Abstract
Background
Cystic fibrosis (CF) is an autosomal recessive disease characterized by recurrent lung infections. Studies of the lung microbiome have shown an association between decreasing diversity and progressive disease. 454 pyrosequencing has frequently been used to study the lung microbiome in CF, but will no longer be supported. We sought to identify the benefits and drawbacks of using two state-of-the-art next generation sequencing (NGS) platforms, MiSeq and PacBio RSII, to characterize the CF lung microbiome. Each has its advantages and limitations.
Methods
Twelve samples of extracted bacterial DNA were sequenced on both MiSeq and PacBio NGS platforms. DNA was amplified for the V4 region of the 16S rRNA gene and libraries were sequenced on the MiSeq sequencing platform, while the full 16S rRNA gene was sequenced on the PacBio RSII sequencing platform. Raw FASTQ files generated by the MiSeq and PacBio platforms were processed in mothur v1.35.1.
Results
There was extreme discordance in alpha-diversity of the CF lung microbiome when using the two platforms. Because of its depth of coverage, sequencing of the 16S rRNA V4 gene region using MiSeq allowed for the observation of many more operational taxonomic units (OTUs) and higher Chao1 and Shannon indices than the PacBio RSII. Interestingly, several patients in our cohort had Escherichia, an unusual pathogen in CF. Also, likely because of its coverage of the complete 16S rRNA gene, only PacBio RSII was able to identify Burkholderia, an important CF pathogen.
Conclusion
When comparing microbiome diversity in clinical samples from CF patients using 16S sequences, MiSeq and PacBio NGS platforms may generate different results in microbial community composition and structure. It may be necessary to use different platforms when trying to correctly identify dominant pathogens versus measuring alpha-diversity estimates, and it would be important to use the same platform for comparisons to minimize errors in interpretation.
Keywords: Lung microbiome, Cystic fibrosis, MiSeq, PacBio RSII, 16S rRNA, Next generation sequencing
1. Introduction
Cystic fibrosis (CF) is an autosomal recessive disease that affects 30,000 people in the United States and is characterized by recurrent lung infections, with mortality primarily due to progressive lung disease [1]. Culture based diagnostics have been the standard of care for identifying pathogens, but are often limited in scope [2,3]. The lung microbiome in CF is of interest as there has been an association between a loss of bacterial diversity and progression of lung disease [4]. While it is not completely understood, it appears that total antibiotic use is one of the strongest predictors of loss of diversity [5]. Before information from the lung microbiome can be utilized as a biomarker of disease progression and may assist clinicians in antibiotic selection, determining the best available sequencing platform is necessary.
In much of the CF literature, the mainstay of taxonomic identification has been next-generation 454 pyrosequencing of different variable regions within the 16S rRNA gene [4–7]. Since Roche will be discontinuing the 454 platform [8], two commercially available state-of-the-art next-generation sequencing (NGS) platforms have been proposed to characterize the CF lung microbiome: MiSeq (Illumina, San Diego CA) and PacBio RSII (Pacific Biosciences, Menlo Park CA). Each has its advantages and limitations.
The MiSeq platform typically yields shorter read lengths than the 454 (although latest implementation allows for 350 bp paired-reads [9]), but can produce a far higher number of reads, at least 1.5 billion per run [10]. The PacBio produces much longer reads which can readily cover the full length of the 16S rRNA gene (~1450 bp), but has a lower accuracy with random base call ambiguity and an observed error rate of 1.8% [11]. To overcome this limitation, the use of circular consensus sequencing allows for sequencing by synthesis to occur on circularized amplicons, providing high single-molecule coverage [12]. Recent comparisons of PacBio and MiSeq at 16S metataxonomics [13] demonstrated a low percentage of non-matching reads if a high number of circular consensus cycles on the PacBio were performed and a similar correlation of abundances of bacteria in even and uneven microbial mock communities [8]. However, other studies have suggested that the long reads attainable with PacBio provide higher quality operational taxonomic units (OTUs) in a sample [14].
We sought to determine if the MiSeq and PacBio NGS platforms could equivalently identify microbial communities within CF sputum and provide similar measures of alpha-diversity. Understanding these methodological aspects is essential to making technical decisions in future metataxonomic studies of the airway microbiome.
2. Materials and methods
2.1. Ethics, consent and permissions
Before beginning the study the protocol was submitted for review to the IRB at Children’s National Health System (CNHS). As the samples obtained for the study would be de-identified, exempt certification was received.
2.2. Sample collection
De-identified sputum samples from patients with CF were obtained from the clinical microbiology lab at CNHS. The Cystic Fibrosis Center at CNHS cares for approximately 150 children with CF from Maryland, Virginia, and Washington DC.
2.3. Specimen preparation and DNA extraction
Sputum specimens were aliquoted into volumes of 250 to 1000 μL. Sputum pellets were prepared by washing 1:1 (v/v) with normal saline, adding 1:1 (v/v) Sputasol (Oxoid, Thermo Fischer Scientific, Waltham MA), vortexing for 1 min, incubating at 37 °C for 10 min, vortexing for 1 min, pelleting by centrifugation at 2000 g for 10 min, and then removing the supernatant. To study the DNA extraction yield in a previous experiment, samples f and g had 100 μL TRIzol (Ambion, Life Technologies, Grand Island NY) added, while samples d and e did not [15]. All samples were then frozen at −80 °C until they underwent DNA extraction.
Samples were thawed at 4 °C overnight before undergoing DNA extraction. Buffer ATL (900 μL) (Qiagen, Valencia CA) was added to the sputum pellets. To homogenize the samples, they were mixed by vortexing for 1 min, incubated at 37 °C for 10 min, then mixed by vortexing for an additional minute. We then utilized the DSP Virus/Pathogen kit on the QIAsymphony SP (Qiagen, Valencia CA) to systematically extract bacterial DNA. Again, to study DNA extraction yield using different protocols, RNA carrier (Qiagen, Valencia CA) was added to samples d and f, but not to samples e and g [15]. DNA concentrations were measured using NanoDrop (Thermo Fischer Scientific, Waltham MA), and were in the range of 13–571 ng/μL in a volume of 60 μL.
2.4. Next generation sequencing
Twelve samples of extracted bacterial DNA were sequenced on both PacBio NGS and MiSeq platforms. For the PacBio RSII sequencing platform at CNHS (Washington DC), DNA (500 ng) was used for library prep and the full 16S rRNA gene (approximately 1450 bp) was sequenced with an average of 16–17 passes using circular consensus sequencing. DNA (300 ng) was amplified for the V4 region (approximately 250 bp) of the 16S rRNA gene and libraries were sequenced on the MiSeq sequencing platform at the University of Michigan (Ann Arbor MI). DNA (200 ng) was also amplified for the V3/V4 region (approximately 450 bp) of the 16S rRNA gene using a MiSeq sequencing platform at CNHS (Washington DC).
2.5. Analyses
For comparison of MiSeq and PacBio, raw FASTQ files generated by the MiSeq V4 and PacBio platforms were processed in mothur v1.35.1 [16]. Default settings were used to minimize sequencing errors as described in Schloss et al. [17] for the MiSeq reads and Schloss et al. [11] for the PacBio reads. Clean sequences were aligned to the SILVA_v123 bacterial reference alignment at http://www.mothur.org. Chimeras were removed using uchime [18] and non-chimeric sequences were classified using the naïve Bayesian classifier of Wang et al. [19]. Sequences were clustered into OTUs at the 0.03 threshold (species level). OTU sequence representatives and taxonomy were imported (BIOM format) into QIIME [20] for subsequent analyses. The mothur OTU table was filtered to a minimum of 2 observations (sequences) per OTU. Samples were subsampled (rarefaction analysis) to the smallest sample size (3590 sequences) to remove the effect of sample size bias on community composition. Taxonomic alpha-diversity was estimated as the number of observed OTUs and by the Chao1, Fisher and Shannon indices, and estimates were compared between gene regions (16S_v4 versus full 16S) using non-parametric versions of the t-test. We used Procrustes analysis to determine whether we would derive the same beta-diversity conclusions, regardless of which gene region was used to compare the samples [21]. We then compared ordinations generated by principal coordinates analysis (PCoA) of Bray-Curtis and weighted Unifrac distances between 16S_V4 and full 16S groups [21]. Significance was estimated by Monte Carlo simulation (10,000 iterations).
The OTUs generated sequencing the V3/V4 region of the 16S rRNA gene and the V4 region of the 16S rRNA gene and Shannon indices were also compared using the 16S Metagenomics application within BaseSpace (Illumina). This application uses an Illumina curated version of the GreenGenes taxonomic database to classify 16S rRNA targeted amplicon reads [22]. Total number of reads, OTUs, and Shannon indices were compared between targeted amplicons (16S_V4 versus full 16S_V3/V4) using the t-test.
3. Results
A total of 24 sputum microbiomes (twelve sample sets) were analyzed via MiSeq and PacBio sequencing of the 16S rRNA V4 (~250 bp) and full 16S (~1450 bp) gene regions, respectively. MiSeq sequencing generated a total of 479,220 sequences ranging from 5657 to 71,193 sequences per sample (mean = 34,230) after quality control analyses and OTU filtering. PacBio sequencing generated a total of 122,526 sequences ranging from 3590 to 12,571 sequences per sample (mean = 8752) after quality control analyses and OTU filtering. From these data, we identified 9–170 OTUs (mean = 117) per sample in the MiSeq data and 2–69 OTUs (mean = 22) per sample in the PacBio data. About 99.3% of the PacBio OTUs were classified to the genus level, while only ~49.4% of the MiSeq OTUs were classified to that level. CF microbiomes sequenced via PacBio were dominated by the following genera: Escherichia or Shigella (34%), Pseudomonas (23%), Streptococcus (12%), Burkholderia (8%), Haemophilus (7%), Prevotella (6%), Staphylococcus (6%), and Gemella (3%). Similarly, CF microbiomes sequenced via MiSeq were dominated by the following genera: Enterobacteriaceae unclassified genus (33%), Pseudomonas (16%), Prevotella (15%), Streptococcus (4%), Haemophilus (3%), Staphylococcus (2%), Bacteroides (2%), and Gemella (1%). The dominant genera reported for each of the 12 samples are shown in Table 1.
Table 1.
Dominant genera identified using MiSeq (16S_V4) and PacBio RSII (full 16S) sequencing data.
| MiSeq (16S_V4) | PacBio RSII (full 16S) | |
|---|---|---|
| S1e | Pseudomonas (64%) | Pseudomonas (84%) |
| S2d | Prevotella (42%) | Streptococcus (36%) |
| S4e | Staphylococcus (11%) | Staphylococcus (43%) |
| S5e | Enterobacteriaceae unclassified genus (16%) | Burkholderia (63%) |
| S6e | Enterobacteriaceae unclassified genus (76%) | Escherichia or Shigella (97%) |
| S9d | Prevotella (38%) | Haemophilus (83%) |
| S11e | Pseudomonas (30%) | Pseudomonas (70%) |
| S11f | Pseudomonas (41%) | Pseudomonas (69%) |
| S11g | Pseudomonas (39%) | Pseudomonas (43%) |
| S12e | Enterobacteriaceae unclassified genus (87%) | Escherichia or Shigella (99%) |
| S12f | Enterobacteriaceae unclassified genus (98%) | Escherichia or Shigella (99%) |
| S12g | Enterobacteriaceae unclassified genus (99%) | Escherichia or Shigella (99%) |
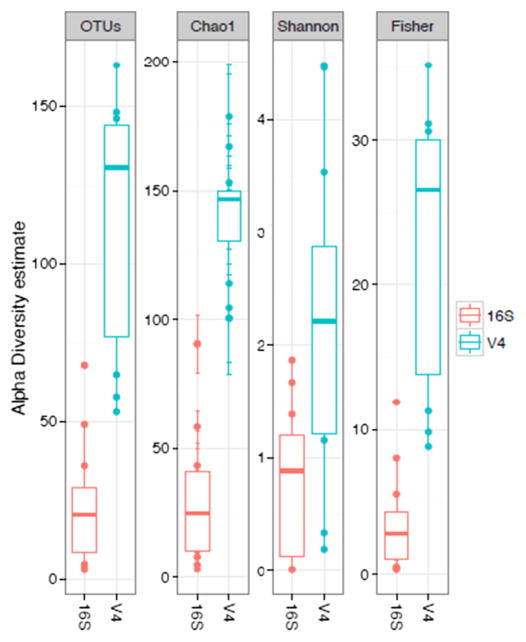
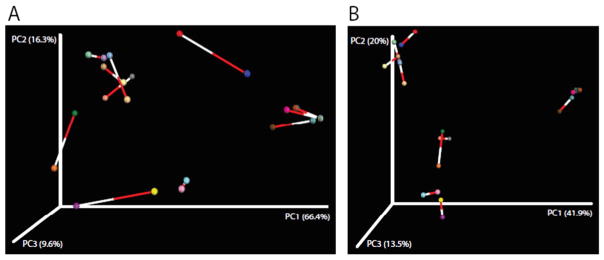
Microbial profiles (Fig. S1) and alpha-diversity estimates (Fig. 1) of observed OTUs, Chao1, Fisher and Shannon indices varied greatly between PacBio and MiSeq groups, with MiSeq showing greater estimates for all indices. These differences were significant in all cases (non-parametric t-test; P < 0.006–0.001). Procrustes analysis comparing PCoA plots of MiSeq and PacBio microbiomes showed greater dissimilarity under Weighted unifrac distances (M2 = 0.220, Fig. 2A) than under Bray-Curtis distances (M2 = 0.095, Fig. 2B) (P < 0.001 for both). Nonetheless, the ordination of samples in space resulted similarly for most sample pairs.
Fig. 1.
Box plots of alpha-diversity indices comparing 24 microbiomes from 12 paired samples sequenced in the MiSeq (16S_V4) and PacBio (whole 16S) platforms. The thick horizontal line within each box represents the median estimate.
Fig. 2.
Procrustes analysis of 24 microbiomes from 12 paired samples sequenced in the MiSeq (16S_V4) and PacBio (whole 16S) platforms. Samples collected from the same patient are connected by a line, the white end indicating microbiome generated via MiSeq and the red end indicating microbiome generated via PacBio. A) Weighted unifrac distances, M2 = 0.220, P < 0.001. B) Bray-Curtis, M2 = 0.095, P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
MiSeq V4 was chosen as the comparison to PacBio because the sequence data was of higher quality when we only sequenced the V4 region compared to the V3/V4 region. Specifically, the MiSeq analyses of the V3/V4 region produced paired reads with low Phred Quality Scores (<10) in the middle section (120 bp). As such, the bioinformatic analysis in mothur was more precise with increased overlap of the paired sequences using V4 alone. Additionally, because of the low Phred Quality Scores in the middle section, the V3/V4 sequence data could not be properly analyzed using mothur as it was unable to create appropriate contigs. BaseSpace, which uses a different algorithm, was still effective in performing the bioinformatic analysis of both regions, and the V3/V4 reads were analyzed and compared to V4 reads.
As fewer samples were sequenced when using the V3/V4 amplicon, more reads were devoted to each sample, thereby significantly increasing the number of OTUs identified compared to the sequences generated using the V4 amplicon (Table S1). However, minimal differences were noted in identification of the dominant pathogen (Table S2). Specifically, in sample S2d the dominant pathogen was different, but the same top four pathogens were identified in each sample (MiSeq V4: Prevotella 41%, Streptococcus 19%, Gemella 11%, Pseudomonas 9%; MiSeq V3/V4: Streptococcus 20%, Gemella 13%, Pseudomonas 12%, and Prevotella 11%). There were also no significant differences in the Shannon indices between the two sequencing methodologies (Table S1).
4. Discussion
Our study revealed significant discordance in alpha-diversity indices and subsequent taxonomic identification based in two different next-generation sequencing platforms. Since sample collection and handling have been shown to influence sequencing results [7,23,24], the extracted DNA used for sequencing was taken from the same aliquot to eliminate any potential upstream variations. Likewise, the same computational pipeline and 16S reference database to analyze sequencing results were used to limit any variations in analysis. As such, the only source of variability was limited to the sequencing platform used.
In our study, we used the MiSeq to amplify the V4 region of the 16S rRNA gene prior to sequencing. Prior studies have suggested that V4 provides the closest analogue to sequencing the full 16S rRNA gene [8, 25]. However, the sequencing information obtained from the V4 region alone was not able to assign a genus-level taxonomic rank to ~50% of the observed OTUs. The PacBio sequence data, however, was able to assign almost all the reads to a microbial genus. PacBio longer sequences accounted for at least 16 passes of the full 16S rRNA gene. Given the randomness of sequencing errors in the PacBio platform, that level of coverage and read length would have greatly reduced total error rate and increase accuracy of OTU assignment, a definite advantage. However, the total sequencing yield depth of sequencing of the PacBio remains much lower than that of the MiSeq and less abundant OTUs may have been missed by the former, which is an advantage of MiSeq. Lastly, the decreased cost to sequence the 16S rRNA gene using MiSeq compared to PacBio is also an advantage of MiSeq.
All genera reported here for CF patients have been detected in other microbiome studies of sputum, although in different proportions (see Table 2) [3–7]. When comparing the taxonomic profiles obtained by both techniques, there were key OTUs assigned to genera using the PacBio platform that were not able to be assigned using the MiSeq platform. One example was the difference in identifying Escherichia or Shigella in samples S6d, S12e, S12f, and S12g using PacBio while the OTU remained classified as Enterobacteriacae unclassified genus when using MiSeq. In the CF human population, the inability for the MiSeq to differentiate Burkholderia from other Pseudomonal taxa is very concerning, as this is a rare but important pathogen associated with severe clinical disease [6,26]. This limitation is likely due to our choosing only to sequence V4, and may be overcome with either broader 16S rRNA sequencing or shotgun sequencing. Unfortunately, the sample S5e that identified Burkholderia as a dominant pathogen using PacBio sequences did not sequence successfully using the MiSeq V3/V4 amplicon, so additional comparisons cannot be made. There were also occurrences where the dominant genus on the two platforms did not align, despite the ability of OTUs from both data sets to be assigned to those genera. Specifically, Prevotella was assigned as the dominant genera in samples S2d and S9d when using MiSeq V4 sequencing data. Using PacBio sequencing data, they were assigned as Streptococcus and Haemophilus, respectively. This finding is unique to our study, as other studies have shown MiSeq and PacBio data to correlate well with the abundances of bacteria in synthetic communities [8]. Another interesting finding was the frequency Escherichia was identified in our patient cohort. While this is a pathogen that has been described in the CF airway, it is by no means common [3,27].
Table 2.
| Authors | Lim et al. [3] | Carmody et al. [4] | Zhao et al. [5] | Fodor et al. [6] | Zemanick et al. [7] |
| Number of patients | 3 | 28 | 6 | 23 | 16 |
| Sequencing platform | Roche/454 | Roche/454 | Roche/454 | Roche/454 | Roche/454 |
| Sequencing region | Metagenome | 16S V3–V5 | 16S V3–V5 | 16S V1–V2 | 16S V1–V2 |
| Bioinformatic analysis | BLAST (NCBI) | Mothur (v.1.24) | Mothur (v.1.21) | BLAST (NCBI) | RDP Classifier (v 2.4) |
| Range of identified OTUs per sample | Not mentioned | 3–34 | 5–114 | Not mentioned | 8–98 |
| Commonly identified bacterial taxa | Escherichia, Streoptococcus, Rothia, Klebsiella, Stenotrophomonas, Pseudomonas, Prevotella, Veillonella, Fusobacterium | Psedomonas, Streptococcus, Burkholderia, Haemophilus, Fusobacterium, Veillonella, Staphylococcus, Prevotella, Achromobacter, Gemella, Rothia | Pseudomonas, Staphylococcus, Streptococcus, Prevotella, Leptotrichia, Abiotrophia | Pseudomonas, Burkholderia, Streptococcus, Prevotella, Rothia, Veillonella, Actinomyces, Granulicatella | Staphylococcus, Enterobacteriaceae, Haemophilus, Prevotella, Streptococcus, Veillonella, Rothia, Fusobacterium, Neisseria |
The observed OTUs obtained with the MiSeq sequencing data, and thus the corresponding alpha-diversity indices, were much higher than those obtained with the PacBio sequencing data. In other studies of the CF lung microbiome using 454, the observed OTUs typically were in the range of 3–114 [4,5,7], which are more similar to the observed OTUs we found using PacBio (see Table 2). However, studies comparing 454 to MiSeq, PacBio, and Ion Torrent using a synthetic community have suggested that 454 may underestimate the number of OTUs [8]. Thus, the discrepancies in alpha-diversity observed here between MiSeq and PacBio could result from OTU underestimation in the latter.
Limitations to the study include using de-identified sputum samples; hence no beta-diversity outputs could be compared between NGS platforms. Although in that regard, Procrustes analysis showed short connecting bars (low dissimilarity) for most sample pairs and low to moderately low values of dissimilarity (as indicated by the Procrustes statistic M2) and highly significant result (P < 0.001), which implies that we would draw the same beta diversity conclusions from either data set. It would have been also helpful to compare the sequencing results to bacterial culture results to verify whether bacteria identified by one platform but not the other were truly present. Another limitation was not determining the bacterial load within each sample, as this also could have influenced our sequencing results.
5. Conclusion
MiSeq and PacBio sequencing platforms generate significant discordance in microbial taxonomic profiles and alpha-diversity in CF sputum microbiomes. The longer sequence reads yielded by PacBio allowed for more accurate taxa assignment of OTUs. However, the larger depth of MiSeq likely led to finding more rare OTUs. These benefits and limitations should be considered when choosing the platform best suited for metataxonomic studies. When comparing microbiome diversity in clinical samples from CF patients using 16S sequences, MiSeq and PacBio NGS platforms may generate different results in microbial community composition and structure. It may be necessary to use different platforms when trying to correctly identify dominant pathogens versus measuring alpha-diversity estimates, and it would be important to use the same platform for comparisons to minimize errors in interpretation.
Supplementary Material
Acknowledgments
We thank the GWU Colonial One High Performance Computing Cluster for computational time. AH and MP-L were funded in part by a K12 Career Development Program K12HL119994 award. This publication was also supported by Award Number UL1TR000075 from the NIH National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mimet.2016.09.002.
Footnotes
Availability of the data
Sequence data have been uploaded to the GenBank under SRA accession number SRP070848.
References
- Ramsey BW. Management of pulmonary disease in patients with cystic fibrosis. N Engl J Med. 1996;335(3):179–188. doi: 10.1056/NEJM199607183350307. [DOI] [PubMed] [Google Scholar]
- Burns JL, Rolain JM. Culture-based diagnostic microbiology in cystic fibrosis: can we simplify the complexity? J Cyst Fibros. 2014;13:1–9. doi: 10.1016/j.jcf.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Lim YW, Evangelista JS, Schmieder R, Bailey B, Haynes M, Furlan M, et al. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2014;52(2):425–437. doi: 10.1128/JCM.02204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody LA, Zhao J, Schloss PD, Petrosino JF, Murray S, Young VB, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc. 2013 Jun;10(3):179–187. doi: 10.1513/AnnalsATS.201211-107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One. 2012;7(9):e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanick ET, Wagner BD, Robertson CE, Stevens MJ, Szefler S, Accurso FJ, et al. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. AnnalsATS. 2015;12(2):221–229. doi: 10.1513/AnnalsATS.201407-310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amore R, Ijaz UZ, Schirmer M, Kenney JG, Gregory R, Darby AC, et al. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiing. BMC Genomics. 2016;17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang CH, Chang YJ, Chung WC, Hsieh PH, Lin CY, Ho JM. Subset selection of high-depth next generation sequencing reads for de novo genomoe assembly using MapReduce framework. BMC Genomics. 2015;16(Suppl 12):S9. doi: 10.1186/1471-2164-16-S12-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Zhou D, Caporaso JG, Knight R, Angenent LT. Comparison of Illumina paired-end and single-direction sequencing for microbial 16S rRNA gene amplicon surveys. ISME J. 2012;6(7):1273–1276. doi: 10.1038/ismej.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Jenior ML, Highlander SK. Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ PrePrints. 2015;3:e778v1. doi: 10.7717/peerj.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichot EB, Norman RS. Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome. 2013;1:10. doi: 10.1186/2049-2618-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen O, Hu J, Bao X, Itzkowitz SH, Peter I, Bashir A. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome. 2015;3:43. doi: 10.1186/s40168-015-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Perez G, Colberg-Poley AM, Campos J, Bryant J, Rose M. Systematic extraction of bacterial DNA for lung microbiome analysis from cystic fibrosis sputum samples. Posters Pediatr Pulmonol. 2015;50:S77–S107. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6(12):e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC. Generalized Procrustes analysis. Psychometrika. 1975;40(1):33–51. [Google Scholar]
- 16S metagenomics. BaseSpace Sequencing Hub. (Retrieved from https://basespace.illumina.com/apps/593593/16S-Metagenomics?preferredversion on August 31 2016)
- Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hafiz T, Hoffman LR, et al. Time between collection and storage significantly influences bacterial sequence composition in sputum samples from cystic fibrosis respiratory infections. J Clin Microbiol. 2014;52(8):3011–3016. doi: 10.1128/JCM.00764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hoffmant LR, Carroll MP, et al. Implications of multiple freeze-thawing on respiratory samples for culture-independent analyses. J Cyst Fibros. 2015;14(4):464–467. doi: 10.1016/j.jcf.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soergel DA, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6(7):1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlosnick JE, Costa PS, Brant R, Mori PY, Hird TJ, Fraenkel MC, et al. Mucoid and nonmucoid Burkholderia cepacia complex bacteria in cystic fibrosis infections. Am J Respir Crit Care Med. 2011;183(1):67–72. doi: 10.1164/rccm.201002-0203OC. [DOI] [PubMed] [Google Scholar]
- Barillova P, Tchesnokova V, Dubbers A, Kuster P, Peters G, Dobrindt U, Sokurenko EV, Kahl BC. Prevalence and persistence of Escherichia coli in the airways of cystic fibrosis patients – an unrecognized CF pathogen? Int J Med Microbiol. 2014;304:415–421. doi: 10.1016/j.ijmm.2014.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.