Abstract
Ebola virus continues to be problematic as sporadic outbreaks in Africa continue to arise, and as terrorist organizations have considered the virus for bioterrorism use. Several proteins within the virus have been targeted for antiviral chemotherapy, including VP35, a dsRNA binding protein that promotes viral replication, protects dsRNA from degradation, and prevents detection of the viral genome by immune complexes. To augment the scope of our antiviral research, we have now employed molecular modeling techniques to enrich the population of compounds for further testing in vitro. In the initial docking of a static VP35 structure with an 80,000 compound library, 40 compounds were selected, of which four compounds inhibited VP35 with IC50 < 200uM, with the best compounds having an IC50 of 20 uM. By superimposing 26 VP35 structures, we determined four aspartic acid residues were highly flexible and the docking was repeated under flexible parameters. Of 14 compounds chosen for testing, five compounds inhibited VP35 with IC50 < 200uM and one compound with an IC50 of 4 uM. These studies demonstrate the value of docking in silico for enriching compounds for testing in vitro, and specifically using multiple structures as a guide for detecting flexibility and provide a foundation for further development of small molecule inhibitors directed towards VP35.
Keywords: Ebola, VP35, Inhibitor, Innate Immune Response, Molecular Dynamic Modeling
Graphical Abstract
Introduction
Although the 2014 ebola virus outbreak has been largely contained, sporadic cases are still occurring two years later 1. The continued emergence of new cases and the absence of either a vaccine or approved antiviral therapeutic is a great concern at both local and global levels. Several experimental treatments have been administered to a select few individuals. However, the low number of study participants has made the efficacy of these treatments difficult to assess 2–4. Furthermore, many of these treatments involve siRNA or monoclonal antibody cocktails that are difficult and expensive to procure 5, 6. There are several small molecule therapies that are in human studies, however, none of these compounds were designed specifically for ebola virus 7–9.
Ebola virus is negative-sense single strand RNA virus that encodes a polyprotein that is cleaved into seven proteins 10. Nearly every ebola virus protein has been characterized for therapeutic targeting potential 11–15. However, the highly mutable genome of ebola virus reduces the development of a drug target to a few highly conserved regions 16. One such region is the interferon inhibitory domain of VP35, (VP35 IID) 17.
As is expected of viral proteins, Ebola virus VP35 IID has multiple functions 18. As a dsRNA binding protein, VP35 IID protects the replicative dsRNA form of the viral genome from degradation 19. The coating of dsRNA by VP35 IID also prevents endogenous dsRNA binding proteins, such as RIG-I and MDA5, from recognizing and binding the viral dsRNA and initiating an innate antiviral immune response 20–23. VP35-RNA interactions occur through the central basic patch of VP35 IID 19, 24, 25. VP35 IID also participates as a cofactor of the viral polymerase, protein L, which binds the first basic patch of VP35 IID, which is obverse to the central basic patch 26, 27. VP35 IID interacts with L protein and was the impetus for an initial search for VP35 IID inhibitors 15. In this study, the authors modeled the first basic patch for inhibition against a 5.4 million molecule library in silico. From these results, 25 compounds were tested, and derivatives of these compounds were found to bind VP35 at micromolar levels 15.
Herein, we docked an 80,000 compound library against the central basic patch of VP35 IID using a static model of VP35 IID, as well as an ensemble-flexible model of VP35 IID based on the 26 known crystal structures of VP35 IID. Both models yielded several positive hits in vitro, with the ensemble-flexible modeling strategy showing improved hit rates and superior quality of hits.
Materials and methods
Molecular Docking
In silico docking was performed using Molegro Docking Software (CLC Bio) using default parameters. VP35 IID structure 3FKE:A (Protein Data Bank) was utilized as the docked protein. Ligands for in silico docking were curated from the ZINC database (http://zinc.docking.org/catalogs/ncip). The center of the docking sphere was set at x=17.0,y=30.0,z=3.0 for structure 3FKE, chain A. The size of the sphere was 23 angstroms. The software was run on a virtualized Windows 8.1 client with 64 gigabytes of RAM and 34 virtual processors hosted within a Server 2012 R2 Hyper-V failover cluster of 4 Dell R620/R720 Servers. LogP scores of selected compounds were predicted by the Molinspiration Property Calculation Service (molinspiration.com).
Vector construction, protein and dsRNA probe purification
cDNAs coding for the IID of VP35 (Genbank Accession Number AIG96632.1) were synthesized by IDT. The cDNA was designed with EcoRI/KpnI ends and cloned into pRSFDuet-1(Novagen), creating a fusion protein with an N-terminal His-tag. After subcloning, purified plasmids were transformed into BL-21 cells, and then incubated overnight at 37°C in a 1 L LB culture without shaking. After 16 h, cells were then shaken at 220 rpm at 37°C until the culture reached an OD600 of 0.8, after which 300 μM IPTG was added and incubation was continued for an additional 2.5 h. The cells were centrifuged and resuspended in lysis buffer (PBS, 100 μM PMSF, 10 mM imidazole). After sonication, cells were centrifuged at 22,000 g for 10 m, and the supernatant was added to an FPLC column containing a 5 ml bed volume of NTA-Ni Superflow agarose (Qiagen). Columns were rinsed sequentially in lysis buffer containing 20 mM and 50 mM imidazole, before elution in lysis buffer containing 250 mM imidazole. For the creation of dsRNA probes, a 5′ CY5.5 labeled ssRNA (CY5.5-CACUGCGACC ) was annealed to a non-labeled or 3′ Iowa Black RQ quencher (IBRQ) labeled ssRNA ( GGUCGCAGUG-IBRQ) in annealing buffer (50mM Tris HCl (pH 7.5), 150mM NaCl). After separation by PAGE, the band corresponding to annealed dsRNA was excised and incubated overnight at 4 °C in 1 ml annealing buffer. RNA probes were purchased from IDT.
Electrophoretic Mobility Shift Assays (EMSA)
Assays were performed in EMSA buffer (10 mM Tris HCl (pH 7.5), 10 mM NaCl, 10% glycerol). Compound was added to 10 μl of 400 nM VP35, mixed and incubated for five min before adding 10 ul of 60 nM dsRNA probe. After successive five min incubations at room temperature and on ice, reactions were run on 1% agarose gels in chilled TAE buffer, then scanned on an infrared scanner (LI-COR).
Structural Protein Alignment
Available ebola virus VP35 IID structures (PDB: 3FKE, 3L25, 3L26, 4IBB, 4IBC, 4IBD, 4IBE, 4IBF, 4IBG, 4IBI, 4IBJ, 4IBK) were downloaded and individual chains separated into separate files. Each structure contained two VP35 IID chains, with the exception of 3L25, which had four. 25 of the structures were each aligned pairwise with the ‘A’ chain of structure 3FKE utilizing the ‘Structure Protein Alignment’ tool of the Molegro Docking Software. Once all structures were aligned pairwise, all 26 structures were imported into the Molegro Docking Software workspace and saved as one integrated .pdb file. This file was then further analyzed by the UCSF Chimera package 28.
Results
Static modeling of VP35 IID
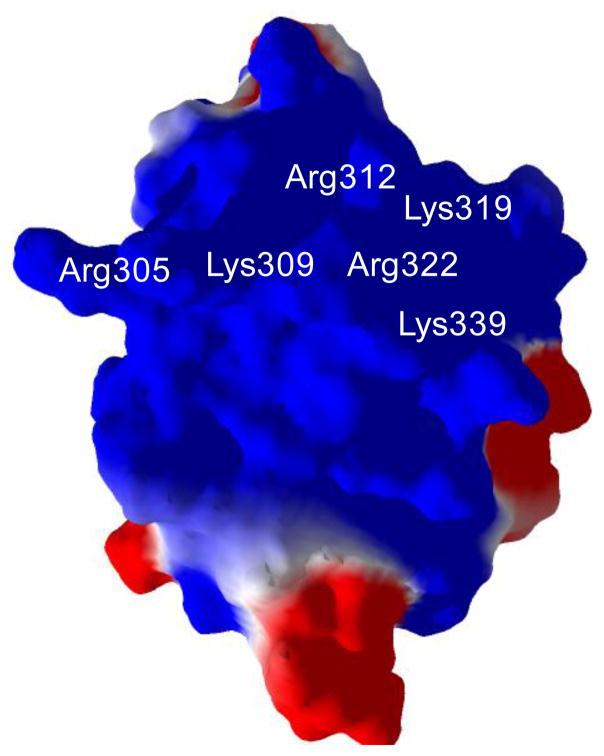
In the initial attempt to screen compounds in silico for VP35 IID inhibitors, the static structure of 3FKE:A was used to dock an 80,000 compound library. This library was chosen for the diversity of compounds, the availability of compound structure files and compounds for experimental use. The search space for docking included the entire central basic cleft of VP35 IID, and specifically surrounded the residues known to be essential for competent dsRNA binding: R305, K309, R312, K319, R322 and K339 (Fig. 1). Because of the spherical requirements of the search space by the docking software and the narrowness of the VP35 IID, a portion of the basic cleft on the reverse side was also included in the search space.
Figure 1.
Structure of the VP35 IID with positive (blue) and negative (red) charged surfaces. Residues essential for competent dsRNA binding are labeled.
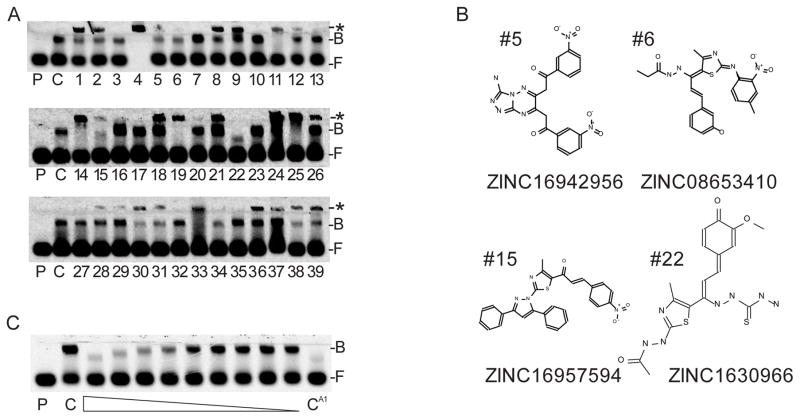
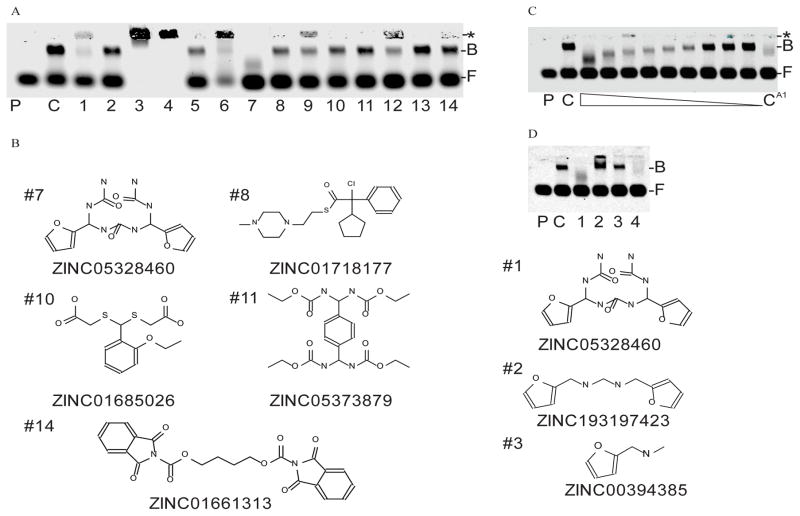
After docking, the compounds were sorted by predicted affinity as described by a ‘rerank’ score. The top 39 scoring compounds were acquired and then tested for their ability to inhibit binding of a fluorescent dsRNA probe to recombinant VP35 IID and to dsDNA and dsRNA (Fig. 2A and Supplementary Fig.1). At 200 μM, four of the 39 compounds were able to inhibit dsRNA binding at greater than 50% of control with nearly no evidence of high-molecular weight complex formation and no indication of DNA and RNA binding (Fig. 2B and Supplementary Fig. 1). Interestingly, these compounds are structurally unique and showed minimal similarity in docking. Of these four compounds, ZINC16957594, showed the greatest amount of inhibition and was tested further in a serial dilution series. The IC50 of ZINC1630966 was calculated to approximately 20 μM (Fig. 2C). The change in mobility of the bound fraction observed may be due to differences in the number of VP35 IID units bound to the substrate in the presence of ZINC16957594. We used coumermycin A1 as a positive control. The discovery of coumermycin A1 came from an initial identification of the aminocoumarin clorobiocin in a high throughput screen (HTS) of 2000 compounds, followed by the testing of a pool of aminocoumarin derivatives that included coumermycin A1.
Figure 2.
(A) Compounds tested for VP35 IID inhibition via EMSA. P=probe only, C=negative control, F=free probe, B=protein bound probe *=complexation of protein-dsRNA and SMI. Compound concentration is 200 μM. (B) Identity and structures of top performing compounds. (C). Dose response of VP35 IID inhibition by ZINC1630966. Compound was serially diluted two fold from 200 μM. CA1=200 μM coumermycin A1.
Alignment of VP35 IID structures reveals flexibility of key residues
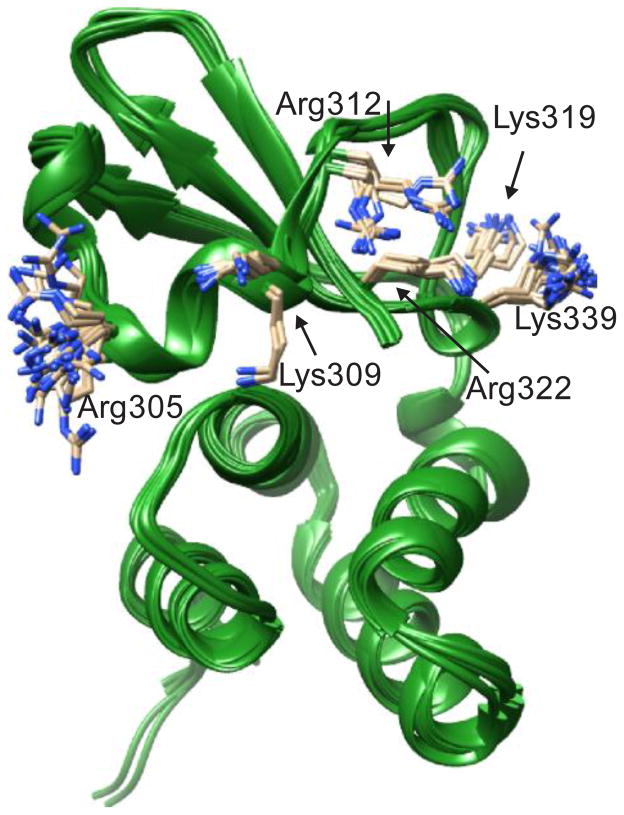
The addition of residue flexibility as a parameter in a docking strategy can add to the overall accuracy of the docking simulation 29, 30. However, each addition of a flexible residue profoundly increases the computing power and time needed for docking 31. Therefore, it is important to select for flexibility only those residues essential for dsRNA binding and are known to have large degrees of freedom. To determine the flexibility of key amino acids, we aligned the 26 available structures of ebola virus VP35 (Fig. 3). Surprisingly, the backbones of the aligned structures are nearly identical, with the exception of cis/trans conformation differences at proline 316. Of the six lysine and arginine residues essential for dsRNA binding, large degrees of freedom were seen in R305 and K339. K309 and R312 residues were each essentially confined to two separate conformations, each with small degrees of freedom. K319 and R322 residues were relatively static. These results suggest that only residues R305, K339, K309 and R312 are flexible and should be modeled as such in the docking strategy.
Figure 3.
Overlay of 26 crystal structures of VP35 IID. Residues essential for dsRNA binding are shown. Arg305 and Lys339 show large degrees of freedom, whereas Lys319 and Arg322 present low degrees of freedom. Lys309 and Arg312 both show a bimodal appearance.
Flexible docking yields increased percentage and quality of hits
Utilizing the flexible docking aspects of the modeling software, 3FKE:A was redocked to the 80,000 compounds library with R305,K339,K309 and R322 designated as flexible. Fourteen of the top scoring compounds were acquired and tested for inhibition of VP35 IID to dsRNA (Fig. 4A). Of the 14 compounds, 5 inhibited binding at greater than 50% without evidence of high-molecular weight complex formation (Fig. 4B). However, one of these compounds, ZINC01661313, was positive for DNA and RNA binding (Supplementary Fig. 1). Of these compounds, ZINC05328460 showed the greatest amount of inhibition and was tested further in a serial dilution series (Fig. 4C). The IC50 of ZINC05328460 was calculated to approximately 4 μM. In an initial test to determine the essential structural aspects of ZINC05328460 for VP35 inhibition, we compared the inhibition of ZINC05328460 to a compound with the ketone and both nitrous moieties removed (ZINC19319423) and a further deletion of one of the 5 member rings (ZINC00394385) (Fig. 4D). Both derivatives showed essentially no inhibition of VP35. As to differences in mobility in the bound fraction in Figures 2C and 4C, we assume that the binding of VP35 to the dsRNA substrate may require anywhere from one to four protein subunits. It is possible to have 4 VP35s bound to the dsRNA substrate. One to each end cap and two on the adjacent strands. Therefore, differences in mobility in the gel of substrate bound VP35 may be due to the prevention of binding of one or more protein subunits by the SMI.
Figure 4.
(A) EMSA of compounds from ensemble-flexible docking. P=probe only, C=negative control, F=free probe, B=protein bound probe. *=complexation of protein-dsRNA and SMI. Compound concentration is 200 μM. (B) Identity, corresponding lane # from (A) and structures of top performing compounds. (C) Dose response of VP35 IID inhibition by ZINC05328460. Compound was serially diluted two fold from 200 μM. CA1=200 μM coumermycin A1. (D) EMSA of ZINC05328460 and partial fragments of ZINC05328460. Lane 1-ZINC05328460. Lane 2-ZINC19319423. Lane 3-ZINC00394385. Lane 4-Coumermycin A1. All compounds were used at 200 μM.
Comparison of static modeling to ensemble-based flexible modeling
Both static modeling and ensemble-flexible modeling of VP35 IID were successful in identifying inhibitors from a relatively small sample. Overall, 16 of 39 (40%) statically modeled compounds profoundly inhibited formation of the VP35 bound to dsRNA band seen in the control sample, whereas 12 of 14 (86%) compounds in the ensemble-flexible strategy did so (Table 1). In both strategies, nearly half of the compounds induced band shifts of high molecular weight, likely due to complex formation with probe, protein and compound. Compound 37 (Fig. 2) also appeared to interact specifically with the dsRNA probe. Complex formation indicates that while it is likely that the compound is interacting with VP35 IID and inhibiting its function, the true nature of the interaction is unknown.
Table 1.
Comparison of static and ensemble-flexible docking strategies
| Strategy | Total Compounds | Inhibition | ||
|---|---|---|---|---|
| <25% | 25–50% | 50–100% | ||
| Static | ||||
| Non-complexing | 21 | 15 | 2 | 4 |
| All | 39 | 20 | 3 | 16 |
| Ensemble-Flexible | ||||
| Non-complexing | 7 | 0 | 2 | 5 |
| All | 14 | 0 | 2 | 12 |
For compounds that show no evidence of complex formation, 4 of 21 statically modeled compounds, and 5 of 7 ensemble-flexible modeled compounds profoundly inhibited dsRNA-VP35 IID interactions at 200 μM. We also note that all ensemble-flexible modeled compounds exhibited at least some (>25%) inhibition of VP35IID, whereas nearly over half of the statically modeled compounds did not.
These results suggest that the ensemble-flexible strategy of molecular docking is considerably more accurate in predicting in vitro inhibition of VP35 IID.
Discussion
Previously we had identified eight compounds in an initial HTS as inhibiting VP35 IID at 200 μM, none inhibited VP35 IID greater than 50% at 100 μM (unpublished observation). Therefore, the high percentage of inhibiting compounds in this study suggest that prescreening compounds in silico greatly enriches the selected pool for quality VP35 inhibitors.
On the other side of VP35 IID, the first basic cleft has also been targeted for protein-protein inhibition 15. In this study, through multiple filtering steps and docking in silico, 25 compounds from a 5.4 million compound database were selected for in vitro testing for VP35 IID ligands, the viral polymerase, L protein and the viral nucleoprotein, NP. Although this strategy did not initially find compounds of moderate affinity to VP35, subsequent derivatives of the pyrrolidinone scaffold containing compound GA017 resulted in the identification of multiple VP35 inhibitors in the mid to low micromolar range. In the current study, no attempt was made to prescreen compounds, such as having a logP score <5 as stated in Brown et al. 15. Interestingly though, all 14 compounds selected by ensemble-flexible docking had a logP score of less than 5 (Table S1). Two compounds selected had negative logP scores, including the high affinity compound, ZINC05328460 (logP=−.48). In contrasting the two VP35 IID strategies, in silico studies were considerably more favorable for the discovery of VP35-dsRNA inhibitors than VP35-protein inhibitors. However, we note that the affinity of VP35 IID to dsRNA is considerably lower than the Kd of VP35 IID to NP and likely L protein 15, 24. Therefore, inhibitors for VP35-dsRNA may not need to be of high affinity to show activity. This advantage may be attenuated by specificity, as these inhibitors may also inhibit the handful of mammalian proteins known to bind dsRNA.
By overlaying an ensemble of available VP35 IID structures, and looking at the orientation of essential residues, we identified four amino acids that acted with considerable freedom, and were designated as flexible for docking purposes. Compounds procured based on this docking method were considerably more active than compounds that were docked from a single protein structure. Future refinements in flexible docking parameters should yield more accurate outcomes, resulting in the discovery of higher quality inhibitors with less time and monetary inputs.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Amarnath Natarajan and Dr. Yogesh Sonawane for their helpful comments. This work was supported by grants from the National Institutes of Health (P20 RR018759-08), the American Cancer Society (RSG-10-031-01-CCG) and the Nebraska Department of Human and Health Services (Grant no. 2025).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Spengler JR, Ervin ED, Towner JS, Rollin PE, Nichol ST. Emerging infectious diseases. 2016:22. doi: 10.3201/eid2206.160021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Griensven J, Edwards T, de Lamballerie X, Semple MG, Gallian P, Baize S, Horby PW, Raoul H, Magassouba N, Antierens A, Lomas C, Faye O, Sall AA, Fransen K, Buyze J, Ravinetto R, Tiberghien P, Claeys Y, De Crop M, Lynen L, Bah EI, Smith PG, Delamou A, De Weggheleire A, Haba N, Ebola-Tx C. The New England journal of medicine. 2016;374:33. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Konde MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Keita S, Kieny MP, Rottingen JA. Lancet. 2015;386:857. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 4.Heald AE, Iversen PL, Saoud JB, Sazani P, Charleston JS, Axtelle T, Wong M, Smith WB, Vutikullird A, Kaye E. Antimicrobial agents and chemotherapy. 2014;58:6639. doi: 10.1128/AAC.03442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunning J, Sahr F, Rojek A, Gannon F, Carson G, Idriss B, Massaquoi T, Gandi R, Joseph S, Osman HK, Brooks TJ, Simpson AJ, Goodfellow I, Thorne L, Arias A, Merson L, Castle L, Howell-Jones R, Pardinaz-Solis R, Hope-Gill B, Ferri M, Grove J, Kowalski M, Stepniewska K, Lang T, Whitehead J, Olliaro P, Samai M, Horby PW team, R.-T. t. PLoS medicine. 2016;13:e1001997. doi: 10.1371/journal.pmed.1001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, Shi W, Choe M, Marcus H, Thompson EA, Cagigi A, Silacci C, Fernandez-Rodriguez B, Perez L, Sallusto F, Vanzetta F, Agatic G, Cameroni E, Kisalu N, Gordon I, Ledgerwood JE, Mascola JR, Graham BS, Muyembe-Tamfun JJ, Trefry JC, Lanzavecchia A, Sullivan NJ. Science. 2016;351:1339. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 7.Sissoko D, Laouenan C, Folkesson E, M’Lebing AB, Beavogui AH, Baize S, Camara AM, Maes P, Shepherd S, Danel C, Carazo S, Conde MN, Gala JL, Colin G, Savini H, Bore JA, Le Marcis F, Koundouno FR, Petitjean F, Lamah MC, Diederich S, Tounkara A, Poelart G, Berbain E, Dindart JM, Duraffour S, Lefevre A, Leno T, Peyrouset O, Irenge L, Bangoura N, Palich R, Hinzmann J, Kraus A, Barry TS, Berette S, Bongono A, Camara MS, Chanfreau Munoz V, Doumbouya L, Souley H, Kighoma PM, Koundouno FR, Rene L, Loua CM, Massala V, Moumouni K, Provost C, Samake N, Sekou C, Soumah A, Arnould I, Komano MS, Gustin L, Berutto C, Camara D, Camara FS, Colpaert J, Delamou L, Jansson L, Kourouma E, Loua M, Malme K, Manfrin E, Maomou A, Milinouno A, Ombelet S, Sidiboun AY, Verreckt I, Yombouno P, Bocquin A, Carbonnelle C, Carmoi T, Frange P, Mely S, Nguyen VK, Pannetier D, Taburet AM, Treluyer JM, Kolie J, Moh R, Gonzalez MC, Kuisma E, Liedigk B, Ngabo D, Rudolf M, Thom R, Kerber R, Gabriel M, Di Caro A, Wolfel R, Badir J, Bentahir M, Deccache Y, Dumont C, Durant JF, El Bakkouri K, Gasasira Uwamahoro M, Smits B, Toufik N, Van Cauwenberghe S, Ezzedine K, Dortenzio E, Pizarro L, Etienne A, Guedj J, Fizet A, Barte de Sainte Fare E, Murgue B, Tran-Minh T, Rapp C, Piguet P, Poncin M, Draguez B, Allaford Duverger T, Barbe S, Baret G, Defourny I, Carroll M, Raoul H, Augier A, Eholie SP, Yazdanpanah Y, Levy-Marchal C, Antierrens A, Van Herp M, Gunther S, de Lamballerie X, Keita S, Mentre F, Anglaret X, Malvy D Group, J. S. PLoS medicine. 2016;13:e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor R, Kotian P, Warren T, Panchal R, Bavari S, Julander J, Dobo S, Rose A, El-Kattan Y, Taubenheim B, Babu Y, Sheridan WP. Journal of infection and public health. 2016 doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turone F. Bmj. 2014;349:g7198. doi: 10.1136/bmj.g7198. [DOI] [PubMed] [Google Scholar]
- 10.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. The Journal of pathology. 2015;235:153. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 11.Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, Shabman RS, Chiu W, Davey RA, Otwinowski Z, Basler CF, Amarasinghe GK. Cell reports. 2015;11:376. doi: 10.1016/j.celrep.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson JA, Bray M, Bakken R, Hart MK. Virology. 2001;286:384. doi: 10.1006/viro.2001.1012. [DOI] [PubMed] [Google Scholar]
- 13.Bornholdt ZA, Ndungo E, Fusco ML, Bale S, Flyak AI, Crowe JE, Jr, Chandran K, Saphire EO. mBio. 2016;7:e02154. doi: 10.1128/mBio.02154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Hemert FJ, Zaaijer HL, Berkhout B. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2015;73:89. doi: 10.1016/j.jcv.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Brown CS, Lee MS, Leung DW, Wang T, Xu W, Luthra P, Anantpadma M, Shabman RS, Melito LM, MacMillan KS, Borek DM, Otwinowski Z, Ramanan P, Stubbs AJ, Peterson DS, Binning JM, Tonelli M, Olson MA, Davey RA, Ready JM, Basler CF, Amarasinghe GK. Journal of molecular biology. 2014;426:2045. doi: 10.1016/j.jmb.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. Science. 2014;345:1369. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grifoni A, Lo Presti A, Giovanetti M, Montesano C, Amicosante M, Colizzi V, Lai A, Zehender G, Cella E, Angeletti S, Ciccozzi M. Asian Pacific journal of tropical medicine. 2016;9:337. doi: 10.1016/j.apjtm.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Leung DW, Prins KC, Basler CF, Amarasinghe GK. Virulence. 2010;1:526. doi: 10.4161/viru.1.6.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, Basler CF, Amarasinghe GK. Nature structural & molecular biology. 2010;17:165. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Muhlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. Journal of virology. 2003;77:7945. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12289. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. Journal of virology. 2006;80:5168. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen B, Mulder LC, Martinez O, Basler CF. Journal of virology. 2014;88:12500. doi: 10.1128/JVI.02163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung DW, Borek D, Farahbakhsh M, Ramanan P, Nix JC, Wang T, Prins KC, Otwinowski Z, Honzatko RB, Helgeson LA, Basler CF, Amarasinghe GK. Acta crystallographica. Section F, Structural biology and crystallization communications. 2010;66:689. doi: 10.1107/S1744309110013266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, Otwinowski Z, Liu G, Huh J, Basler CF, Amarasinghe GK. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20661. doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Journal of virology. 2010;84:10581. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, Basler CF. Journal of virology. 2010;84:3004. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. Journal of computational chemistry. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Kokh DB, Wenzel W. Journal of medicinal chemistry. 2008;51:5919. doi: 10.1021/jm800217k. [DOI] [PubMed] [Google Scholar]
- 30.Sivanesan D, Rajnarayanan RV, Doherty J, Pattabiraman N. Journal of computer-aided molecular design. 2005;19:213. doi: 10.1007/s10822-005-4788-9. [DOI] [PubMed] [Google Scholar]
- 31.Forli S. Molecules. 2015;20:18732. doi: 10.3390/molecules201018732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.