Abstract
We have analyzed a set of quinolinequinones with respect to their reactivities, cytotoxicities, and anti-HIV-1 properties. Most of the quinolinequinones were reactive with glutathione, and several acted as sulfhydryl crosslinking agents. Quinolinequinones inhibited binding of the HIV-1 matrix protein to RNA to varying degrees, and several quinolinequinones showed the capacity to crosslink HIV-1 matrix proteins in vitro, and HIV-1 structural proteins in virus particles. Cytotoxicity assays yielded quinolinequinone CC50 values in the low micromolar range, reducing the potential therapeutic value of these compounds. However, one compound, 6,7-dichloro-5,8-quinolinequinone potently inactivated HIV-1, suggesting that quinolinequinones may prove useful in the preparation of inactivated virus vaccines or for other virucidal purposes.
Graphical Abstract
1. INTRODUCTION
Quinolinequinones and analogous napthoquinones have been reported to have a variety of potentially useful biomedical properties (1–15). Notably, the 5,8-quinolinequinone, streptonigrin and its derivatives have demonstrated anti-cancer activities (1). Similar 5,8-quinolinequinones have been reported to have anti-bacterial, anti-malarial, anti-fungal, and anti-inflammatory activities (2–7). Another quinolinequinone, LY83583 (6-anilino-5,8-quinolinequinone), has been shown to inhibit soluble guanylate cyclase and leukotriene synthesis with IC50 concentrations in the 1–2 micromolar range (8–9). Studies on related napthoquinones have identified a number of 1,4-napthoquinones with anti-cancer, anti-bacterial, anti-fungal, and anti-parasitic effects (10–15). In terms of chemical reactivity, quinolinequinones have been shown to react with sulfhydryls, and primary and secondary amines (1), but it is not clear how these reactions contribute to quinolinequinone biomedical properties.
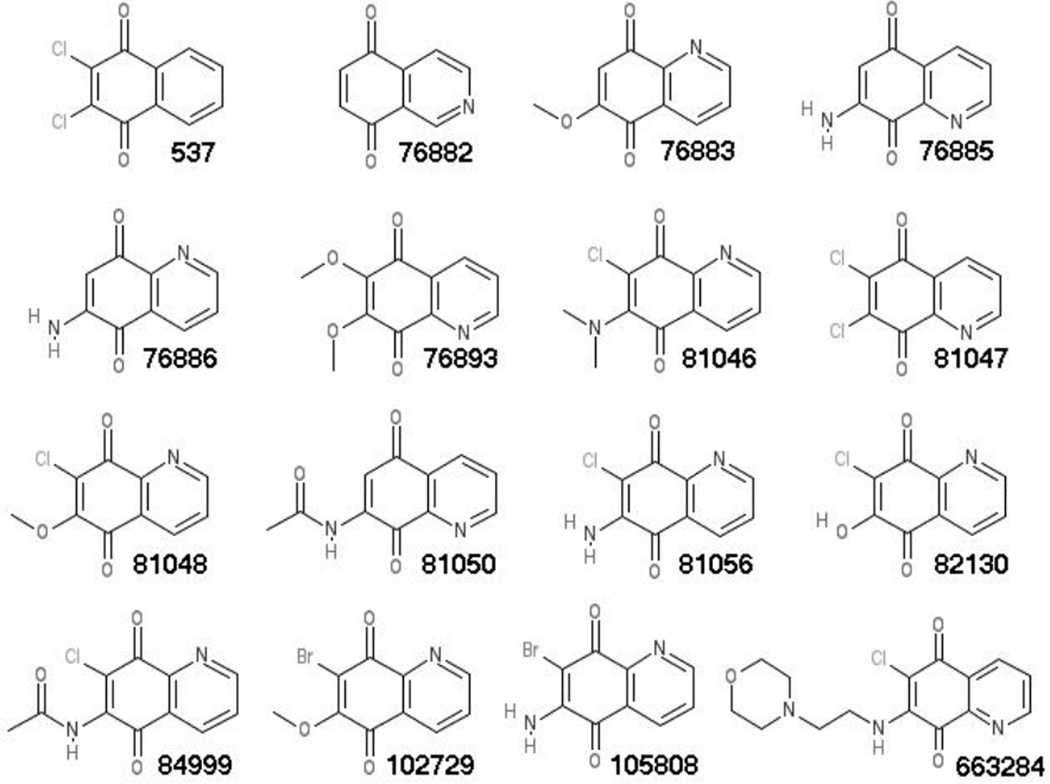
We previousy identified 2,3-dichloronapthoquinone (Figure 1, 537) as an inhibitor of RNA binding to the HIV-1 matrix (MA) protein (16; referred to as DDD). MA is the N-terminal domain of the HIV-1 structural precursor Gag (PrGag) protein, and MA-RNA binding has been proposed to facilitate PrGag delivery to viral assembly sites at the plasma membranes of infected cells (17–20). Thus, a MA-RNA binding inhibitor potentially could impair HIV-1 assembly or perturb HIV-1 replication (16–20). Recently, in a library screen of 50,000 compounds, we found that the quinolinequinone analogue of 537 (Figure 1; 81047; 6,7-dichloro-5,8-quinolinedione) also potently inhibited MA-RNA binding. Based on these results, we obtained a set of related quinolinequinones (Figure 1) and examined their properties. A number of the quinolinequinones were found to inhibit HIV-1 MA-RNA binding, and inhibition appeared to be counteracted by sulfhydryl-containing compounds, including glutathione (GSH). Several of the MA-RNA binding inhibitors had the capacity to crosslink MA proteins, and cysteine-containing peptides, suggesting that their mechanism of action was via sulfhydryl-specific crosslinking. This was supported by the observation that such quinolinequinones could crosslink the structural proteins in HIV-1 viruses, and the related Moloney murine leukemia virus (MLV).
Figure 1. Compounds for analysis.
The compounds employed in our studies are designated according to their NSC numbers, and their chemical names are as follows: 537, 2,3-dichloronaphthoquinone; 76882, 5,8-isoquinolinedione; 76883, 6-methoxy-5,8-quinolinedione; 76885, 7-amino-5,8-quinolinedione; 76886, 6-amino-5,8-quinolinedione; 76893, 6,7-dimethoxy-5,8-quinolinedione; 81046, 7-chloro-6-(dimethylamino)-5,8-quinolinedione; 81047, 6,7-dichloro-5,8-quinolinedione; 81048, 7-chloro-6-methoxy-5,8-quinolinedione; 81050, N-(5,8-dioxo-5,8-dihydro-7-quinolinyl)acetamide; 81056, 6-amino-7-chloro-5,8-quinolinedione; 82130, 7-chloro-6-hydroxy-5,8-quinolinedione; 84999, N-(7-chloro-5,8-dioxo-5,8-dihydro-6-quinolinyl)acetamide; 102729, 7-bromo-6-methoxy-5,8-quinolinedione; 105808, 6-amino-7-bromo-5,8-quinolinedione; 663284, 6-chloro-7-((2-(4-morpholinyl)ethyl)amino)-5,8-quinolinedione.
We tested our most potent MA crosslinker and MA-RNA binding inhibitor, 81047, for inhibition of HIV-1 replication, but although it demonstrated some inhibitory activity at 1 µM, it also showed cytotoxicity in human T cells at that concentration, reducing its therapeutic potential. However, relative to previously studied compounds (21–3), 81047 proved highly effective in inactivation of HIV-1 virus particles, suggesting that quinolinequinones might prove useful the preparation of inactivated virus vaccines or for other virucidal purposes. Overall, our results will help guide new research on the biomedical activities of quinolinequinones.
2. MATERIALS AND METHODS
2.1. Materials
The HIV-1 MA protein was expressed in bacteria and purified by nickel-chelate chromatography as described previously (16, 18, 20). The Sel25 RNA ligand had the sequence of 5'GGACA GGAAU UAAUA GUAGC UGUCC3', and was obtained from Invitrogen, as was the 5'-fluorescein isothiocyanate (FITC) tagged version of Sel25. The cysteine-containing 31 residue peptide had the sequence CSILD IRQGP KEPFR DYVDR FYKTL RAEQA S, was obtained from Peptide Express (Colorado State University) and was described previously (24). The cysteine-minus 33 residue peptide corresponded to a trimerizing domain of the hantavirus nucleocapsid protein (N43-75), and was synthesized and purified at the Portland Shriners Hospital for Children (25). The napthoquinone and quinolinequinones used in this study are shown in Figure 1, referred to by their NSC numbers, and were kindly provided by the National Cancer Institute (NCI)/Developmental Therapeutics Program (DTP): http://dtp.cancer.gov. The thiadiazolane 2-(2-chloro-6-methylphenbyl)-4-(cyclopropylmethyl)-2-1,2,4-thiadiazolane-3,5-dione (TD1) has been described previously (16), and was obtained from Maybridge. Ellman's reagent (5,5'-dithio-bis-[2-nitrobenzoic acid]) was from Sigma-Aldrich, as were 2,2'-dipyridyldisulfide (Aldrithiol-2), 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPS), β-mercaptoethanol (βME), glutathione (GSH), cysteine, glycine, glycylglycine, and Stains-All. Acetonitrile was from EM Science; 1,4-dithiothreitol (DTT) was from BioRad; dimethyl sulfoxide (DMSO) was from Fisher; and 1,6-bis-maleimidohexane (BMH), bis-maleimidoethane (BMOE), and tris(2-carboxyethyl)phosphine (TCEP) were from ThermoFisher.
2.2. MA-RNA binding assays
Fluorescence polarization (FP) MA-RNA binding assays were modified from our previously published protocol (16, 20) for multiwell plate analysis. HIV-1 MA (2 µM) in 25 mM sodium phosphate (pH 6.0), 50 mM NaCl, 0.0025% CHAPS was incubated 30 min at 25°C with 0.25% DMSO or 1–100 µM compound (final concentration) in DMSO. Incubations then were supplemented with FITC-Sel15 RNA to 20 nM and allowed to incubate another 30 min at 25°C. After these incubations, FP measurements were performed on a BioTek Synergy 4 Microplate Reader with 485 nm excitation and 520 nm emission wavelengths.
Electrophoretic mobility shift assays for monitoring MA-RNA binding were performed as described before (16, 20). MA samples (15 µM) in 25 mM sodium phosphate (pH 7.8), 50 mM NaCl were preincubated 30 min at 25° with 5% DMSO or 50–100 µM of the indicated compounds in DMSO. After preincubations, samples were supplemented with 15 µM Sel1 5 RNA, and incubated 30 min at 25°. Bound and free RNAs were separated by electrophoresis, stained with Stains-All, and scanned on an Epson Perfection 1240U scanner, after which band intensities were quantified with Image J software (26). In sulfhydryl inhibition experiments, preincubations also contained 200 µM concentrations of GSH, cysteine, βME, DTT, or TCEP.
2.3. Glutathione reactivity assays
Glutathione (GSH) reactivity assays were performed to assess compound abilities to react with or oxidize GSH. GSH alone (200 µM) in 80 mM sodium phosphate (pH 7.8), or compounds (200 µM) in the presence or absence of 200 µM GSH were incubated for 1 h at 25°C. Incubations then were supplemented either with an equal volume of sodium phosphate or 200 µM Ellman's reagent in sodium phosphate, and reactions were continued an additional 30 min, prior to spectrophotometric monitoring levels of Ellman's reagent product at 412 nm. Residual GSH levels were calculated by subtraction of optical density (OD) 420 nm values of GSH-minus incubations from incubations in the presence of GSH. Percentage of GSH inactivation levels were determined by comparison of GSH levels in the absence of compound versus compound presence and were averaged from two separate experiments.
2.4. Peptide and protein crosslinking
For crosslink analysis of the 31 residue cysteine-containing peptide (Section 2.1.), samples of 100 µM peptide plus 1 mM of the indicated compounds were incubated 2 h at 25°C in 25 mM sodium phosphate (pH 7.8) plus 50 mM NaCl, after which peptide monomers and dimers were separated by Schagger and von Jagow sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions (SDS-PAGE; 25), and visualized by staining with 0.025% Coomassie Blue G-250 in 10% acetic acid (25). Similar experiments were performed with the 33 residue cysteine-minus peptide (Section 2.1.), but no dimer bands were observed.
For MA crosslink analysis, samples of 30 µM MA in 25 mM sodium phosphate (pH 7.8) plus 50 mM NaCl were incubated at 25°C for 2 h in the absence or presence of 50 µM compounds. Incubation products were fractionated on 12% SDS-PAGE gels under reducing conditions (27–8) in parallel with molecular weight size standards, and immunoblotted for MA detection as described previously (27–8), using a primary mouse monoclonal antibody to MA (Capricorn #01848170) at a 1:2000 dilution, an alkaline phosphatase-conjugated anti-mouse secondary antibody (Promega #S3728) at a 1:15,000 dilution, and nitro-blue tetrazolium (NBT; Promega) plus 5-bromo-4-chloro-3'-indolyphosphate (BCIP; Promega) for visualization (27–8). Blots were scanned on and Epson Perfection 1240U scanner, band intensities were obtained with Image J software, and percentage crosslinking levels were determined from dimer divided by dimer plus monomer MA band intensities.
2.5. Mass spectrometry analysis
Analysis of GSH alone, 81047 alone, and GSH plus 81047 reaction products was performed at the OHSU Bioanalytical Shared Resource Pharmacokinetics (BSR/PK) Core. GSH (10 mM) alone, 81047 (5 mM) alone, or 10 mM GSH plus 5 mM 81047 in 50% DMSO were incubated for 3 h at 25°C and then supplemented with a four-fold molar excess (v/v) of acetonitrile, and subjected to mass spectrometry (MS) analysis using electrospray ionization in the positive ion mode on the BSR/PK Applied BioSystems 4000 QTRAP triple-quadrupole, linear ion trap MS. Alternative predicted structures were compared with obtained spectra. In the GSH plus 81047 sample, the 770.3 m/z peak corresponded to the protonated form of 6,7-bis(glutathionyl)-5,8-quinolinedione, and the 792.3 peak corresponded to the sodium ionized form.
2.6. Cell culture and viruses
2.6.1. Cell culture and virus production
Human embryonic kidney HEK293T cells (29) were grown at 37°C in 5% carbon dioxide in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with penicillin, streptomycin, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.4), and 10% fetal calf serum (FCS). Human MT4 (30–31) and CEM-SS (30–31) T cell lines were grown Roswell Park Memorial Institute (RPMI) medium with the same penicillin, streptomycin, HEPES and supplements. Wild type (WT) HIV-1 NL4-3 (32) strain virus was produced by transfection of HEK293T cells as described previously (27, 30–31). Viruses for crosslink analysis also were produced by transfection of HEK293T cells as described previously (27, 30–31): immature HIV-1 viruses were produced by transfection of cells with the HIV-1 Gag-Pol expression construct psPAX2 (33) and treatment of cells with 10 µM ritonavir; immature Moloney murine leukemia virus (MLV) was produced by transfection with the MLV Gag expression construct pXM-2453 (34); and immature MLV with all Gag cysteine residues converted to serines (all C>S) was produced by transfection of cells with the construct pXM-2450(C-) (34). Recombinant HIV-1 viruses that transduce the gene for expression of the green fluorescent protein (GFP) were generated by co-transfection of psPAX2, the vesicular stomatitis virus (VSV) glycoprotein (G) expression vector p-VSVG (27), and the lentivirus GFP reporter construct phage-ubc-nls-tdPCP-GFP (35). Recombinant HIV-1 viruses that transduce the gene for luciferase (Luc) were generated by co-transfection of the HIV-1 expression construct HIV-Luc (30–31) plus p-VSVG.
2.6.2. Virus crosslink analysis
Immature HIV-1, MLV, and MLV all C>S viruses produced as described in Section 2.6.1. were pelleted through 20% sucrose cushions in phosphate-buffered saline (PBS; 27, 30–31, 33–4), and resuspended in PBS as described before (27, 30–31, 33–4). Each virus sample was split into six equal aliquots, and aliquots were treated for 1 h at 25°C with 10% DMSO or 1 mM BMH, 537, 81047, 76882, or 102729. After incubations, samples were fractionated by 10% SDS-PAGE under reducing conditions (27, 30–31, 33–4), and immunoblotted for precursor Gag (PrGag) detection using primary monoclonal antibodies to HIV-1 CA (Hy183, used at 1:10 from cell culture media; 27, 30–31, 33) or MLV CA (Hy187, used at 1:10 from cell culture media; 34), followed by secondary antibody and NBT plus BCIP reactions as described in Section 2.4. PrGag monomer and dimer bands were identified by antibody reactivity and electrophoretic migration relative to molecular weight size standards run in parallel.
2.6.3. Virus infections
Infections of MT4 cells with WT NL4-3 strain HIV-1 were performed with equivalent amounts of input virus (based on capsid [CA] protein quantitation; 30–31) on equal amounts of cells treated either with DMSO, or 1 µM or 10 µM 81047 in DMSO. At 7 d post-infection, cells were collected, washed, and processed for immunoblot detection of HIV-1 PrGag as in Section 2.6.2 or cellular actin, using a primary anti-actin mouse monoclonal antibody (Santa Cruz Biotech. #sc-8432) at a 1:500 dilution. Immunoblot bands were scanned and quantified as in Section 2.4., and band intensities of actin and the HIV-1 Gag protein bands (PrGag, p41, and CA) from 81047-treated cells were normalized to those from DMSO controls.
For analysis of compound effects on purified HIV-1 viruses, GFP- and luciferase-transducing HIV-1 stocks were prepared as in Section 2.6.1., and concentrated by pelleting and resuspension in PBS as in Section 2.6.2. After suspension, identical 20 µl aliquots of each virus type were treated 1 h at 37°C with 5% DMSO, 1 mM Aldrithiol-2, or 0.1–1 mM 81047. Treated viruses each were used to infect 200,000 HEK293T cells in 30 mm dishes in a total volume of 2 ml of DMEM growth media plus 8 µg/ml polybrene (Sigma-Aldrich). Infections were allowed to proceed under cell growth conditions for 48 h at 37°C. For monitoring cells infected with GFP-transducing HIV-1, cells were fixed 20 min at 25°C in 4% paraformaldehyde in PBS, washed in PBS, and imaged by fluorescence microscopy on a Zeiss Axio Observer Z1 inverted microscope equipped with filter set 10 (excitation band-pass range [BP], 450 to 490 nm; emission BP, 515 to 565). For measurement of luciferase activities in cells infected with luciferase-transducing HIV-1, HEK293T cells were infected as above, cell samples were collected, lysed, and subjected to luciferase assays in 100 mM sodium phosphate (pH 8.0), 6 mM MgCl2, 1 mM sodium pyrophosphate, 4 mM ATP, and D-luciferin (BD Biosciences #51-556877) as described previously (30–31).
2.6.4. Cytotoxicity assays
For cytotoxicity assays (16) of HEK293T, MT4, and CEM-SS cells, cells were seeded at 15,000 cells (in 150 µl growth media) per well of a 96 well plate, treated with 1% DMSO or compound dilution in DMSO, and grown 48 h. After 48 h of growth, 30 µl of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; Promega, Celltiter 96) was added to each well, mixed and incubated 60–90 min at 37°C, after which 490 nm light absorbance readings were taken. Fifty percent cytotoxicity (CC50) values were determined from absorbance versus concentration curves for each compound.
3. RESULTS
3.1. Quinolinequinone inhibition of HIV-1 matrix-RNA binding
Previously, we identified 2,3-dichloronapthoquinone (Figure 1, 537) as an inhibitor of RNA binding to the HIV-1 matrix (MA) protein, but the compound proved too cytotoxic to act as an effective antiviral (16). More recently, we modified our previous approach (16), and employed a multiplate FP MA-RNA binding assay (Section 2.2.) to screen a 50,000 member compound library, and found 6,7-dichloro-5,8-quinolinequinone (Figure 1, 81047) as an, effective MA-RNA binding inhibitor. Based on this observation, we obtained fourteen additional quinolinequinones (Figure 1) for examination of their potential antiviral activities.
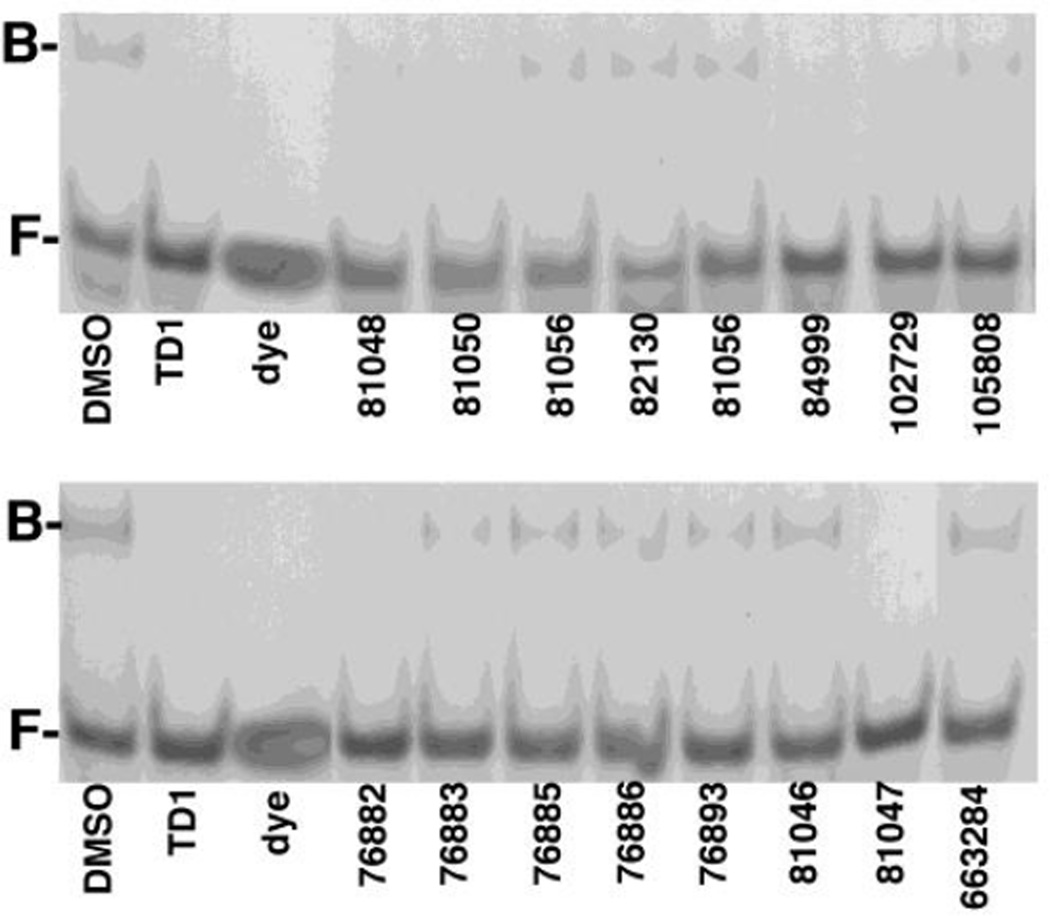
As an initial test, we subjected the quinolinequinones to MA-RNA electrophoretic mobility shift assays (EMSAs; Section 2.2.) as we previously did with 537 (16). For these assays, we used a 15mer RNA oligonucleotide ligand, Sel15 (16, 20), at 15 µM, and purified HIV-1 MA at the same concentration. As shown in Figure 2, lefthand lanes), in the presence of DMSO, bound (B) MA-RNA complexes with electrophoretic mobilities lower than free (F) RNA ligand were observed readily. As a control, we performed binding reactions in the presence of 100 µM of the thiadiazolane TD1 (Section 2.1.; 16), which has been shown to block MA-RNA binding (16). As illustrated (Figure 2, TD1 lanes), the presence of TD1 resulted in a disappearance of the bound MA-RNA complex. Relative to TD1, the quinolinequinones at 100 µM showed varying capacities to inhibit MA-RNA binding. The compound identified in our FP screen, 81047, strongly inhibited MA-RNA binding, as did the isoquinolinequinone, 76882, but we also observed significant inhibition with 84999 and 102729, and varying degrees of inhibition with the other quinolinequinones (Figure 2). The levels of MA-RNA binding inhibition were quantified and are tabulated in Table 1.
Figure 2. Inhibition of HIV-1 MA binding to RNA.
Electrophoretic mobility shift assays were performed to assess compound effects on HIV-1 MA binding to the Sel15 RNA oligonucleotide. MA (15 µM) samples were preincubated with 100 µM of the indicated compounds, then incubated with 15 µM RNA, after which free (F) and bound (B) RNAs were separated by electrophoresis and visualized by staining with Stains-All. Note that TD1 2-(2-chloro-6-methylphenyl)-4-(cyclopropylmethyl)-1,2,4-thiadiazolane-3,5-dione) was used as a postitive control for inhibition of MA-RNA binding, and that "dye" indicates a dyeonly lane, used to monitor electrophoresis.
Table 1. Quinolinequinone activities.
Compounds are as shown in Figure 1, and listed in accordance with their NSC numbers. Percentage RNA binding inhibition values were determined in electrophoretic mobility gel shift assays with 15 µM MA, 15 µM Sell5 RNA ligand, and 100 µM compounds, as shown in Figure 2. Percentage glutathione (GSH) inactivation values were determined by measurement of GSH free sulfhydryl levels after incubation of 200 µM GSH with 200 µM compound for 1 h at 25°C. Percentage MA crosslinking values were determined after 2 h, 25°C incubations of 30 µM MA with 50 µM compound, and measurement of dimer and monomer bands as shown in Figure 5. Fifty percent cytotoxic (CC50) concentrations (in µM) are shown for human embryonic kidney HEK293T (293) cells and for the human T cell lines CEM-SS (CEM) and MT4. Values were determined by MTS assay at 2 d post-treatment with compounds. Values are averages from three (GSH inactivation) or two (all other assays) independent experiments, with standard deviations as shown.
| Compound | %RNA binding | % GSH | % MA | CC50 values (µM) | ||
|---|---|---|---|---|---|---|
| inhibition | inactivation | X-linking | 293 | CEM | MT4 | |
| 76882 | 69±26 | 96±1 | 47±17 | 8±6 | 0.6±0.1 | 0.7±0.1 |
| 76883 | 36±16 | 33±12 | 10±2 | 22±20 | 2±0 | 2±0 |
| 76885 | 22±16 | 54±23 | 6±1 | 7±3 | 2±0 | 1±1 |
| 76886 | 28±1 | 95±0 | 10±4 | 8±2 | 6±0 | 2±0 |
| 76893 | 26±1 | 50±9 | 13±5 | 9±2 | 2±1 | 2±0 |
| 81046 | 21±6 | 92±3 | 16±11 | 9±1 | 2±0 | 4±3 |
| 81047 | 99±2 | 95±3 | 63±6 | 17±4 | 2±1 | 1±1 |
| 81048 | 50±11 | 50±4 | 29±1 | 18±0 | 5±0 | 3±4 |
| 81050 | 39±30 | 96±2 | 13±10 | 5±4 | 0.7±0.4 | 1±1 |
| 81056 | 26±13 | 53±6 | 11±6 | 4±3 | 0.9±0.2 | 1±1 |
| 82130 | 21±13 | 3±3 | 6±1 | ND | ND | ND |
| 84999 | 76±6 | 30±4 | 20±13 | 7±2 | 5±0 | 3±1 |
| 102729 | 73±1 | 56±1 | 51±11 | 18±2 | 3±0 | 1±1 |
| 105808 | 31±1 | 96±1 | 10±2 | 4±2 | 1±0 | 0.6±0.6 |
| 663284 | 3±3 | 46±5 | 8±2 | 13±5 | 6±1 | 2±0 |
3.2. Sulfhydryl reactivity
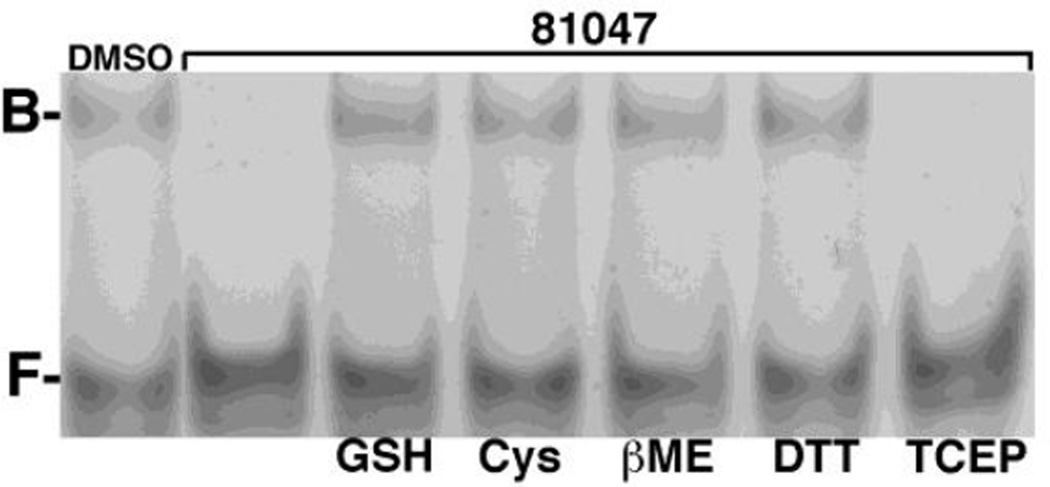
Chemical synthesis studies have shown that a variety of quinolinequinones are reactive to primary amines and sulfhydryls under mild conditions (1, 36–8). Because of this it seemed possible that quinolinequinones might exert their anti-MA activities by covalent reaction with the protein. To test this assumption, we initially examined the effects of primary amines by supplementing MA-RNA binding reactions with 200 µM concentrations of lysine, glycine, and glycylglycine, but found that none of these blocked the effects of quinolinequinones on MA-RNA binding (data not shown). In contrast, we found that sulfhydryl-containing compounds interfered with the anti-MA effects of 81047. This is illustrated in Figure 3. As shown, MA-bound (B) RNA was observed in the presence of DMSO (lefthand lane), but not in the presence of 100 µM 81047 in DMSO (second lane from the left). The effects of 81047 were blocked in the presence of 200 µM glutathione (GSH), cysteine (Cys), β-mercaptoethanol (βME), and dithiothreitol (DTT), but not with the sulfhydryl-free reducing agent tris(2-carboxyethyl)phosphine (TCEP). These results support the notion sulfhydryls readily react with 81047.
Figure 3. Sulfydryl inhibition of anti-MA quinolinequinone activity.
HIV-1 MA-RNA electrophoretic mobility assays were performed as described in Figure 2 to monitor free (F) and MA-bound (B) Sel15 RNA oligonucleotide. In the lefthand lane, MA (15 µM) was preincubated with DMSO prior to the RNA (15 µM) binding incubation. In the righthand lanes, MA was preincubated with 50 µM 81047 alone (second lane from the left), or 50 µM 81047 plus 200 µM glutathione (GSH), cysteine (Cys), β-mercaptoethanol (βME), dithiothreitol (DTT) or Tris(2-carboxyethyl)phosphine hydrochloride (TCEP).
To extend our observations, we performed GSH reactivity assays with all of our quinolinequinones. To do so, equimolar concentrations GSH and the various quinolinequinones were incubated together, after which unreacted GSH levels were quantified in assays with Ellman's reagent (Section 2.3.; 39) to determine percentages of GSH inactivation. Our results (Table 1) show that several quinolinequinones were highly reactive to GSH, including 76882, 76886, 81046, 81047, 81050, and 105808. However, all of the quinolinequinones inactivated at least 20% of the input GSH, with the exception of the 6-hydroxy analogue, 82130.
3.3. Peptide and protein crosslinking
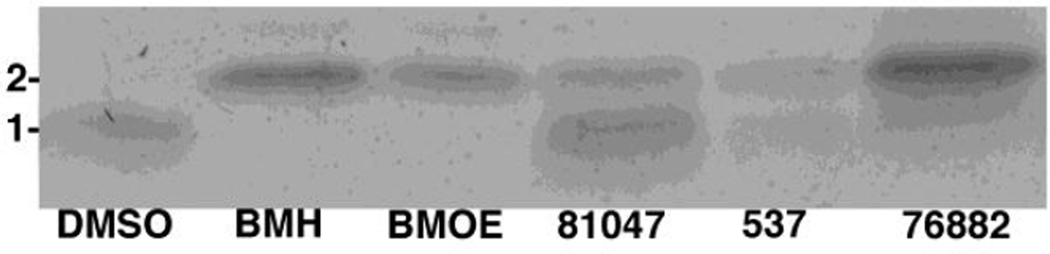
Our sulfhydryl reactivity experiments (Section 3.2.) and results from chemical synthesis studies (1, 36–8) suggested that quinolinequinones and their analogues might have the capacity to react with two separate nucleophiles at their 6 and 7 positions (occupied by chlorines in 81047, Figure 1), and thus potentially act as crosslinking agents. We initially tested this in incubations of quinolinequinones with a lysine-rich, but cysteine-free 33 residue peptide (Section 2.1.), but no crosslinked peptide products were observed (data not shown). Contrasting results were observed when incubations were performed with a 31 residue peptide that carried an amino-terminal cysteine (Section 2.1.). Specifically, the cysteine-containing peptide was incubated either with DMSO, 537, 81047, 76882, or the known sulfhydryl-specific crosslinking agents 1,6-bis-maleimidohexane (BMH) or bis-maleimidoethane (BMOE). After incubations, peptide monomers and dimers were separated by gel electrophoresis and stained. Importantly, while DMSO yielded no peptide dimer band, BMH, BMOE, 81047, 537, and 76882 all yielded dimers (Figure 4). These results indicate that the isoquinolinequinone 76882, and di-chloro napthoquinone (537) and quinolinequinone (81047) analogues all have the capacity to crosslink cysteines, albeit with varying degrees of efficiency. Moreover, because samples were treated with DTT and βME prior to electrophoresis (Section 2.4), quinolinequinone crosslinking does not to simply entail oxidation of cysteines to cystines.
Figure 4. Peptide crosslinking analysis.
Samples of the 31 residue peptide (0.1 mM) with the sequence CSILD IRQGP KEPFR DYVDR FYKTL RAEQA S were incubated with either DMSO, or 1 mM BMH, BMOE, 81047, 537, or 76882. After incubations, monomers (1) and dimers (2) were separated by electrophoresis under reducing conditions and detected via Coomassie blue staining.
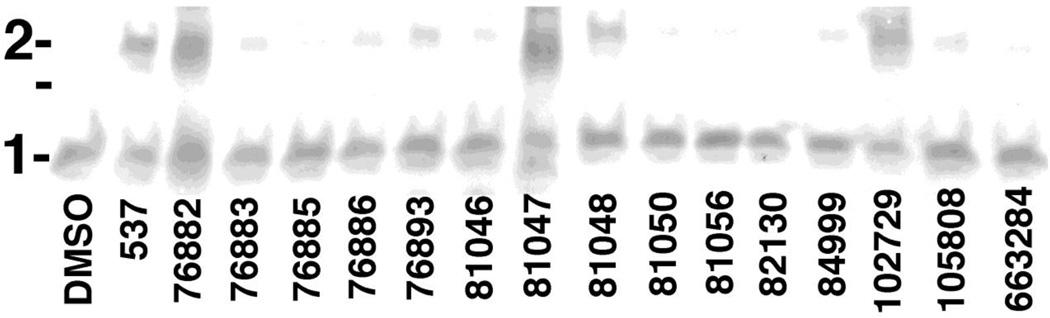
Because HIV-1 MA has two free cysteines, we next examined whether 537, 76882, 81047 and the other quinolinequinones might be able to crosslink the purified HIV-1 MA protein itself. Thus, 30 µM MA samples were incubated in the absence or presence of 50 µM compounds, after which monomers and dimers were separated by electrophoresis and detected by immunoblotting. Our results (Figure 5) were consistent with our peptide crosslinking observations. In particular, DMSO did not yield MA dimers (Figure 5, lefthand lane), while 537, 76882, and 81047 efficiently formed dimers. Of the other quinolinequinones, we observed significant levels of dimer formation for 102729 and 81048, but lower levels for the other analogues. These values were quantified and are listed in Table 1.
Figure 5. HIV-1 MA crosslinking reactions.
Samples of the HIV-1 MA protein (30 µM) were incubated at 25°C for 2 h with DMSO or 50 µM of the indicated compounds. After incubations, MA monomers (1) and dimers (2) were separated by electrophoresis under reducing conditions, and detected by immunoblotting. The unmarked line indicates the migration of a 25 kDa molecular weight marker run in parallel.
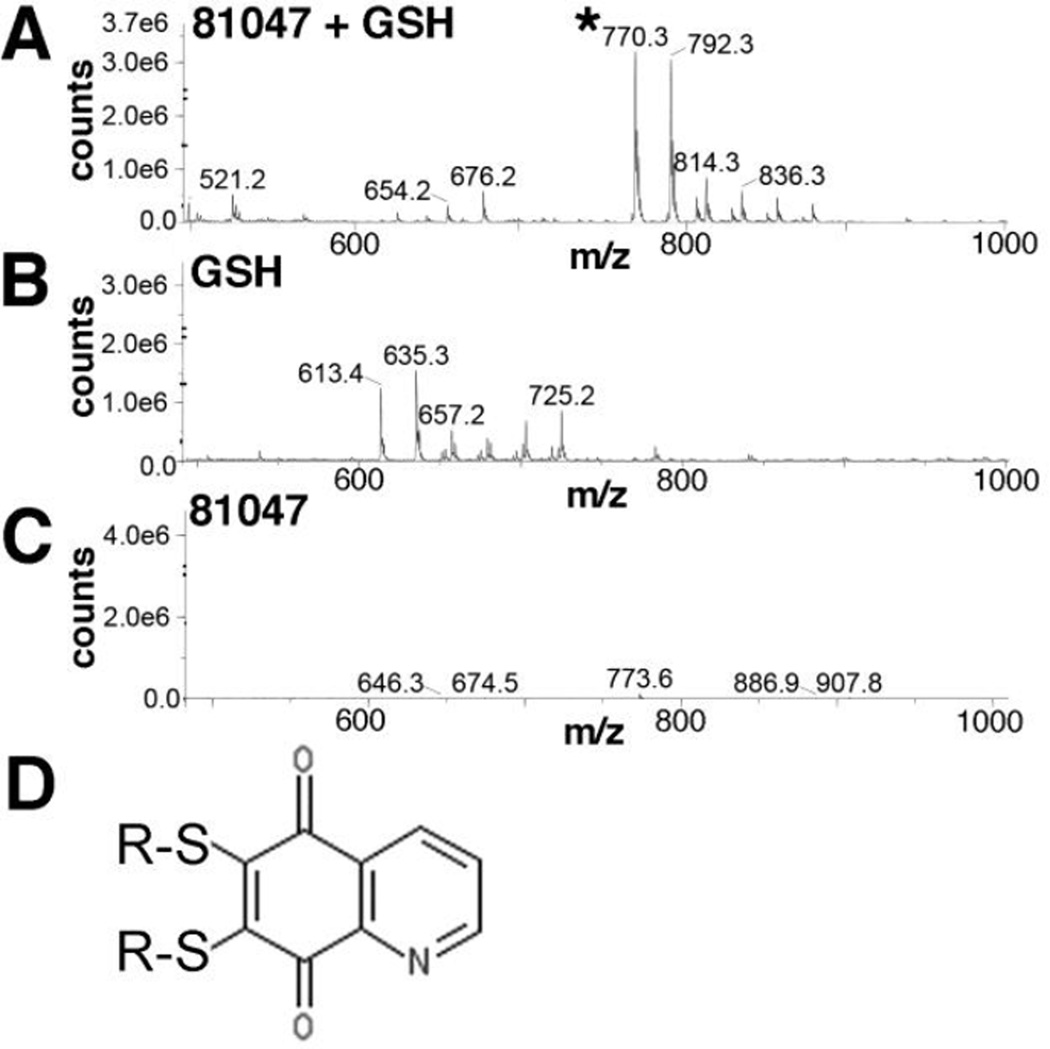
Because both chloro groups of 81047 appeared to be viable targets for sulfhydryl attack, we wished to assess whether 81047 might crosslink peptides and proteins by reacting with cysteines at both the 6- and 7- positions. To do so, we incubated 10 mM GSH with 5 mM 81047, and subjected products to mass spectrometry (MS) analysis. Our results showed clear evidence of a 770.3 m/z proeuct (Figure 6A, asterisked), as well as a 792.3 m/z product (Figure 6A). These values correspond respectively to the singly protonated and sodium-complexed forms of 6,7-bis(glutathionyl)-5,8-quinolinedione (Figure 6D), and no other predicted reaction products matched these spectral peaks. Notably, neither control incubations of GSH alone (Figure 6B) or 81047 alone (Figure 6C) gave these peaks. Thus, our results suggest that 81047 acts as a sulfhydryl specific crosslinker, and can react with sulfhydryls at its 6- and 7- positions.
Figure 6. Analysis of GSH plus 81047 reaction products.
GSH (10 mM) plus 81047 (5 mM) were incubated in 50% DMSO for 3 h at 25°C, supplemented with a fourfold excess (v/v) of acetonitrile, and subjected to mass spectrometry (MS) analysis using electrospray ionization in the positive ion mode. The 770.3 (asterisked) and 792.3 m/z peaks observed in A, but not B or C respectively correspond to the protonated and sodium ion-complexed forms of 6,7-bis(glutathionyl)-5,8-quinolinedione, illustrated in panel D.
3.4. Quinolinequinone effects on cells and viruses
3.4.1. Retrovirus structural protein crosslinking
In infected cells, the HIV-1 MA protein is initially translated as the amino-terminal domain of the structural precursor Gag (PrGag) protein (17–20). During virus assembly and morphogenesis, HIV-1 PrGag proteins direct the assembly of immature virus particles, after which the viral protease processes PrGag proteins into the mature proteins, including MA, capsid (CA), nucleocapsid (NC) and p6 (17–20). Because our quinolinequinone analogues demonstrated the ability to crosslink cysteine-containing peptides and proteins in vitro, it was of interest to assess whether the compounds could act similarly on PrGag proteins in immature virus particles. For our purposes, we employed immature HIV-1 viruses, and immature viruses produced by the related retrovirus, the Moloney murine leukemia virus (MLV; 34). HIV-1 PrGag is 55 kDa and contains ten cysteines (two in MA); MLV PrGag is 65 kDa and encodes five cysteines (one in MA): the reason for including MLV in our analysis was the availability of an assembly competent variant of MLV in which all PrGag cysteines have been mutated to serines (MLV all C>S; Section 2.6.1.; 34).
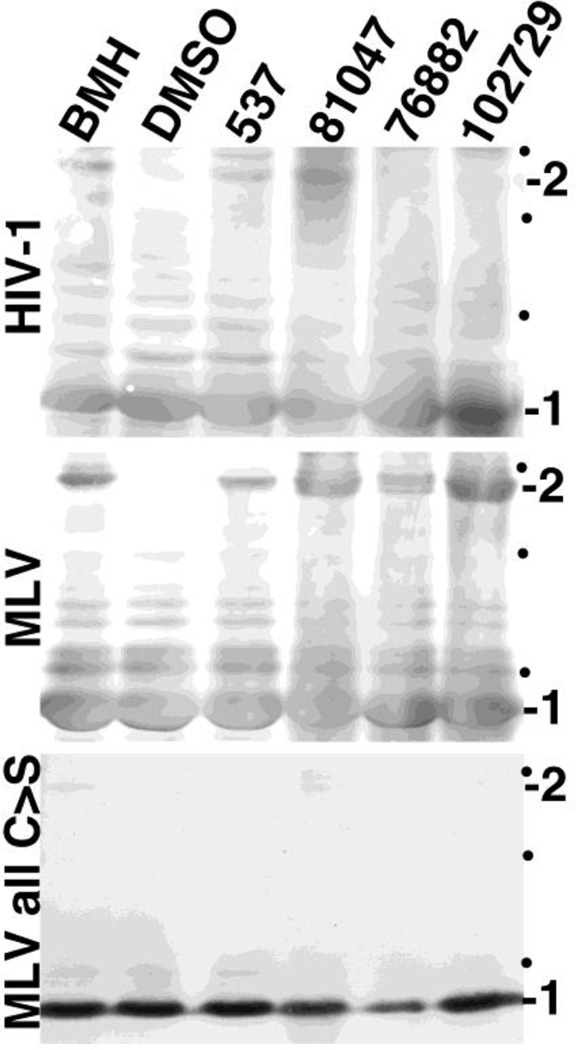
To test compound effects, purified immature HIV-1, MLV and MLV all C>S particles were incubated with DMSO (as a control), 537, 81047, 76882, 102729 or the known sulfhydryl-specific crosslinking reagent, BMH. After incubations, PrGag monomer and dimers were separated by gel electrophoresis and detected by immunoblotting. As illustrated in Figure 7 (top panel), while DMSO treatment of immature HIV-1 did not yield any detectable PrGag dimers, a dimer band was observed with BMH treatment, as were putative higher order crosslink product (data not shown) consistent with previous observations (40). Importantly, 537 and 81047 also showed the HIV-1 PrGag dimer band (Figure 7) and putative higher order crosslink bands (data not shown), as did 76882 and 102729, albeit to lower extents. Results were even more clear with MLV viruses (Figure 7, middle panel), where all compounds gave clear PrGag dimer signals, whereas DMSO alone did not. In contrast, with MLV all C>S viruses (Figure 7, bottom panel), no crosslink products were observed. These results demonstrate that 537, 81047, 76882, and 102729 were able to pass through the membrane envelopes of immature HIV-1 and MLV viruses and crosslink their PrGag proteins via cysteine residues.
Figure 7. Crosslinking of structural proteins in retrovirus particles.
Immature virus particles were prepared for HIV-1, Moloney murine leukemia virus (MLV), and a MLV variant in which all cysteine residues in the structural precursor Gag (PrGag) protein were converted to serines (MLV all C>S). Virus particle preparations were treated with DMSO, or 1 mM BMH, 537, 81047, 76882, or 102729. Viral proteins from reactions were separated by electrophoresis under reducing conditions and immunoblotted to detect either the HIV-1 or MLV PrGag proteins. Monomer (1) and dimer (2) PrGag bands are as indicated, and black dots indicate the mobilities of 150, 100, and 75 kDa marker proteins run in parallel.
3.4.2. Analysis of HIV-1 infection and cytotoxicity
We tested the effects of 81047 by infecting human CEM-SS T cells with wild type (WT) HIV-1 in the presence of DMSO, 1 µM, or 10 µM 81047. To monitor the extent of infection, at 7 d post-infection, cell samples were collected and processed for immunoblot detection of the HIV-1 PrGag, p41, and capsid (CA) proteins (Figure 8). As compared to DMSO controls, 10 µM 81047 dramatically reduced HIV-1 protein levels, and 1 µM 81047 reduced HIV-1 protein levels to a signficant, but lesser extent. However, when we monitored actin levels in the same cells (Figure 8, bottom panel), we observed that actin signals in 10 µM 81047-treated cells were barely detectable, while 1 µM 81047-treated cells showed slightly reduced actin levels. These results suggest that the antiviral effects of 81047 were due wholly or in part to cytotoxicity.
Figure 8. Effects of 81047 on HIV-1 replication.
Human CEM T cells were treated with DMSO or the indicated concentrations of 81047 and infected with the NL4-3 strain of HIV-1. At 7 d post-infection, cell samples were collected, proteins were fractionated by electrophoresis. Actin levels (bottom panel) and HIV-1 PrGag, p41 and capsid (CA) levels (top panel) were determined by immunoblotting. Quantitation indicated that 10 µM 81047 treatment reduced protein levels (as percentages of untreated samples) as follows: PrGag, 17%; p41, 5%; CA, 18%; actin, 14%. Quantitation of 1 µM 81047 treatment levels was as follows: PrGag, 36%; p41, 35%; CA, 54%; actin, 83%.
To determine whether any of our compounds might possess low enough cytotoxicities to be good antiviral candidates, we performed compound cytotoxicity assays on human embryonic kidney HEK293T cells, and in the human CEM-SS and MT4 T cell lines (Table 1). The general trend of our results was that quinolinequinones appeared more toxic to T cell lines than the HEK293T cells. Indeed, CC50 values in T cells for all of the compounds were less than 10 µM.
3.4.3. Virus inactivation studies
Although the quinolinequinones we studied appear too toxic to be used as anti-HIV-1 therapeutics, previous studies have validated the use of cysteine-reactive reagents as potential virucidal agents for the preparation of killed virus vaccines or other biomedical purposes (21–3). One such agent is 2,2'-dipyridyldisulfide (Aldrithiol-2), which was shown to inactivate HIV-1 at 300–1000 µM concentrations (21–3). We thus examined the ability of 81047 to inactivate HIV-1. To do so, HIV-1 viruses that transduce expression of the green fluorescent protein (GFP) or luciferase were produced (Section 2.6.3.). The infectivity of these viruses can be sensitively assayed by infecting target cells and monitoring for either GFP or luciferase expression (Section 2.6.3.; 30, 31, 35).
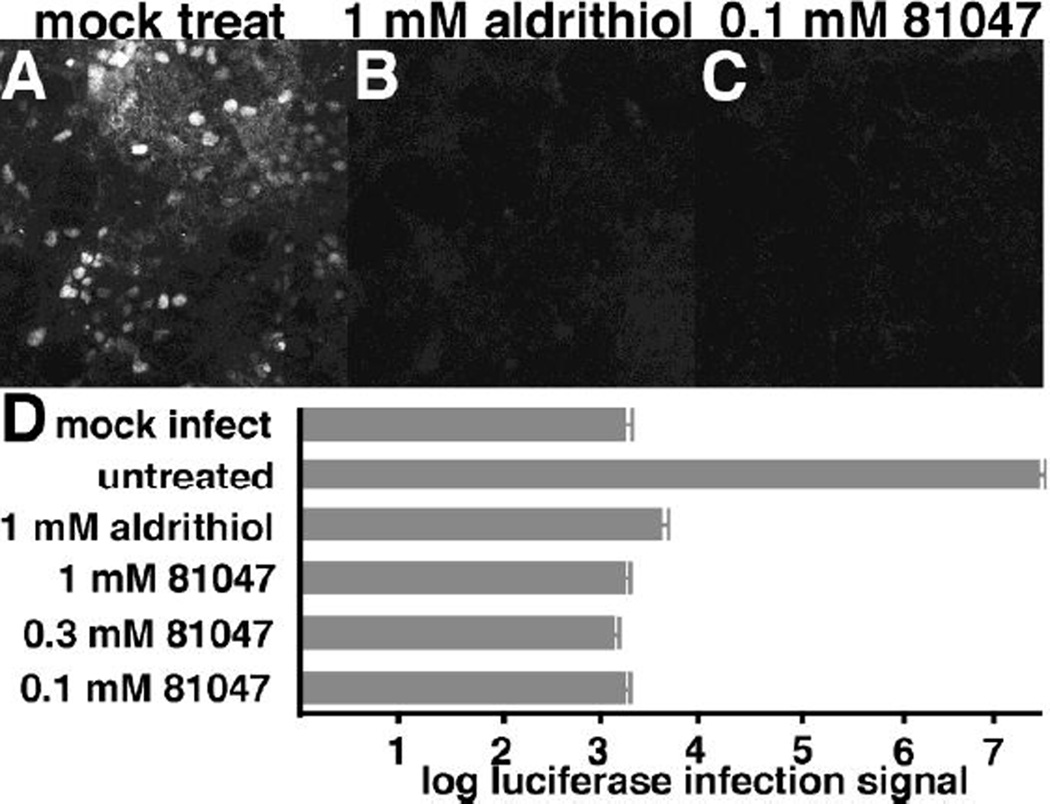
The virucidal effects of 81047 first were tested on GFP-transducing HIV-1 virus. Viruses were mock-treated (Figure 9A), treated with 1 mM Aldrithiol-2 (Figure 9B) or 0.1 mM 81047 (Figure 9C) and used to infect target cells. Virus infection was monitored by fluorescent visualization of GFP in successfully infected cells. As is clearly evident (Figure 9A-C), 0.1 mM 81047 and 1 mM Aldrithiol-2 dramatically reduced virus infection levels. To examine compound virucidal effects in a different assay, luciferase-transducing HIV-1 viruses were mock-treated, or treated with Aldrithiol-2, or 81047, and infection rates were scored via luciferase assays of target cells. In these assays, luciferase levels in mock-infected cells were reduced over four logs from infected, but mock-treated cells (Figure 9D). As expected, 1 mM Aldrithiol-2 treatment reduced virus infectivity levels to nearly mock-infection levels. Significantly, 0.1, 0.3, and 1 mM 81047 treatments reduced infectivity levels over 10,000-fold to mock-infection levels, and below the levels achieved with the Aldrithiol-2 control (Figure 9D). These results suggest that quinolinequinones might prove useful as crosslinking agents, or in the preparation of inactivated virus vaccines, or for other virucidal purposes.
Figure 9. Compound inactivation of HIV-1.
Recombinant HIV-1 viruses expressing the green fluorescent protein (GFP) or the luciferase protein were prepared as described in the Materials and Methods. In panels A-C, the GFP-encoding viruses were mock-treated, or treated with 1 mM 2,2'-dithiodipyridine (aldrithiol) or 0.1 mM 81047 for 1 h at 37°C, and then used to infect human HEK293T cells. At 48 h post-infection, cells were fixed and imaged for visualization of GFP-positive (white) cells. Note that panels B and C had approximately the same number of cells as panel A, but they were not GFP-positive. In panel D, luciferase-encoding viruses were untreated, or treated for 1 h at 37°C with the indicated concentrations of aldrithiol or 81407. The treated and untreated viruses were then used to infect HEK293T cells in parallel with a mock (no virus) control. At 48 h post-infection, cells were collected and processed to measure luciferase signals that are indicative of infection levels. Note that a logarithmic scale was used, and that 81047 signals were the same as mock infected cells, approximately 10,000 times lower than the untreated infection control. Values are the averages of two separate experiments with standard deviations as shown.
4. DISCUSSION
Previous research has established the biomedical potential of napthoquinone and quinolinequinone derivatives (1–15). Originally of interest as dyes, napthoquinones and their derivatives have been investigated with respect to their potential as anti-cancer and microbiocidal agents (10–15). Quinolinequinones also have been reported to have such properties (1–9). Notably, streptonigrin and its derivatives have been studied for more than five decades as potential anti-cancer agents (1). The structurally related lavendamycin has been the subject of investigation for both its anti-cancer and antibiotic activities (2–3). Similar quinolinequinones have been reported to have potent anti-inflammatory and anti-fungal properties (1–9), and LY83583 has been identified as a specific inhibitor of soluble guanylate cyclase (7–9).
We identified the napthoquinone 537 and its quinolinequinone homologue 81047 as inhibitors of HIV-1 MA binding to RNA (16; Figure 2, Table 1). Based on these observations, we decided to examine the properties of a panel of quinolinequinones (Figure 1), and found that several, including 76882, 84999, and 102729 also inhibited MA-RNA binding. Because reports showed that quinolinequinones could react with compounds containing primary amines and sulfhydryls in chemical syntheses (1, 36–8), we tested reactivities of our quinolinequinones under more physiological conditions. Although we did not observe reactions with lysine, glycine, or glycylglycine (data not shown), we found that the majority of our compounds reacted with GSH, and that sulfhydryl-containing compounds blocked the ability of 81047 to inhibit MA-RNA binding. Moreover, several quinolinequinones, notably 81047, 76882, and 102729, efficiently crosslinked cysteine-containing peptides, MA proteins and/or PrGag proteins in HIV-1 and MLV virus particles (Table 1, Figures 4–7). Given the correlation of quinolinequinone crosslinking activities and their abilities to block MA-RNA binding, we believe it likely that quinolinequinone inhibition of MA-RNA binding results from the intra- or intermolecular crosslinking between one or both of the two cysteine residues on each MA protein. In particular, we speculate that crosslinking of MA cysteines, which appear partially solvent-exposed, results in protein misfolding and loss of RNA binding.
Our cytotoxicity analyses (Table 1) demonstrated that quinolinequinones are significantly toxic to human T cell lines. We speculate that the reduced toxicity observed in HEK293T cells may be related an increased sensitivity of the T cells lines to oxidative and electrophilic stress, leading to cell death via necrotic and/or apoptotic pathways (41). In any case, the low CC50 values for quinolinequinones in T cells markedly decreases the potential antiviral value of these compounds, and our studies with 81047 (Figure 8) would appear to support this conclusion. However, 81047 proved extremely potent in the inactivation of HIV-1 virus particle infectivity (Figure 9)--possibly even moreso than the previously characterized (21–3) Aldrithiol-2. Consequently, we believe that quinolinequinones may find applications as microbiocides of contaminated instruments, surfaces and fluids; and possibly in protein studies as membranepermeable, cysteine-specific crosslinking agents.
Acknowledgments
We are grateful to Dr. Dennis Koop and the OHSU Bioanalytical Shared Resource Pharmacokinetics Core for help with the MS analysis, to the Oregon Translational Research and Drug Development Institute (OTRADI) for assistance and financial support, and to the NIH for support through grants R01 GM060170 and R01 GM101983.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mulchin B, Newton C, Baty J, Grasso C, Martin W, Walton M, Dangerfield E, Plunkett C, Berridge M, Harper J, Timmer M, Stocker B. Bioorg. Med Chem. 2010;18:3238. doi: 10.1016/j.bmc.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Hassani M, Cai W, Holley D, Lineswala J, Maharjan B, Ebrahimian G, Seradj H, Stocksdale M, Mohammadi F, Marvin C, Gerdes J, Beall H, Behforouz M. J. Med. Chem. 2005;48:7733. doi: 10.1021/jm050758z. [DOI] [PubMed] [Google Scholar]
- 3.Musiol R, Serda M, Hensel-Bielowka S, Polanski J. Curr. Med. Chem. 2010;17:1960. doi: 10.2174/092986710791163966. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, An LK, Wu N, Wang XD, Bu XZ, Huang ZS, Gu LQ. Bioorg. Med. Chem. 2008;16:4617. doi: 10.1016/j.bmc.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 5.Bolognese A, Correale G, Manfra M, Esposito A, Novellino E, Lavecchia A. J. Med. Chem. 2008;51:8148. doi: 10.1021/jm8007689. [DOI] [PubMed] [Google Scholar]
- 6.Valderrama J, Ibacache J, Arancibia V, Rodriquez J, Theoduloz C. Bioorg. Med. Chem. 2009;17:2894. doi: 10.1016/j.bmc.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Fleisch J, Haisch K, Spaethe S, Rinkema L, Cullinan G, Schmidt M, Marshall W. J. Pharmacol. Exp. Ther. 1984;229:681. [PubMed] [Google Scholar]
- 8.Mulsch A, Busse R, Liebau S, Forstermann UJ. Pharmacol. Exp. Ther. 1988;247:283. [PubMed] [Google Scholar]
- 9.Hasegawa T, Bando A, Tsuchiya K, Abe S, Okamoto M, Kirima K, Ueno S, Yoshizumi M, Houchi H, Tamaki T. Biochim. Biophys. Acta. 2004;1670:19. doi: 10.1016/j.bbagen.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Lopez Lopez L, Nery Flores S, Silva Belmares S, Saenz Galindo A Vitae. Revista de la Facultad de Quimica Farmaceutica. 2014;21:248. [Google Scholar]
- 11.Delarmelina M, Daltoe R, Cerri M, Madiera K, Rangel L, Lacerda V, Jr, Romao W, Taranto A, Greco S. J. Braz. Chem. Soc. 2015;26:1804. [Google Scholar]
- 12.Perez E, Diaz R, Estevez A, Ravelo A, Garcia J, Pardo L, Campillo M. J. Med. Chem. 2007;50:696. doi: 10.1021/jm060849b. [DOI] [PubMed] [Google Scholar]
- 13.Oramas-Royo S, Torrejon C, Cuadrado I, Hernandez-Molino R, Hortelano S, Estevez-Braun A, de las Heras B. Bioorg. Med. Chem. 2013;21:2471. doi: 10.1016/j.bmc.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Bhasin D, Chettiar S, Etter J, Mok M, Li P. Bioorg. Med. Chem. 2013;21:4662. doi: 10.1016/j.bmc.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasiri H, Madej M, Panisch R, Lafontaine M, Bats J, Lancaster C, Schwalbe H. J. Med. Chem. 2013;56:9530. doi: 10.1021/jm400978u. [DOI] [PubMed] [Google Scholar]
- 16.Alfadhli A, McNett H, Eccles J, Tsagli S, Noviello C, Sloan R, Lopez C, Peyton D, Barklis E. J. Biol. Chem. 2013;288:666. doi: 10.1074/jbc.M112.399865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed E. Virology. 1998;251:1. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 18.Alfadhli A, Still A, Barklis E. J. Virol. 2009;83:12196. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chukkapalli V, Oh S, Ono A. Proc. Natl. Acad. Sci. USA. 2010;107:1600. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfadhli A, McNett H, Tsagli S, Bachinger H, Peyton D, Barklis E. J. Mol. Biol. 2011;410:653. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossio J, Esser M, Suryanarayana K, Schneider D, Bess J, Vasquez G, Wiltrout T, Chertova E, Grimes M, Sattentau O, Arthur L, Henderson L, Lifson L. J. Virol. 1998;72:7992. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott D, Hewes S, Alvord WG, Henderson L, Arthur L. Virology. 1998;243:283. doi: 10.1006/viro.1998.9062. [DOI] [PubMed] [Google Scholar]
- 23.Rutebemberwa A, Bess J, Brown B, Arroyo M, Eller M, Slike B, Polonis V, McCutchan F, Currier J, Birx D, Robb M, Marovich M, Lifson J, Cox J. AIDS Res. Hum. Retroviruses. 2007;23:532. doi: 10.1089/aid.2006.0136. [DOI] [PubMed] [Google Scholar]
- 24.Clish C, Peyton D, Barklis E. Eur. J. Biochem. 1998;257:69. doi: 10.1046/j.1432-1327.1998.2570069.x. [DOI] [PubMed] [Google Scholar]
- 25.Alfadhli A, Steel E, Finlay L, Bachinger HP, Barklis E. J. Biol. Chem. 2002;277:27103. doi: 10.1074/jbc.M203395200. [DOI] [PubMed] [Google Scholar]
- 26.Schneider C, Rasband W, Eliceiri K. Nature Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie C, Cylinder I, Platt E, Barklis E. J. Virol. 2015;89:3988. doi: 10.1128/JVI.03584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alfadhli A, Mack A, Ritchie C, Cylinder I, Harper L, Tedbury P, Freed E, Barklis E. J. Virol. 2016 doi: 10.1128/JVI.00509-16. pii: JVI00509-16 [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DuBridge R, Tang P, Hsia H, Leong P, Miller J, Calos M. Mol. Cell. Biol. 1987;7:379. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noviello C, Lopez C, Kukull B, McNett H, Still A, Eccles J, Sloan R, Barklis E. J. Virol. 2011;85:4730. doi: 10.1128/JVI.00099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez C, Tsagli S, Sloan R, Eccles J, Barklis E. Virology. 2013;447:95. doi: 10.1016/j.virol.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adachi A, Gendelman H, Koenig S, Folks T, Willey R, Rabson A, Martin M. J. Virol. 1986;59:284. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez C, Sloan R, Cylinder I, Kozak S, Kabat D, Barklis E. Virology. 2014;462–463:126. doi: 10.1016/j.virol.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen M, Barklis E. J. Virol. 1995;69:1150. doi: 10.1128/jvi.69.2.1150-1159.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu B, Chao J, Singer R. Biophys. J. 2012;102:2936. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt Y. J. Org. Chem. 1962;27:3905. [Google Scholar]
- 37.Tandon V, Maurya H. Tetrahedron Lett. 2009;50:5896. [Google Scholar]
- 38.Egu S, Okoro U, Wirth T. ScienceOpen Res. 2015 [Google Scholar]
- 39.Ellman G. Arch. Biochem. Biophys. 1959;82:70. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 40.McDermott J, Farrell L, Ross R, Barklis E. J. Virol. 1996;70:5106. doi: 10.1128/jvi.70.8.5106-5114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morales MC, Perez-Yarza G, Nieto-Rementeria N, Boyano MD, Jangi M, Atencia R, Asumendi A. Anticancer Res. 2005;25:1945. [PubMed] [Google Scholar]