2010; Cohen and Walker 2011). Recently, it has been reported that UvrD is a major player in this response. UvrD binds RNAP and travels with it as it elongates along the DNA template. When the RNAP stalls, UvrD can actively pull the polymerase back, using its helicase activity to unwind the duplex DNA “behind” the transcription bubble (Epshtein et al. 2014). This activity of UvrD would be in addition to its role in dissociating the oligonucleotide resulting from incision during NER. It has been reported that a uvrD mutant that retained some capacity for GGR was deficient in TCR of CPDs (Crowley and Hanawalt 2001). Although it is difficult to evaluate TCR when GGR is impaired, this report is consistent with the idea that UvrD can participate in some form of TCR. According to the new scenario, NusA facilitates the backtracking mediated by UvrD, whereas Mfd counteracts it (Epshtein et al. 2014). The interactions of NusA with UvrA, and UvrD with UvrB suggest that UvrD and NusA may recruit NER enzymes to the site of damage after removing the polymerase.
The existence of these 2 pathways that deal with arrested transcription may have resulted from the need to respond to various situations. Lesions that occur at low frequencies and result in a few arrested transcription complexes may be efficiently removed via Mfd-mediated TCR, whereas genotoxic stresses that induce the SOS response, increasing the levels of UvrD by 3-fold, may favor the NusA-UvrD-mediated TCR. However, the idea of a TCR pathway independent of Mfd is difficult to reconcile with the observation that mfd mutations eliminate TCR of CPDs in vitro and in vivo (Manelyte et al. 2010; Mellon and Champe 1996; Selby and Sancar 1993; Selby et al. 1991). Further analyses will be needed to reconcile these observations and to identify other factors that may be involved. For example, a domain of NusG shows a structural similarity to the RNAP binding domain of Mfd (Deaconescu et al. 2006) and, like Mfd (Wimberly et al. 2013), NusG can interact with R-loops resulting from transcription (Leela et al. 2013).
TCR was initially defined as “faster” repair of UV-induced cyclobutane pyrimidine dimers on template DNA strands than in non-transcribed DNA sequences. What determines the rate of repair in TCR compared to GGR? The search for DNA damage by the pre-incision complex might be the rate determinant, considering the vast amount of undamaged DNA that must be scanned for relatively rare lesions (reviewed in Kad and Van Houten 2012) whereas the 1-dimensional scanning by the elongation complex might lead to more rapid encounters with lesions in transcribed strands than 3D scanning by the UvrAB pre-incision complex. Another consideration might be which mode of TCR operates on a particular DNA sequence and for a particular type of lesion: several lines of evidence indicate that Mfd interacts with the polymerase only after the elongation complex is formed (reviewed in Spivak and Ganesan 2014). Furthermore, the interaction appears to require that the complex be stalled, possibly because, at least in vitro, Mfd makes several attempts before it interacts productively with the complex (Howan et al. 2012). In principle, this should have important implications for the rate of repair. The NusA-UvrD-directed TCR mode might be stimulated through the SOS response when higher levels of genomic damage require faster interactions with arrested transcription complexes.
An important difference between the 2 modes of TCR is the fate of the nascent transcript: Mfd-dependent TCR may result in transcription termination if the RNAP is dislodged from the DNA; if the RNAP is pushed forward but remains active, implying bypass of the lesion, elongation may proceed to completion producing a normal or a mutated mRNA depending on whether the correct or the wrong ribonucleotide is incorporated opposite the lesion, a phenomenon termed transcriptional mutagenesis (Bregeon et al. 2003) consistent with the finding that Mfd promotes mutagenesis of active genes in stationary phase B. subtilis (Martin et al. 2011; Ross et al. 2006). As for the second mode, UvrD/NusA-dependent TCR results in backtracked RNAP that can resume forward elongation via anti-backtracking factors such as GreA and GreB and including Mfd itself, which cleaves the transcript and reactivate the backtracked RNAP; these events would presumably occur after repair of the lesion, thus the transcripts would be correct. The potential advantage of this mode of TCR is that the transcript need not be discarded during the repair process.
Global and transcription-coupled repair in human cells
Various recent review articles have covered the roles of human GGR and TCR factors and presented hypothetical models for the overall mechanisms and their steps, and for relationships between mutant NER factors and the phenotypes of syndromes resulting from the mutations (Gaillard and Aguilera 2013; Iyama and Wilson 2013; Mullenders 2015; Spivak 2015; Spivak and Ganesan 2014; Spivak and Hanawalt 2015; Vermeulen and Fousteri 2013). Since the discovery of TCR in the 1980s in the Hanawalt laboratory, many of the underlying genetic and biochemical details have been elucidated, but several areas still need clarification. Although the factors involved in moving the stalled RNAP forward or backward have been recently described for E. coli (see above), the mechanism(s) for removing the polymerase to allow repair in human cells are not clear; moreover, the process is more complicated and the number of proteins involved is much larger than in E. coli, as shown in Fig 2 and 3. Importantly, in E. coli all the NER factors are required for GGR and for Mfd-mediated or UvrD-mediated TCR, while in humans TCR can take place in the absence of the GGR factors XPE, XPC or hRAD23b (Fig 2). Furthermore, E. coli has only one RNAP whereas in human cells there are 3 nuclear RNAPs; RNAPII is the only one that has been reported to participate in TCR to date.
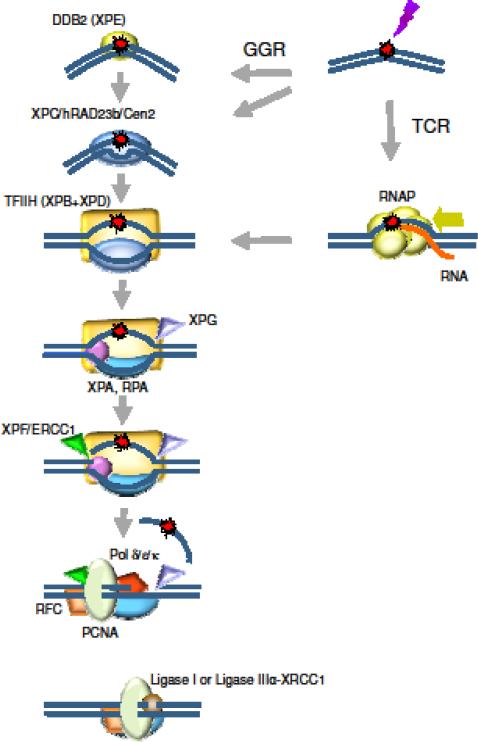
Fig. 2. NER in humans.
Higher eukaryotes utilize different, independent mechanisms for detecting DNA alterations in actively transcribed and in silent DNA strands.
In TCR, RNAPII arrested at a lesion constitutes the signal for recruitment of TCR factors, and the polymerase is removed or backtracked to allow access to TFIIH and other NER repair enzymes.
In GGR, a helix distorting lesion or structure can be directly recognized by the XPC/hRAD23b/Cen2 complex, or it is first recognized by DDB2, the XPE factor. The XPC complex melts the DNA around the lesion and attracts the multiunit complex TFIIH. TCR and GGR converge; the XPB and XPD helicases within TFIIH unwind the DNA to create a ~30 nucleotide bubble, while XPA and RPA verify the lesion containing strand and bind to the opposite strand. The XPG flap endonuclease binds the 3’ single/double strand junction and the XPF-ERCC1 flap endonuclease complex incises the 5’ junction; the DNA replication machinery pol δ/ε/κ-PCNA-RFC synthesizes a patch displacing the damaged strand, XPG incises the 3’ junction, and ligases seal the DNA
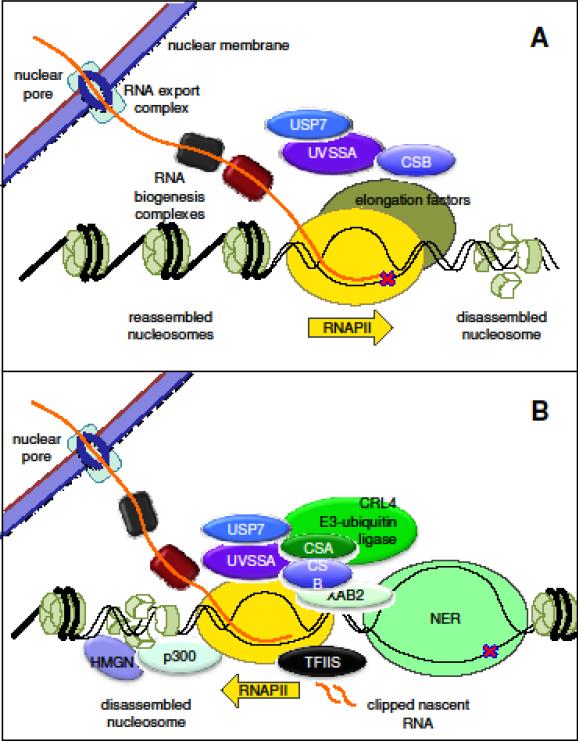
Fig 3. TCR in humans: a hypothesis.
As RNAPII translocates, nucleosomes ahead of the polymerase must slide or disassemble, and then regenerate behind the transcription complex. Protein complexes that process mRNA and export it to the cytoplasm function in coordination with transcription elongation. When RNAPII is blocked, CSB binds the polymerase and recruits the CSA E3 ubiquitin-ligase complex, NER proteins and p300. UVSSA and USP7, TFIIS, HMGN1 and XAB2 are recruited by CSA. The FACT protein SPT16 is also recruited to TCR sites. The chromatin modifiers p300, HMGN1 and perhaps SPT16 may loosen nucleosomes behind the polymerase to allow it to backtrack, an activity facilitated by TFIIS-dependent activation of nascent RNA cleavage by RNAPII. The vacated transcription bubble around the lesion is now accessible to TFIIH and downstream NER factors. Upon completion of the repair reaction, CSB is ubiquitinated and degraded, and transcription can resume
A lesion or structure that causes a significant distortion of the DNA double helix can be directly recognized by the XPC/hRAD23b/centrin2 complex; minor distortions that are not detectable by the XPC complex are recognized by DDB2 (XPE) in complex with DDB1, enhancing the disruption of the DNA duplex and recruiting the XPC complex. DDB1 and DDB2 are part of the CUL4-ROC1 ubiquitin ligase complex that ubiquitinates DDB2, XPC and histones when DNA is damaged. XPC binds the DNA strand opposite the lesion; this explains the ability of NER to recognize a wide array of lesions, adducts and abnormal structures in DNA (Lee et al. 2014). The XPC/hRAD23b/Cen2 complex melts the DNA around the lesion and recruits the transcription/repair factor TFIIH. TFIIH is a basal transcription initiation complex that comprises 10 proteins; two of them are the ATPases/helicases XPB and XPD. Only the ATPase activity of XPB is required for NER, while XPD must be active as both ATPase and helicase, suggesting that XPD translocates along the DNA and detects lesions when its motion is impeded (Mathieu et al. 2013). XPB and XPD unwind the DNA to create a ~30-nucleotide bubble. Other components of TFIIH participate in NER: p8, the smallest subunit, is an absolute requirement for NER and it is defective in trichothiodystrophy complementation group A (TTD-A); p52 stimulates XPB, and p44 stimulates XPD (Singh et al. 2015). Once the pre-incision complex is assembled, XPA, RPA and XPG are recruited. XPA binds the DNA near the 5″ side of the bubble, where it interacts with TFIIH, RPA, PCNA, XPC, DDB2 and ERCC1-XPF. RPA is composed of three subunits, and binds the single-stranded DNA opposite the lesion, protecting it from degradation and helping to coordinate excision and repair events (Scharer 2013). XPG associates with TFIIH and lends structural support, but its endonuclease activity is not triggered until ERCC1-XPF has been recruited by XPA to the 5″ end of the bubble. The order in which the dual incisions are made had been the subject of discussion; Fagbemi and colleagues demonstrated that ERCC1-XPF makes the first incision, and that repair synthesis can be initiated and will proceed for several nucleotides in the absence of XPG incision (Fagbemi et al. 2011). The DNA replication machinery pol δ/ϵ/κ-PCNA–RFC–RPA synthesizes a patch displacing the damaged strand and TFIIH; pol ϵ is active for NER in replicating cells and pol δ and κ are the main NER polymerases in non-replicating cells (Lehmann 2011; Ogi et al. 2010). XPG incises the 3′ single/double strand junction, and ligase I seals the DNA in replicating cells, while ligase IIIα and its cofactor XRCC1 carry out this step in quiescent cells (Scharer 2013).
In TCR, the arrested RNAPII constitutes the universal signal for recruitment of TCR factors such as CSB, CSA, XAB2, UVSSA, USP7, plus histone-remodeling factors described below. The lesion recognition complexes XPC/hRAD23b/Cen2 and DDB1/DDB2 that are essential for GGR are absolutely dispensable for initiating TCR.
CSB (ERCC6), a transcription elongation factor that translocates along template DNA with RNAPII, strongly binds to the polymerase when it is blocked at a lesion, and changes the DNA conformation by wrapping the DNA around the protein itself, altering the interface between RNAPII and DNA (Beerens et al. 2005). CSB recruits the CSA complex, NER factors and chromatin remodelers such as p300 to sites of arrested RNAPII, and it has been considered the master coordinator of TCR in humans, with roles similar to those carried out by Mfd in E. coli. The C-terminal region of CSB, essential for TCR, is required for interaction with RNAPII and for translocation of CSA to the nucleus, and is modified by small ubiquitin-like modifier (SUMO) 2/3 (Sin et al. 2016). CSB colocalizes with lesions other than those induced by UV, such as interstrand crosslinks induced by trioxalen, angelicin monoadducts, double-strand breaks, and oxidative damage (Iyama and Wilson 2016), and is involved in the repair of endogenously generated cyclopurines in mouse tissues (Kirkali et al. 2009).
CSA (ERCC8) is the substrate recognition factor in the DCX E3 ubiquitin ligase complex, which contains CSA, RBX1, and CUL4A. Like CSB, CSA localizes to sites containing double-strand breaks, interstrand crosslinks and angelicin adducts, but not to oxididation damage (Iyama and Wilson 2016). CSA is required for the recruitment of XAB2, HMGN1 and TFIIS to sites of arrested RNAPII; CSA also recruits UVSSA to chromatin following UV irradiation (Fei and Chen 2012). CSA-dependent degradation of CSB is required for recovery of RNA synthesis after UV damage (Brooks 2013).
The UVSSA protein and its partner USP7 (Nakazawa et al. ; Schwertman et al. 2012; Zhang et al. 2012) are loosely associated with elongating RNAPII. Upon transcription arrest, these factors bind strongly to RNAPII, and USP7 deubiquitinates CSB to stabilize it. Together UVSSA and USP7 prevent DNA damage-induced degradation of CSB, facilitate CSA and CSB-dependent ubiquitination of arrested RNAPIIo (the phosphorylated form of RNAPII during elongation), its remodeling and USP7-dependent deubiquitination, and its recycling to non-phosphorylated RNAPIIa for transcription initiation (reviewed in Schwertman et al. 2013). XAB2 (XPA-binding protein 2) is a protein involved in transcription and splicing of pre-mRNA, and it is required for TCR but not for GGR of UV-induced photolesions. XAB2 interacts with XPA, RNAPII and RNA; these interactions are enhanced upon DNA damage. Moreover, chromatin immunoprecipitation studies have shown that XBA2 coprecipitates with stalled RNAPII, CSB, CSA and other TCR factors, but the specific role of XAB2 in TCR is not known (Kuraoka et al. 2008; Nakatsu et al. 2000)
Chromatin remodeling
Nucleosome-bound DNA poses significant barriers to RNA polymerases. Tightly controlled and extremely complex sets of histone modifiers have evolved to regulate each step during transcription: binding of RNAPs to promoters, initiation, elongation and termination, all in a chromatin context. As the elongating RNAPII translocates along the DNA of a gene, histones in front of the polymerase are acetylated by histone acetyltransferases (HATs), displaced and redeposited behind RNAPII by histone chaperones, including FACT, SPT6 and Asf1. The hyperacetylated histones are methylated by Set2 and then deacetylated by the Rpd3S complex. The methyl groups are eliminated by a histone demethylase when the gene is to be derepressed (reviewed in Li et al. 2007).
Repair of damaged DNA also requires chromatin remodeling for access to repair complexes, and it has been suggested that certain chromatin components prepare a repair-competent region and signal specific repair pathways (reviewed in Czaja et al. 2012; Soria et al. 2012). Things become more complicated when repair occurs in conjunction with transcription. In addition to HATs, histone chaperones, deacetylases (HDACs) and other chromatin modifying enzymes that regulate transcription, some factors are specifically recruited by TCR proteins, or act exclusively during TCR. For example, the HAT p300 and the nucleosomal nonhistone binding protein HMGN1, which stimulates HAT activity and unwinds chromatin, are recruited to TCR sites in a CSA and CSB-dependent manner (Fousteri et al. 2006), and UVSSA interacts with HMGN1 (Schwertman et al. 2012; Vermeulen and Fousteri 2013). CAF-1 is involved in chromatin assembly associated with DNA replication and repair, but to our knowledge it has not been found in association with TCR complexes.
The newly identified FACT (facilitates chromatin transcription) subunit SPT16 specifically binds to TCR sites in chromatin in UV-irradiated cells, whereas RNAPII stalling per se does not elicit binding of SPT16. A knock-down of SPT16 resulted in decreased or delayed recovery of RNA synthesis after UV (Dinant et al. 2013).
Resolving transcription complexes blocked by DNA lesions
Stalled RNAPII has a ~35 nucleotide footprint, occupying 10 nucleotides in front and 25 nucleotides behind a lesion (Tornaletti et al. 1999). The lesion may be trapped within or near the active site of the polymerase. What happens to this polymerase, and how does the NER complex find the lesion? Several scenarios have been proposed:
Reverse translocation or backtracking by the RNA polymerase occurs normally in the absence of DNA damage when the transcription complex encounters certain sequence contexts. Resumption of elongation requires degradation of the nascent RNA to reposition its 3’ end in the active site of the polymerase. This reaction is facilitated by the elongation factor TFIIS, which stimulates the transcript cleavage activity of RNAPII. In the context of TCR, backtracking can occur to allow space for the repair complex to operate, as shown in a proof-of-principle experiment in which a photolyase gained access to a CPD upon TFIIS-mediated backtracking of the transcription elongation complex (Tornaletti et al. 1999). The involvement of TFIIS is underlied by its requirement for recovery of RNA synthesis after UV-induced damage, and its recruitment to sites of damage by CSA (Fousteri et al. 2006). RNAPII backtracking would result in reannealing of the bubble around the lesion. In GGR, XPC or XPE initiate the denaturation of DNA at damaged sites. XPC and XPE are not required for TCR, thus bringing up the question of how is this step is carried out in TCR: is the transcription bubble kept open? Is the DNA denatured again? Another issue is how to deal with the nucleosomes that reassemble behind RNAPII. The p300 and HMGN1 chromatin factors, which have been found to co-precipitate with stalled RNAPII complexes, might facilitate nucleosome sliding upstream of RNAPII so that the polymerase can backtrack (Fig. 3) (Hanawalt and Spivak 2008; Lans et al. 2012).
Elements of the NER complex might access and remove the offending lesion within a remodeled polymerase, as suggested by Sarker et al. (Sarker et al. 2005). The multistep NER process, which must include recruitment of the 10-unit TFIIH, endonucleases XPG and XPF-ERCC1, DNA synthesis machinery and the ligase complex, is difficult to envision with the polymerase in place.
Although some DNA lesions are absolute blocks to transcription, at least in vitro, other lesions can be bypassed by RNAPII. Lesion bypass can be stimulated by CSB (Selby and Sancar 1997), TFIIF, Elongin and TFIIS (Charlet-Berguerand et al. 2006) and may result in transcriptional mutagenesis (Saxowsky et al. 2008). This mechanism for avoidance of stalled transcription complexes might constitute the sole equivalent of E. coli's “push forward” Mfd-mediated TCR, except that in humans it does not result in TCR, but rather it clears the way for GGR to find and repair the lesion post-trancriptionally, a situation analogous to post-replication repair that occurs when lesions are bypassed during replication with the aid of TLS polymerases.
Persistent stalling of RNAP can have catastrophic consequences, including cell death. As a last resort, the arrested RNAPII is tagged for degradation, thus aborting transcription and presumably degrading the transcript. Stalled RNAPII is first mono-ubiquitinated by Nedd4, and subsequently poly-ubiquitinated by the ElonginA/B/C-Cullin5 complex, targeting the polymerase for disassembly and proteasomal degradation (Harreman et al. 2009).
As the nascent RNA exits RNAP, various protein complexes process the RNA readying it for export to the cytoplasm. Of these, the THO, TREX and THSC/TREX-2 complexes have been shown to be required for TCR, suggesting that proper biogenesis of export-competent mRNA is important for transcription processivity (Gaillard et al. 2007). Other factors that modulate transactions between transcription and mRNA biogenesis also play a role in TCR; these include the PAF/Paf1 and the Ccr4-Not complexes (reviewed in (Gaillard and Aguilera 2013)). After repair of the transcription blocking lesion is completed, CSB is ubiquitinated by CSA, targeting it for proteasomal degradation as mentioned above, and independently by BRCA1 (Wei et al. 2011), probably to restore its pre-TCR conformation, thus returning it to its role as an elongation factor.
TCR of non-bulky or non-distorting lesions
Bulky, DNA distorting lesions that are recognized by NER generally obstruct elongation by RNA polymerases, eliciting TCR. Smaller adducts and nucleotide modifications have been shown to arrest transcription under certain conditions, including sequence context, promoter strength, and distance from the promoter, both in vitro and in vivo.
Whether oxidation products of DNA, in particular 8-oxoGuanine (8-oxoG), are subject to TCR has interested researchers for many years; these lesions are efficiently removed from the genome through the base excision repair (BER) pathway. Several groups have presented direct and indirect evidence of preferential repair of oxidized bases (Banerjee et al. 2010; Reis et al. 2012; Spivak and Hanawalt 2006). Menoni et al. developed a novel laser-directed method to inflict oxidative DNA damage locally in living cells and observed a clear transcription-dependent recruitment of CSB to these lesions (Menoni et al. 2007); they have subsequently documented the localization of XPC, but not of XPA or XPB, to oxidatively-damaged spots, which suggests that “canonical NER” is not involved in genomic repair of those lesions (Menoni et al. 2012). We have recently shown that 8-oxoG is preferentially repaired in the transcribed strand of the ataxia telangiectasia mutated (ATM) gene in human cells, and that this activity requires CSB, UVSSA, hOGG1, XPA and RNAPII in its active elongation mode. The fact that both hOGG1 (a BER factor) and XPA (an NER factor) are needed led us to speculate that a crossover occurs between these distinct DNA repair pathways. Our hypothesis is that RNAPII is not arrested by oxidized bases but rather by products of their recognition, including depurination and/or the resulting single-nucleotide gap resulting from the activities of specialized glycosylases and endonucleases that initiate BER; the blocked RNAPII recruits TCR factors thus feeding into the NER pathway (Guo et al. 2013). In E. coli, lack of Mfd activity does not affect lesion removal, survival or recovery of RNA synthesis after treatment with H2O2 (Schalow et al. 2012) indicating that in this organism there is no TCR of oxidized bases, or that TCR is not important for coping with oxidative damage; a role for UvrD/NusA remains to be elucidated. Oxidized guanine bases in DNA can be subject to further oxidation, leading to the formation of hydantoin lesions, which are highly mutagenic (McKibbin et al. 2013). The human homologs of the E. coli endonuclease eight (Nei), NEIL1, 2 and 3, have been shown to remove these lesions from DNA. To our knowledge, TCR of these lesions has not been demonstrated to date.
NER in yeast
NER has been extensively studied in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe; the overall mechanism is very similar to that in mammals ((Prakash and Prakash 2000; and Li 2011)). The names of the proteins involved differ between the 2 strains (Table 1). In S. cerevisiae, a bulky lesion is recognized by Rad14, RPA, and the Rad4/Rad23 and Rad7/Rad16/Elc1 complexes. It has been suggested that Rad7/Rad16/Elc1 bind the lesion site first and recruit other NER factors; no human homologs of these proteins have been found although they may play a role similar to that of DDB2 (XPE) (Reed 2005). An important difference between yeast and mammalian NER is that the human lesion recognition factor XPC is dispensable for TCR, whereas its yeast homolog Rad4 is required for both GGR and TCR. TFIIH is recruited, and the Rad3 and Rad25 helicases unwind the DNA. Incisions are performed at the 5’ junction of the bubble by the Rad1/Rad10 complex, and by Rad2 at the 3’ junction. As in humans, synthesis of the repair patch is carried out by DNA polymerases, PCNA, RPA and RFC, and the resulting gap is sealed by the Cdc9 DNA ligase; no homologs of human ligase III or XRCC1 have been found in yeast. As in humans, GGR requires chromatin remodeling and histone modifications (reviewed in Tatum and Li 2011).
TABLE 1.
NER factors: homologs, orthologs and functional equivalents
| Human | Rodent | S. cerevisiae | S. pombe | Function | Ref |
|---|---|---|---|---|---|
| XPC | Rad4 | rhp41, rhp42 | Lesion recognition and binding complex | 1 | |
| HR23B | Rad23 | rhp23 | 1 | ||
| Centrin2 | 2 | ||||
| DDB1 | Rad7 | ddb1 | Lesion recognition complex | 1 | |
| DDB2, XPE | Rad16 | rph16 | 1 | ||
| Elongin C | Elc1 | Lesion recognition complex with Rad16 and Rad7 | 1 | ||
| XPA | Rad14 | rhp14 | Lesion recognition, complex stabilization | 1 | |
| XPB | ERCC3 | SSL2, Rad25 | ercc3sp | Helicases; TFIIH factor | 1 |
| XPD | ERCC2 | Rad3 | rad15 | 1 | |
| TTDA | TFIIH factor | 2 | |||
| MMS19L | Mms19 | Stabilizes XPD | 1 | ||
| RPA | RFA | ssb | Stabilizes open single stranded DNA; damage recognition | 1 | |
| XPF | ERCC4 | Rad1 | rad16 | Catalyzes 5’ incision | 1 |
| ERCC1 | ERCC1 | Rad10 | swi10 | 1 | |
| XPG | ERCC5 | Rad2 | rad13 | Catalyzes 3’ incision; stabilizes open complex | 1 |
| DNA pol δ | DNA pol δ | Gap-filling repair synthesis | 1 | ||
| DNA pol ε | DNA pol ε | Gap-filling repair synthesis | 1 | ||
| DNA pol κ | DNA pol κ | 2 | |||
| PCNA | PCNA | Sliding clamp for DNA polymerases | 1 | ||
| RFC | 2 | ||||
| Ligase I | Cdc9 | cdc17 | Ligation | 1 | |
| Ligase III | Ligation complex | 2 | |||
| XRCC1 | |||||
| RNAPII | 2 | ||||
| TFIIS | 2 | ||||
| CSB | ERCC6 | Rad26 | TCR-specific DNA-dependent ATPase | 1 | |
| Rpb9 | Rpb9 | TCR-specific subunit of RNA polymerase II | 1 | ||
| CSA | ERCC8 | Rad28 | TCR-specific ubiquitin ligase | 2 | |
| UVSSA | TCR-specific de-ubiquitinase | 2 | |||
| USP7 | 2 | ||||
| Senataxin | Sen1 | 3 |
References: 1. Tatum and Li 2011; 2. Spivak 2015; 3. Li et al. 2016
TCR in S. cerevisiae was initially ascribed to Rad26, the homolog of CSB (van Gool et al. 1994). However, Rad26-deficient strains are not particularly UV-sensitive. An alternative TCR pathway was discovered later to depend on Rpb9, a subunit of RNAPII (Li and Smerdon 2002), These seemingly overlapping mechanisms have apparently evolved to deal with differing circumstances; for example Rad26 operates in both the promoter and coding sequences of expressed genes whereas Rpb9 acts only on coding regions; their contributions also depend on the particular gene being analyzed: Rad26 is required for TCR in URA3 and dispensable for TCR in GAL1, which depends on Rpb9. It appears that Rad26 participates in TCR (and in GGR!) of sequences that are repressed or transcribed at low levels, whereas Rpb9 plays a larger role in TCR of highly transcribed genes. Curiously, rad26 Δ rpb9 Δ double mutants are moderately UV sensitive, suggesting that additional pathways or proteins could be involved in TCR in yeast. As recently reported by Li and coworkers, Sen1 might be a candidate participant in TCR. Sen1, the homolog of human senataxin, is an essential transcription termination factor and it resolves naturally formed R-loops. The Sen1 interactions with Rad2 and with the Rpb1 subunit of RNAPII suggested a role in TCR. Indeed, Sen1 deletions or mutations cause UV sensitivity, but the function of Sen1 in TCR has not been elucidated (Li et al. 2016). To complicate things even further, a number of TCR suppressors have been found in yeast, including Spt4, Spt5, PAFc and the subcomplex Rpb4/Rpb7 (reviewed in Tatum and Li 2011).
The functional homolog of CSA in S. cerevisiae is Rad28, but its role in TCR of CPDs is unclear: Rad28 is not necessary for survival, TCR or recovery of RNA synthesis after UV radiation, however the double mutants rad28 Δ rad7 Δ or rad28 Δ rad16 Δ are more UV sensitive than either single mutant, suggesting a role of Rad28 in tolerance of UV damage (Bhatia et al. 1996; Reagan and Friedberg 1997). TFIIS appears not to be required for TCR in S. cerevisiae (Verhage et al. 1997).
General discussion
UV radiation is the most ubiquitous mutagen and carcinogen on Earth, and primordial organisms developed systems that efficiently remove UV-induced photolesions from their DNA, including photoreversal and NER (reviewed in Ganesan and Hanawalt 2016). What is the biological benefit of TCR? Its importance is underlined by the evolution of parallel TCR pathways, in some cases redundant, in diverse species. TCR of UV-induced CPDs is detected in human cells over a vast range of UV doses, from 1000 (Gao et al. 1994) to 0.01 J/m2; the latter induces approximately 1 CPD/megabase, or ~3000 lesions per genome (Guo et al. 2013). TCR-deficient cells exhibit varying degrees of sensitivity to treatments with DNA damaging agents in the laboratory; however, it appears that some TCR-deficient organisms are viable, develop normally and lead productive lives, as illustrated by UVSS individuals, who suffer only from sun sensitivity and accompanying skin hyperpigmentation. However it can be argued that early humans would have suffered a severe disadvantage if they were unable to hunt and gather while exposed to sunlight. As eloquently stated by Brooks (Brooks 2013), the devastating symptoms of Cockayne syndrome are not caused by the TCR defect by itself; thus, we will not consider this disease as the prototype TCR-defective example in humans. Moreover, TCR-deficient mutants of E. coli are only slightly UV-sensitive and grow at a normal rate. So is TCR simply a redundant mechanism to ensure that transcription templates are quickly cleared of damage, particularly when GGR is slow or ineffective? The latter situation occurs for particular classes of lesions that are invisible to GGR but are recognized and removed by TCR. Examples of these are produced by the acylfulvenes Illudin S and irofulven (Koeppel et al. 2004) and references therein) and by aristolochic acid (Sidorenko et al. 2012). In certain organs and tissues of multicellular organisms, terminally differentiated cells exhibit attenuated global NER but retain TCR to repair template strands in active genes, and use GGR to repair only the non-transcribed strands in such genes. This mechanism, dubbed “domain-associated repair”, maintains lesion-free transcription templates and their complementary strands, which are needed for error-free DNA synthesis during repair (Nouspikel et al. 2006). One assumes that in the absence of TCR, both strands in active genes are repaired by GGR in such tissues; to our knowledge this has not been examined. In summary, although the principal enzymatic steps required for TCR are known, many of the details of the protein associations and modifications, the identity of the participants and the highly coordinated succession of steps they perform, remain to be ascertained.
Acknowledgments
Supported by National Institute of Environmental Health Sciences grant 5RO1 CA077712. We apologize to those whose work has not been cited.
References
- Banerjee D, Mandal SM, Das A, et al. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2010;286(8):6006–16. doi: 10.1074/jbc.M110.198796. doi:M110.198796 10.1074/jbc.M110.198796: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J Biol Chem. 2005;280(6):4722–9. doi: 10.1074/jbc.M409147200. doi:M409147200 10.1074/jbc.M409147200: [DOI] [PubMed] [Google Scholar]
- Bhatia PK, Verhage RA, Brouwer J, Friedberg EC. Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J Bacteriol. 1996;178(20):5977–88. doi: 10.1128/jb.178.20.5977-5988.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol Cell. 2003;12(4):959–70. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- Brooks PJ. Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA repair. 2013;12(8):656–71. doi: 10.1016/j.dnarep.2013.04.018. doi:S1568-7864(13)00099-2 10.1016/j.dnarep.2013.04.018: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433(7025):531–7. doi: 10.1038/nature03239. doi:10.1038/nature03239: [DOI] [PubMed] [Google Scholar]
- Charlet-Berguerand N, Feuerhahn S, Kong SE, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25(23):5481–91. doi: 10.1038/sj.emboj.7601403. doi:7601403 10.1038/sj.emboj.7601403: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218(5142):652–6. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Cohen SE, Lewis CA, Mooney RA, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107(35):15517–22. doi: 10.1073/pnas.1005203107. doi:1005203107 10.1073/pnas.1005203107: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SE, Walker GC. New discoveries linking transcription to DNA repair and damage tolerance pathways. Transcription. 2011;2(1):37–40. doi: 10.4161/trns.2.1.14228. doi:10.4161/trns.2.1.14228: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley DJ, Hanawalt PC. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat Res. 2001;485(4):319–29. doi: 10.1016/s0921-8777(01)00068-4. doi:S0921-8777(01)00068-4 [pii]: [DOI] [PubMed] [Google Scholar]
- Czaja W, Mao P, Smerdon MJ. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int J Mol Sci. 2012;13(9):11954–73. doi: 10.3390/ijms130911954. doi:10.3390/ijms130911954: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaconescu AM, Chambers AL, Smith AJ, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124(3):507–20. doi: 10.1016/j.cell.2005.11.045. doi:S0092-8674(06)00064-X 10.1016/j.cell.2005.11.045: [DOI] [PubMed] [Google Scholar]
- Dinant C, Ampatziadis-Michailidis G, Lans H, et al. Enhanced Chromatin Dynamics by FACT Promotes Transcriptional Restart after UV-Induced DNA Damage. Mol Cell. 2013;51(4):469–79. doi: 10.1016/j.molcel.2013.08.007. doi:S1097-2765(13)00580-7 10.1016/j.molcel.2013.08.007: [DOI] [PubMed] [Google Scholar]
- Epshtein V, Kamarthapu V, McGary K, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505(7483):372–7. doi: 10.1038/nature12928. doi:10.1038/nature12928: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA repair. 2011;10(7):722–9. doi: 10.1016/j.dnarep.2011.04.022. doi:10.1016/j.dnarep.2011.04.022: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Chen J. KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J Biol Chem. 2012;287(42):35118–26. doi: 10.1074/jbc.M112.398131. doi:10.1074/jbc.M112.398131: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23(4):471–82. doi: 10.1016/j.molcel.2006.06.029. doi:S1097-2765(06)00465-5 10.1016/j.molcel.2006.06.029: [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Elledge SJ, Lehmann AR, Lindahl T, Muzi-Falconi M. DNA Repair, Mutagenesis, and Other Responses to DNA Damage. Cold Spring Harbor Laboratory Press; New York: 2014. [Google Scholar]
- Gaillard H, Aguilera A. Transcription coupled repair at the interface between transcription elongation and mRNP biogenesis. Biochim Biophys Acta. 2013;1829(1):141–50. doi: 10.1016/j.bbagrm.2012.09.008. doi:S1874-9399(12)00168-X 10.1016/j.bbagrm.2012.09.008: [DOI] [PubMed] [Google Scholar]
- Gaillard H, Wellinger RE, Aguilera A. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 2007;35(12):3893–906. doi: 10.1093/nar/gkm373. doi:gkm373 10.1093/nar/gkm373: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A, Hanawalt P. Photobiological Origins of the Field of Genomic Maintenance. Photochem Photobiol. 2016;92(1):52–60. doi: 10.1111/php.12542. doi:10.1111/php.12542: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. doi:B978-0-12-387665-2.00002-X 10.1016/B978-0-12-387665-2.00002-X: [DOI] [PubMed] [Google Scholar]
- Gao S, Drouin R, Holmquist GP. DNA repair rates mapped along the human PGK1 gene at nucleotide resolution. Science. 1994;263(5152):1438–40. doi: 10.1126/science.8128226. [DOI] [PubMed] [Google Scholar]
- Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41(16):7700–12. doi: 10.1093/nar/gkt524. doi:gkt524 10.1093/nar/gkt524: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–70. doi: 10.1038/nrm2549. doi:nrm2549 10.1038/nrm2549: [DOI] [PubMed] [Google Scholar]
- Harreman M, Taschner M, Sigurdsson S, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A. 2009;106(49):20705–10. doi: 10.1073/pnas.0907052106. doi:0907052106 10.1073/pnas.0907052106: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howan K, Smith AJ, Westblade LF, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490(7420):431–4. doi: 10.1038/nature11430. doi:nature11430 10.1038/nature11430: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA repair. 2013;12(8):620–36. doi: 10.1016/j.dnarep.2013.04.015. doi:S1568-7864(13)00096-7 10.1016/j.dnarep.2013.04.015: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Wilson DM., 3rd Elements That Regulate the DNA Damage Response of Proteins Defective in Cockayne Syndrome. J Mol Biol. 2016;428(1):62–78. doi: 10.1016/j.jmb.2015.11.020. doi:10.1016/j.jmb.2015.11.020: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283(5404):1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Kad NM, Van Houten B. Dynamics of lesion processing by bacterial nucleotide excision repair proteins. Prog Mol Biol Transl Sci. 2012;110:1–24. doi: 10.1016/B978-0-12-387665-2.00001-8. doi:B978-0-12-387665-2.00001-8 10.1016/B978-0-12-387665-2.00001-8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarthapu V, Nudler E. Rethinking transcription coupled DNA repair. Curr Opin Microbiol. 2015;24:15–20. doi: 10.1016/j.mib.2014.12.005. doi:10.1016/j.mib.2014.12.005: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkali G, de Souza-Pinto NC, Jaruga P, Bohr VA, Dizdaroglu M. Accumulation of (5'S)-8,5′-cyclo-2′-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA repair. 2009;8(2):274–8. doi: 10.1016/j.dnarep.2008.09.009. doi:10.1016/j.dnarep.2008.09.009: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harbor perspectives in biology. 2013;5(3):a012591. doi: 10.1101/cshperspect.a012591. doi:5/3/a012591 10.1101/cshperspect.a012591: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel F, Poindessous V, Lazar V, Raymond E, Sarasin A, Larsen AK. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin Cancer Res. 2004;10(16):5604–13. doi: 10.1158/1078-0432.CCR-04-0442. doi:10.1158/1078-0432.CCR-04-0442 10/16/5604. [DOI] [PubMed] [Google Scholar]
- Kuraoka I, Ito S, Wada T, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J Biol Chem. 2008;283(2):940–50. doi: 10.1074/jbc.M706647200. doi:M706647200 10.1074/jbc.M706647200: [DOI] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4. doi: 10.1186/1756-8935-5-4. doi:1756-8935-5-4 10.1186/1756-8935-5-4: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Cai Y, Mu H, et al. The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: is there a correlation? DNA repair. 2014;19:55–63. doi: 10.1016/j.dnarep.2014.03.026. doi:10.1016/j.dnarep.2014.03.026: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leela JK, Syeda AH, Anupama K, Gowrishankar J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110(1):258–63. doi: 10.1073/pnas.1213123110. doi:1213123110 10.1073/pnas.1213123110: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR. DNA polymerases and repair synthesis in NER in human cells. DNA repair. 2011;10(7):730–3. doi: 10.1016/j.dnarep.2011.04.023. doi:10.1016/j.dnarep.2011.04.023: [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–19. doi: 10.1016/j.cell.2007.01.015. doi:S0092-8674(07)00109-2 10.1016/j.cell.2007.01.015: [DOI] [PubMed] [Google Scholar]
- Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21(21):5921–9. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Selvam K, Rahman SA, Li S. Sen1, the yeast homolog of human senataxin, plays a more direct role than Rad26 in transcription coupled DNA repair. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw428. doi:10.1093/nar/gkw428: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40(5):714–24. doi: 10.1016/j.molcel.2010.11.012. doi:S1097-2765(10)00850-6 10.1016/j.molcel.2010.11.012: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HA, Pedraza-Reyes M, Yasbin RE, Robleto EA. Transcriptional de-repression and Mfd are mutagenic in stressed Bacillus subtilis cells. J Mol Microbiol Biotechnol. 2011;21(1-2):45–58. doi: 10.1159/000332751. doi:10.1159/000332751: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Kaczmarek N, Ruthemann P, Luch A, Naegeli H. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Current biology : CB. 2013;23(3):204–12. doi: 10.1016/j.cub.2012.12.032. doi:10.1016/j.cub.2012.12.032: [DOI] [PubMed] [Google Scholar]
- McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J Am Chem Soc. 2013;135(37):13851–61. doi: 10.1021/ja4059469. doi:10.1021/ja4059469: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93(3):1292–7. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–9. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Menoni H, Gasparutto D, Hamiche A, et al. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol Cell Biol. 2007;27(17):5949–56. doi: 10.1128/MCB.00376-07. doi:MCB.00376-07 10.1128/MCB.00376-07: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menoni H, Hoeijmakers JH, Vermeulen W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J Cell Biol. 2012;199(7):1037–46. doi: 10.1083/jcb.201205149. doi:10.1083/jcb.201205149: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33(1):97–108. doi: 10.1016/j.molcel.2008.12.021. doi:S1097-2765(08)00891-5 10.1016/j.molcel.2008.12.021: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders L. DNA damage mediated transcription arrest: Step back to go forward. DNA repair. 2015;36:28–35. doi: 10.1016/j.dnarep.2015.09.005. doi:10.1016/j.dnarep.2015.09.005: [DOI] [PubMed] [Google Scholar]
- Nakatsu Y, Asahina H, Citterio E, et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem. 2000;275(45):34931–7. doi: 10.1074/jbc.M004936200. doi:10.1074/jbc.M004936200: [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Sasaki K, Mitsutake N, et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nature genetics. 44(5):586–92. doi: 10.1038/ng.2229. doi:ng.2229 10.1038/ng.2229: [DOI] [PubMed] [Google Scholar]
- Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Mol Cell Biol. 2006;26(23):8722–30. doi: 10.1128/MCB.01263-06. doi:MCB.01263-06 10.1128/MCB.01263-06: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–27. doi: 10.1016/j.molcel.2010.02.009. doi:10.1016/j.molcel.2010.02.009: [DOI] [PubMed] [Google Scholar]
- Pettijohn D, Hanawalt P. Evidence for Repair-Replication of Ultraviolet Damaged DNA in Bacteria. J Mol Biol. 1964;9:395–410. doi: 10.1016/s0022-2836(64)80216-3. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451(1-2):13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Reagan MS, Friedberg EC. Recovery of RNA polymerase II synthesis following DNA damage in mutants of Saccharomyces cerevisiae defective in nucleotide excision repair. Nucleic Acids Res. 1997;25(21):4257–63. doi: 10.1093/nar/25.21.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SH. Nucleotide excision repair in chromatin: the shape of things to come. DNA repair. 2005;4(8):909–18. doi: 10.1016/j.dnarep.2005.04.009. doi:10.1016/j.dnarep.2005.04.009: [DOI] [PubMed] [Google Scholar]
- Reis AM, Mills WK, Ramachandran I, Friedberg EC, Thompson D, Queimado L. Targeted detection of in vivo endogenous DNA base damage reveals preferential base excision repair in the transcribed strand. Nucleic Acids Res. 2012;40(1):206–19. doi: 10.1093/nar/gkr704. doi:gkr704 10.1093/nar/gkr704: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol. 2006;188(21):7512–20. doi: 10.1128/JB.00980-06. doi:10.1128/JB.00980-06: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker A, Tsutakawa S, Kostek S, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH insights for transcription-coupled repair and Cockayne syndrome. Mol Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(48):18877–82. doi: 10.1073/pnas.0806464105. doi:0806464105 10.1073/pnas.0806464105: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalow BJ, Courcelle CT, Courcelle J. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J Bacteriol. 2012;194(10):2637–45. doi: 10.1128/JB.06725-11. doi:JB.06725-11 10.1128/JB.06725-11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harbor perspectives in biology. 2013;5(10):a012609. doi: 10.1101/cshperspect.a012609. doi:10.1101/cshperspect.a012609: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P, Lagarou A, Dekkers DH, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nature genetics. 2012;44(5):598–602. doi: 10.1038/ng.2230. doi:ng.2230 10.1038/ng.2230: [DOI] [PubMed] [Google Scholar]
- Schwertman P, Vermeulen W, Marteijn JA. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma. 2013;122(4):275–84. doi: 10.1007/s00412-013-0420-2. doi:10.1007/s00412-013-0420-2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260(5104):53–8. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94(21):11205–9. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991;88(24):11574–8. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Carrier WL. The Disappearance of Thymine Dimers from DNA: An Error-Correcting Mechanism. Proc Natl Acad Sci U S A. 1964;51:226–31. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2012;40(6):2494–505. doi: 10.1093/nar/gkr1095. doi:gkr1095 10.1093/nar/gkr1095: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin Y, Tanaka K, Saijo M. The C-terminal Region and SUMOylation of Cockayne Syndrome Group B Protein Play Critical Roles in Transcription-coupled Nucleotide Excision Repair. J Biol Chem. 2016;291(3):1387–97. doi: 10.1074/jbc.M115.683235. doi:10.1074/jbc.M115.683235: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Compe E, Le May N, Egly JM. TFIIH subunit alterations causing xeroderma pigmentosum and trichothiodystrophy specifically disturb several steps during transcription. Am J Hum Genet. 2015;96(2):194–207. doi: 10.1016/j.ajhg.2014.12.012. doi:10.1016/j.ajhg.2014.12.012: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46(6):722–34. doi: 10.1016/j.molcel.2012.06.002. doi:S1097-2765(12)00491-1 10.1016/j.molcel.2012.06.002: [DOI] [PubMed] [Google Scholar]
- Spivak G. Nucleotide excision repair in humans. DNA repair. 2015;36:13–8. doi: 10.1016/j.dnarep.2015.09.003. doi:10.1016/j.dnarep.2015.09.003: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA repair. 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. doi:S1568-7864(14)00096-2 10.1016/j.dnarep.2014.03.025: [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA repair. 2006;5(1):13–22. doi: 10.1016/j.dnarep.2005.06.017. doi:S1568-7864(05)00195-3 10.1016/j.dnarep.2005.06.017: [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC. Photosensitive human syndromes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2015;776:24–30. doi: 10.1016/j.mrfmmm.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stantial N, Dumpe J, Pietrosimone K, Baltazar F, Crowley DJ. Transcription-coupled repair of UV damage in the halophilic archaea. DNA repair. 2016;41:63–8. doi: 10.1016/j.dnarep.2016.03.007. doi:10.1016/j.dnarep.2016.03.007: [DOI] [PubMed] [Google Scholar]
- Tatum D, Li S. Nucleotide Excision repair in S. cerevisiae. In: Storici F, editor. DNA repair - on the pathway to fixing DNA damage and errors. InTech; 2011. www.intechopen.com/books/dna-repair-onthe-pathways-to-fixing-dna-damage-and-errors/nucleotide-excision-repair-in-s-cerevisiae. [Google Scholar]
- Tornaletti S, Reines D, Hanawalt PC. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J Biol Chem. 1999;274(34):24124–30. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool AJ, Verhage R, Swagemakers SM, et al. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13(22):5361–9. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage RA, Heyn J, van de Putte P, Brouwer J. Transcription elongation factor S-ii is not required for transcription-coupled repair in yeast. Mol Gen Genet. 1997;254(3):284–90. doi: 10.1007/s004380050417. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harbor perspectives in biology. 2013;5(8) doi: 10.1101/cshperspect.a012625. doi:5/8/a012625 10.1101/cshperspect.a012625: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Lan L, Yasui A, et al. BRCA1 contributes to transcription-coupled repair of DNA damage through polyubiquitination and degradation of Cockayne syndrome B protein. Cancer Sci. 2011;102(10):1840–7. doi: 10.1111/j.1349-7006.2011.02037.x. doi:10.1111/j.1349-7006.2011.02037.x: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun. 2013;4:2115. doi: 10.1038/ncomms3115. doi:ncomms3115 10.1038/ncomms3115: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Horibata K, Saijo M, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nature genetics. 2012;44(5):593–7. doi: 10.1038/ng.2228. doi:ng.2228 10.1038/ng.2228: [DOI] [PubMed] [Google Scholar]