Abstract
Various chemicals, i.e., furfural, vanillin, 4-hydroxybenzaldehyde and acetate produced during the pretreatment of biomass affect microbial fermentation. In this study, effect of vanillin, 4-hydroxybenzaldehyde and acetate on antibiotic production in Streptomyces coelicolor is investigated. IC 50 value of vanillin, 4-hydroxybenzaldehyde and acetate was recorded as 5, 11.3 and 115 mM, respectively. Vanillin was found as a very effective molecule, and it completely abolished antibiotic (undecylprodigiosin and actinorhodin) production at 1 mM concentration, while 4-hydroxybenzaldehyde and acetate have little effect. Microscopic analysis with field emission scanning electron microscopy (FESEM) showed that addition of vanillin inhibits mycelia formation and increases differentiation of S. coelicolor cells. Vanillin increases expression of genes responsible for sporulation (ssgA) and decreases expression of antibiotic transcriptional regulator (redD and actII-orf4), while it has no effect on genes related to the mycelia formation (bldA and bldN) and quorum sensing (scbA and scbR). Vanillin does not affect the glycolysis process, but may affect acetate and pyruvate accumulation which leads to increase in fatty acid accumulation. The production of antibiotics using biomass hydrolysates can be quite complex due to the presence of exogenous chemicals such as furfural and vanillin, and needs further detailed study.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-016-0539-y) contains supplementary material, which is available to authorized users.
Keywords: Antibiotic, Biomass, Furfural, Streptomyces coelicolor, Vanillin
Introduction
Lignocellulose is a complex-structured biomass composed of cellulose, hemicelluloses, and lignin. It cannot be used as carbon source directly and it should be available as fermentable sugar (Bhatia et al. 2015a, 2016; Verma et al. 2016). Various pretreatment methods using acids (such as sulfuric acid and phosphoric acid) or bases (such as ammonium hydroxide, sodium hydroxide, and potassium hydroxide) have been reported to improve the accessibility of enzymes to the polysaccharides embedded inside the cell wall and to release free sugars (Laurens et al. 2015; Patel et al. 2012; Verma et al. 2013). Lignocellulose pretreatment using different methods also lead to the release of different inhibitors, i.e., furfural, vanillin, 4-hydroxybenzaldehyde, and acetate, which further affects the ability of microbes to utilize free sugar as a carbon source (Allen et al. 2010). Furfural and vanillin are more toxic compounds beyond certain concentration and affect microbial growth (Zhang et al. 2014). A number of approaches have been used to investigate the mechanism of furfural action as this inhibits growth by damaging DNA and chemically reacting with cellular constituents (Akillioglu et al. 2011). Vanillin is one of the most prevalent phenolic compounds found in various lignocellulosic hydrolysates, e.g., from spruce, pine, poplar, corn stover and sugarcane bagasse, and acts as a quorum sensing (QS) blocker (Lv et al. 2014).
Streptomyces coelicolor is a commercially important actinomycete having the potential to produce two chemically distinct pigments as secondary metabolite, i.e., actinorhodin (Act, diffusible blue pigment) and undecylprodigiosin (Red), a cell wall-associated red pigment (Bhatia et al. 2016; Gomez-Escribano and Bibb 2014). The production of antibiotics is regulated by nutrients, growth rate, quorum sensing, transcriptional regulators and other pleiotropic genes (Liu et al. 2013). Regulation is influenced by various low molecular mass compounds, transfer RNA, sigma factors and gene products formed during post-exponential development. These events generate signals which affect a cascade of regulatory events resulting in chemical differentiation (secondary metabolism) and morphological differentiation (morphogenesis). The transcriptional regulation of each antibiotic’s biosynthetic gene cluster depends on a cluster-linked, antibiotic-specific and transcriptional regulator genes. Extensive classical and molecular genetic studies have led to the identification and characterization of numerous developmental genes, the bld and whi, and antibiotic-specific regulators, actII-orf4 for Act and redD for Red (Bush et al. 2013; Lee et al. 2012; Price et al. 1999). Various genes responsible for the physiological controls which operate on pigment production in S. coelicolor are unknown. Molecular biology of antibiotic production is still not understood to a great degree and thus provides an opportunity for further investigation. Most of the research groups have reported that biomass-derived molecules act as an inhibitor, but interestingly in our previous research we found that furfural can elicit antibiotic production in S. coelicolor and it can be used to increase undecylprodigiosin production (Bhatia et al. 2016). Without considering various chemicals exist in biomass hydrolyzate, it is quite risky to use this as a carbon source. In this work, effect of other inhibitory molecules (vanillin, 4-hydroxybenzaldehyde and acetate) on antibiotic production and expression of various regulatory genes were studied.
Materials and methods
Chemicals
All the chemicals for media were purchased from Difco laboratories (Becton–Dickinson, Franklin Lakes, NJ, USA) and other chemicals, e.g., vanillin, 4-hydroxybenzaldehyde and acetate, were from Sigma-Aldrich (St. Louis, MO, USA). Agarose and bacterial agar were supplied by the Microbial carbohydrate resource bank at Konkuk University, Korea.
Microorganism and seed culture
Streptomyces coelicolor A3 (2) M145 used in this study for secondary metabolite production was purchased from the Korean Culture Type Collection (KCTC), South Korea. Streptomyces coelicolor spores were cultivated on R5 agar plates for 72 h, harvested by scraping and suspended in 20 % (v/v) glycerol and stored at −80 °C (Kieser 2000). Streptomyces coelicolor seed culture was prepared by inoculating spores in 50 mL of LB liquid medium, with five glass beads of 3 mm size, and incubated at 30 °C under shaking condition (200 rpm). The germinated spores were harvested by centrifugation (3200×g, 4 °C, 10 min) and resuspended in 5 mL of ion-free water. 0.1 mL (2 × 106 CFU) of germinated seed culture was used as inocula for further experiments in M9 minimal media of Difco laboratories with 1 % glucose as carbon source.
Antibiotic extraction and quantification
For estimation of undecylprodigiosin (red) and actinorhodin (blue) antibiotics, 2 mL of culture samples was taken and divided into two aliquots. Actinorhodin estimation was performed by adding an equal volume of 1 M NaOH into one of the aliquot, centrifuged for 5 min at 4000g, and absorbance was taken at 633 nm. Undecylprodigiosin is a membrane-associated red pigment, so culture was harvested by centrifugation (4000g for 5 min) and cell pellet was suspended in methanol and incubated at 37 °C in a shaking incubator (200 rpm) for 1 h. Cells were removed by centrifugation at 4000g for 5 min; then 0.1 M HCl was added to the supernatant to adjust its pH and absorbance was measured at 533 nm. The concentration of actinorhodin and undecylprodigiosin was calculated as described already (Horinouchi and Beppu 1984).
Inhibitors effect on S. coelicolor antibiotic production
In this study, the effect of biomass-derived inhibitors, vanillin, 4-hydroxybenzaldehyde and acetate, was investigated on growth and secondary metabolite production in S. coelicolor. To check the effect of various inhibitors, S. coelicolor was cultured in M9 minimal media of Difco laboratories, with 1 % glucose as carbon source and different concentrations of vanillin (0–1 mM), 4-hydroxybenzaldehyde (0–8 mM) and acetate (0–80 mM), for 72 h at 30 °C under shaking condition (200 rpm) at 10-mL scale. After 72 h, 2 mL of the culture sample was taken and biomass and antibiotics, i.e., undecylprodigiosin (Red) and actinorhodin (Blue), were estimated as mentioned above.
Field emission scanning electron microscopy (FESEM)
Vanillin was observed as the most effective molecule which drastically changes the antibiotic production in S. coelicolor at very low concentration. To study the effect of this compound further, S. coelicolor was cultured using minimum effective concentration of vanillin (1 mM) as mentioned above and monitored for morphological change. Samples were prepared for FESEM analysis using methods as already reported (Ishii et al. 2004). FESEM was performed by SUPRA 55VP, CarlZeiss, Oberkochen, Germany. The samples were monitored with a 15-kV accelerating voltage and photographic images were captured digitally at different magnification.
Lipids and metabolite quantification
Antibiotic and fatty acid production pathways are interrelated (Revill et al. 1996), so total fatty acid of S. coelicolor cultured with and without vanillin was extracted and analyzed for composition as described already (Bhatia et al. 2015b). For metabolite analysis, S. coelicolor was cultured with vanillin at 30 °C for 72 h. On completion of growth 1 mL of sample was collected, centrifuged at 12,000g and supernatant was analyzed using an HPLC system equipped with a Bio-Rad Aminex HPX-87H column (Bio-Rad Co., Hercules, CA, USA). A mobile phase of 5 mM H2SO4 at a flow rate of 0.6 mL/min was used and the column temperature was maintained at 50 °C. Various organic acids were quantified at 210 nm.
Antibiotic regulatory genes study
There are various genes of S. coelicolor already reported having a role in quorum sensing, secondary metabolite and morphology development, which altogether affects antibiotic production (Table 1). To study the mRNA expression level of these genes in the presence of vanillin, RT-PCR analysis was performed. For mRNA extraction, S. coelicolor spores were germinated for 5 h in LB broth at 30 °C and further used as seed. Streptomyces coelicolor culture was grown in the presence of vanillin (1.0 mM) at 10-mL scale for 72 h at 30 °C and samples were withdrawn at different time intervals. Collected samples were rapidly cooled on ice in pre-chilled Falcon tubes after which they were centrifuged at 4000g for 10 min at 4 °C. The supernatant was discarded and the pellet was added with 0.5 mL of RNA protect reagent (Qiagen, Valencia, CA, USA). The mixture was then incubated at 25 °C for 5 min. The RNeasy mini kit (Qiagen) was used for extracting total RNA from the control and vanillin-affected samples. The RNA was quantified using NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) by measuring the absorbance at 260 and 280 nm. Super-script II reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) was used for cDNA synthesis from the extracted RNA.
Table 1.
Various genes targeted for the mRNA expression and primers designed for their amplification to study the effect of vanillin on their expression level
| Gene ID | Gene | Primer |
|---|---|---|
| Antibiotic synthesis genes | ||
| SCO5877 | redD | RT_redD_F: CCCGACAACGTCCTCAAC RT_redD_R: CGAGACGAGTCTCAGGAAGC |
| SCO5085 | actII-orf4 | RT_actII4_F: AGAATAGGGCCGATGATTCC RT_actII4_R: CCCAGTTCGTCGGACAGTAT |
| Morphological genes | ||
| SCOt24 | bldA | RT_bldA_F: GCCCGGATGGTGGAATGCAG RT_bldA_R: TGGTGCCCGGAGCCGGACTT |
| SCO3323 | bldN | RT_bldN_F: CCTCGAGTCCCTCTCCAAC RT_bldN_R: CGGTACTGGAGCGTTTTGAT |
| SCO3926 | ssgA | RT_ssgA_F: CCTTTCATCTGCCCGGAGAC RT_ssgA_R: CGACCTGAAGTCGGATCAGC |
| SCO1541 | ssgB | RT_ssgB_F: TCGTGTGCATCGCTCTCAG RT_ssgB_R: CTAGCTTTCCGCCAGGATGT |
| SCO3925 | ssgR | RT_ssgR_F: GGCTGTTCTTCCTCGGTGAG RT_ssgR_R: GAGACGCACATGACCTCGAT |
| SCO2082 | fstz | RT_ftsZ_F: GTTCATCGCCATCAACACCG RT_ftsZ_R: TGTCACGAAGACCATGTCGG |
| Quorum sensing and pleiotropic genes | ||
| SCO6266 | scbA | RT_scbA_F: ACTACACCTGCCACCTCGAC RT_scbA_R: GCCGGTAGACTTGAGGACTG |
| SCO6265 | scbR | RT_scbR_F: TCTTCGAGAAGCAGGGCTAC RT_scbR_R: GCCCATGTCGATGAGTTCTT |
| SCO4425 | afsS | RT_afsS_F: ATGAGCGACAAGATGAAGGA RT_afsS_R: GGTTGTCCATCGTGGTGAT |
| SCO4426 | afsR | RT_afsR_F: GGCTGCTGGACTTCTACCTG RT_afsR_R: CCTCCGTGTACAGCCAGTC |
Reverse transcription polymerase chain reaction (RT-PCR)
The primers specific for various genes and for the endogenous control (16S rRNA) were designed using the Primer Express software® (Applied Biosystems, Foster City, CA, USA) based on S. coelicolor genome published in the NCBI database. Custom-synthesized primers for each gene were obtained from Integrated DNA Technologies (Foster City, CA, USA). The primers used in the study are provided in Table 1. RT-PCR was done with the LifePro thermal cycler by custom thermal cycling conditions with the normalized cDNA as a template. The samples were analyzed in the duplicates and standardized against 16S rRNA gene expression. The relative changes in mRNA expression levels were determined using comparative band density between the vanillin and control S. coelicolor.
Results and discussion
Effect of inhibitors on antibiotic production and morphology
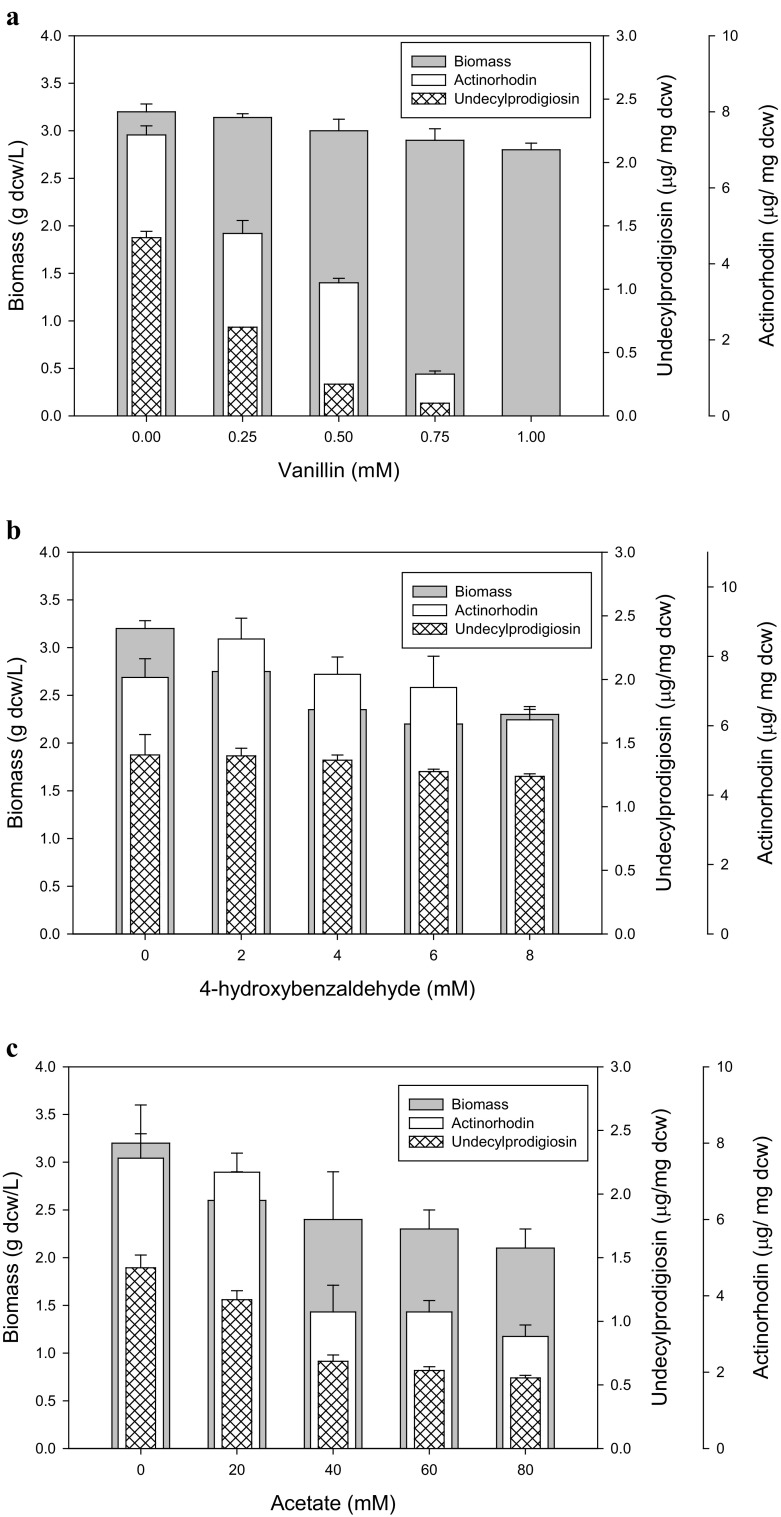
Furfural, vanillin, 4-hydroxybenzaldehyde and acetate are chemical molecules produced during the pretreatment of biomass (Allen et al. 2010). To know the effect of vanillin, 4-hydroxybenzaldehyde and acetate on antibiotic production in S. coelicolor, different concentrations were investigated. Vanillin had little effect on biomass production while a rapid decrease in antibiotic (undecylprodigiosin and actinorhodin) production was recorded with the increase of its concentration. There was no antibiotic production observed above 0.75 mM vanillin (Fig. 1a). 4-Hydroxybenzaldehyde and acetate have a mild effect on biomass and antibiotic production in S. coelicolor (Fig. 1b, c). IC 50 value for vanillin, 4-hydroxybenzaldehyde and acetate was calculated as 5.0, 11.3 and 115 mM, respectively. Streptomyces coelicolor cells were cultured in the presence of various inhibitors at their IC 50 concentration and antibiotics (undecylprodigiosin and actinorhodin) were extracted. In the presence of vanillin, no antibiotic was observed; however, in case of 4-hydroxybenzaldehyde and acetate, reduction in both antibiotics’ production was observed as compared to control (Fig. S1). Other biomass-derived molecule such as furfural enhanced undecylprodigiosin production and inhibits actinorhodin production in S. coelicolor as already reported (Bhatia et al. 2016). In this study, vanillin was found as a most effective molecule which affected antibiotic production in S. coelicolor dramatically (Fig. S1). It was not easy to expect why this phenomena happened because there is no report on vanillin effect.
Fig. 1.
Effect of biomass-derived chemicals a vanillin, b 4-hydroxybenzaldehyde (4-HB) and c acetate on S. coelicolor growth and antibiotic production. S. coelicolor was cultured in M9 media with 1 % glucose and various concentrations of inhibitors
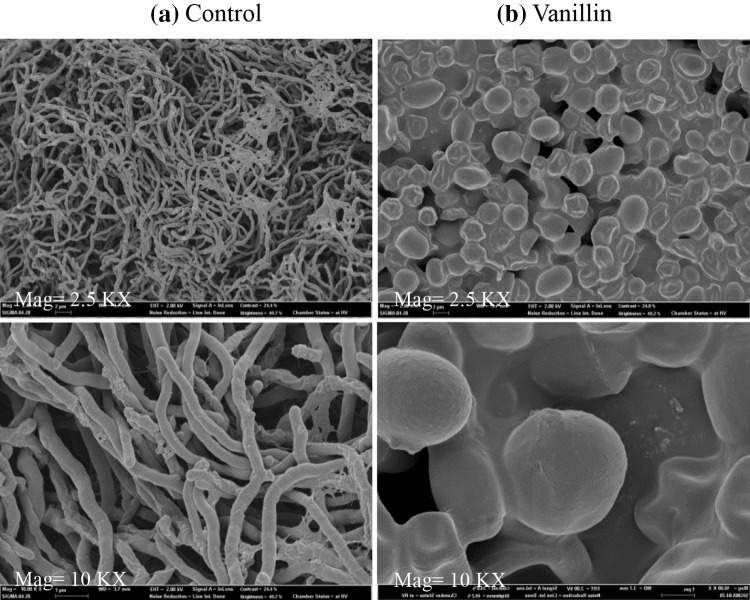
Among examined inhibitors, as vanillin showed inhibitory effects on antibiotic production at low concentration, any morphological change was further investigated. Field emission scanning electron microscopy (FESEM) of S. coelicolor cells cultured in the presence of vanillin was performed. Streptomyces coelicolor cells without any inhibitors showed normal mycelial growth and morphology (Fig. 2a). Streptomyces coelicolor cells grown in the presence of vanillin showed inhibition of mycelia formation (Fig. 2b), and cells had a round structure. In the presence of vanillin, S. coelicolor cells are unable to produce antibiotics due to the lack of mycelia. Mycelia formation is required for polyketide antibiotic production in S. coelicolor as already reported (Gehring et al. 2001).
Fig. 2.
Field emission scanning electron microscopy (FESEM) of S. coelicolor. a Control: normal cell morphology with mycelia; b with vanillin: cells are round shaped and mycelia formation is inhibited
Transcriptional analysis of antibiotic regulatory gene
Streptomyces coelicolor cultured in the presence of vanillin completely abolished antibiotic production. mRNA expression profile of redD and actII-orf4 was analyzed at 48- and 72-h intervals and reduction in expression level was observed (Fig. S2). redD and actII-orf4 are transcriptional activator genes of the undecylprodigiosin and actinorhodin biosynthetic pathway, respectively, and regulate antibiotic production (Fujii et al. 1996; Wang et al. 2014). Bhatia et al. reported that increase in undecylprodigiosin and reduction in actinorhodin production in S. coelicolor under the effect of furfural is due to altered expression of redD and actII-orf4 (Bhatia et al. 2016). There was no change in the mRNA expression level of genes responsible for mycelia formation (bldA, bldN) analyzed in the presence of vanillin during all growth phases (Fig. S2). BldA has a role in mycelia development in S. coelicolor and it codes tRNA for leucine codon UUA required for undecylprodigiosin production at higher phosphate concentrations (White and Bibb 1997). BldN gene codes for sigma factor (BldN) required for the formation of specialized spore-bearing aerial hyphae during differentiation in the mycelial bacterium S. coelicolor (Bibb and Buttner 2003). Genes responsible for sporulation and differentiation of S. coelicolor were investigated for mRNA expression level. An increase in the ssgA mRNA expression level (1.6-fold) was recorded. The ssgA gene involved in cell division and development as already reported (van Wezel et al. 2000). Other genes responsible for sporulation, i.e., ssgB, ssgR and ftsZ, were not affected by vanillin (Fig. S2). Various genes involved in quorum sensing which may affect antibiotic production were also studied. Vanillin had no effect on scbA and scbR expression levels (Fig. S2). Change in scbA expression has no role in secondary metabolite production as already reported (D’Alia et al. 2011); an scbA mutant that failed to produce gamma-butyrolactones can still produce antibiotics, i.e., actinorhodin (Act) and undecylprodigiosin (Red). The decrease in scbR expression leads to a delay in undecylprodigiosin antibiotic production, as scbR failed to make gamma-butyrolactones (D’Alia et al. 2011). AfsS and afsR are pleiotropic genes that regulate undecylprodigiosin and actinorhodin synthesis pathways (Horinouchi 2003; Lian et al. 2008). The increase in afsR copy number can stimulate both Act and Red production (Floriano and Bibb 1996), but no changes in the mRNA expression level of afsS and afsR were recorded in the presence of vanillin. From mRNA expression data of morphological genes, it could be concluded that vanillin is affecting genes involved in sporulation and enhances differentiation of cells shown by FESEM analysis result.
Total lipid and metabolite profiling
Streptomyces coelicolor cells were cultured with and without inhibitors in the above-mentioned conditions. Streptomyces coelicolor without any inhibitors was able to accumulate fatty acid (6.3 µg/mg dcw), while vanillin addition resulted in an increase in fatty acid accumulation (19 µg/mg dcw) (Table 2). Vanillin inhibited antibiotic production and resulted in an increase in fatty acid accumulation in S. coelicolor, as antibiotic and fatty acid synthesis pathways are interrelated (Revill et al. 1996). Metabolite concentrations in S. coelicolor culture supernatant were quantified at 72 h. Glucose was consumed almost completely in control while 6 % reduction in glucose utilization was recorded in the presence of vanillin. Increase in acetate (2.6 mM) and pyruvate (4.7 mM) accumulation was observed in the presence of vanillin (Fig. S3). This observation suggests that glycolysis pathway of S. coelicolor in control and vanillin-treated cell is working properly as there was little change in glucose consumption. Vanillin represses the expression of antibiotic synthesis genes and enhances acetate accumulation which further reduces antibiotic production as observed above with the external addition of acetate. Organic acid content, i.e., acetate and pyruvate, affects the pH of fermentation broth which further changed antibiotic synthesis in S. coelicolor as already explained (Yang et al. 2010).
Table 2.
Total fatty acid profile of S. coelicolor under the effect of vanillin
| Fatty acid | Control (%) | Vanillin (%) |
|---|---|---|
| C12:0-3OH | 3.3 ± 0.1 | 4.6 ± 0.4 |
| C14:0-13M | 22.0 ± 2.3 | 18.3 ± 3.4 |
| C15:1 | 2.0 ± 0.2 | 1.76 ± 0.05 |
| C14:0-2OH | 0.62 ± 0.04 | 0.49 ± 0.02 |
| C14:0-3OH | 29.2 ± 4.0 | 33.54 ± 2.9 |
| C15:0-14M | 3.0 ± 0.07 | 2.26 ± 0.7 |
| C16:1-n9 | 19.0 ± 3.3 | 22.6 ± 1.3 |
| C16:0 cyclo | 18.7 ± 2.7 | 14.30 ± 2.1 |
| C16:0-15M | 1.30 ± 0.06 | 1.0 ± 0.06 |
| C16:0-2OH | 0.45 ± 0.02 | 0.28 ± 0.02 |
| C18:3-n6,9,12 | 0.21 ± 0.01 | 0.58 ± 0.04 |
| C18:1-n9t | 0.36 ± 0.05 | 0.27 ± 0.01 |
| Total fatty acid (µg/mg dcw) | 6.2 ± 0.8 | 19 ± 2.3 |
Conclusion
Biomass is an abundantly available raw material and can be used to develop an economic bioprocess for the production of industrial valuable compounds. Use of biomass hydrolysate without knowing its composition may lead to adverse effects on microbial fermentation and productivity. Cultivation of S. coelicolor in the presence of biomass-derived molecules affects metabolite pool and morphology of S. coelicolor, which further leads to change in antibiotic production. Microbes show different behavior against each inhibitory compound; therefore, there is a need to study the role of such type of compounds to use biomass as a potential carbon source.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1A2A2A04006014) and (NRF-2015M1A5A1037196), R&D Program of MOTIE/KEIT (10048350, 10049674), Korea Institute of Energy Technology Evaluation and Planning (KETEP), and the Ministry of Trade, Industry & Energy (MOTIE) of the Republic of Korea (No. 20163010092150). This article was also supported by the KU research Professor Program and KU Brain Pool Fellowship Program of Konkuk University, Seoul, South Korea.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Akillioglu HG, Mogol BA, Gokmen V. Degradation of 5-hydroxymethylfurfural during yeast fermentation. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2011;28:1629–1635. doi: 10.1080/19440049.2011.609491. [DOI] [PubMed] [Google Scholar]
- Allen SA, et al. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels. 2010;3:1–10. doi: 10.1186/1754-6834-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SK, Kumar N, Bhatia RK. Stepwise bioprocess for exopolysaccharide production using potato starch as carbon source. 3 Biotech. 2015;5:735–739. doi: 10.1007/s13205-014-0273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SK, et al. Development of semi-synthetic microbial consortia of Streptomyces coelicolor for increased production of biodiesel (fatty acid methyl esters) Fuel. 2015;159:189–196. doi: 10.1016/j.fuel.2015.06.084. [DOI] [Google Scholar]
- Bhatia SK, et al. Medium engineering for enhanced production of undecylprodigiosin antibiotic in Streptomyces coelicolor using oil palm biomass hydrolysate as a carbon source. Bioresour Technol. 2016;27:30182–30188. doi: 10.1016/j.biortech.2016.02.055. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Buttner MJ. The Streptomyces coelicolor developmental transcription factor sigmaBldN is synthesized as a proprotein. J Bacteriol. 2003;185:2338–2345. doi: 10.1128/JB.185.7.2338-2345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio. 2013 doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2) Microb Biotechnol. 2011;4:239–251. doi: 10.1111/j.1751-7915.2010.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- Fujii T, Gramajo HC, Takano E, Bibb MJ (1996) redD and actII-ORF4, pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing sigma hrdD. J Bacteriol 178:3402–3405. https://www.ncbi.nlm.nih.gov/pubmed/8655533 [DOI] [PMC free article] [PubMed]
- Gehring AM, Yoo NJ, Losick R. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J Bacteriol. 2001;183:5991–5996. doi: 10.1128/JB.183.20.5991-5996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Indus Microbiol Biotechnol. 2014;41:425–443. doi: 10.1007/s10295-013-1348-5. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2) J Indus Microbiol Biotechnol. 2003;30:462–467. doi: 10.1007/s10295-003-0063-z. [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Beppu T. Production in large quantities of actinorhodin and undecylprodigiosin induced by afsB in Streptomyces lividans. Agric Biol Chem. 1984;48:2131–2133. [Google Scholar]
- Ishii S, Koki J, Unno H, Hori K. Two morphological types of cell appendages on a strongly adhesive bacterium, Acinetobacter sp. strain Tol 5. Appl Environ Microbiol. 2004;70:5026–5029. doi: 10.1128/AEM.70.8.5026-5029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T (2000) Practical streptomyces genetics. John Innes Foundation. https://www.jic.ac.uk/science/molmicro/Strepmanual/Manual.htm. Accessed 5 July 2016
- Laurens LML, Nagle N, Davis R, Sweeney N, Van Wychen S, Lowell A, Pienkos PT. Acid-catalyzed algal biomass pretreatment for integrated lipid and carbohydrate-based biofuels production. Green Chem. 2015;17:1145–1158. doi: 10.1039/C4GC01612B. [DOI] [Google Scholar]
- Lee HN, Kim HJ, Kim P, Lee HS, Kim ES. Minimal polyketide pathway expression in an actinorhodin cluster-deleted and regulation-stimulated Streptomyces coelicolor. J Ind Microbiol Biotechnol. 2012;39:805–811. doi: 10.1007/s10295-011-1083-8. [DOI] [PubMed] [Google Scholar]
- Lian W, et al. Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2) BMC Genom. 2008;9:1471–2164. doi: 10.1186/1471-2164-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Wang Y, Zhong C, Li Y, Hao W, Zhu J. The effect of quorum sensing and extracellular proteins on the microbial attachment of aerobic granular activated sludge. Bioresour Technol. 2014;152:53–58. doi: 10.1016/j.biortech.2013.10.097. [DOI] [PubMed] [Google Scholar]
- Patel SK, Singh M, Kumar P, Purohit HJ, Kalia VC. Exploitation of defined bacterial cultures for production of hydrogen and polyhydroxybutyrate from pea-shells Biomass Bioenerg. 2012;36:218–225. [Google Scholar]
- Price B, Adamidis T, Kong R, Champness W (1999) A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J Bacteriol 181:6142–6151. https://www.ncbi.nlm.nih.gov/pubmed/10498729. Accessed 7 July 2016 [DOI] [PMC free article] [PubMed]
- Revill WP, Bibb MJ, Hopwood DA (1996) Relationships between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J Bacteriol 178:5660–5667. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC178404/. Accessed 9 July 2016 [DOI] [PMC free article] [PubMed]
- van Wezel GP, van der Meulen J, Kawamoto S, Luiten RG, Koerten HK, Kraal B (2000) ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J Bacteriol 182:5653–5662. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC94684/. Accessed 7 July 2016 [DOI] [PMC free article] [PubMed]
- Verma ML, Rajkhowa R, Wang X, Barrow CJ, Puri M. Exploring novel ultrafine Eri silk bioscaffold for enzyme stabilisation in cellobiose hydrolysis. Bioresour Technol. 2013;145:302–306. doi: 10.1016/j.biortech.2013.01.065. [DOI] [PubMed] [Google Scholar]
- Verma ML, Puri M, Barrow CJ. Recent trends in nanomaterials immobilised enzymes for biofuel production. Crit Rev Biotechnol. 2016;36:108–119. doi: 10.3109/07388551.2014.928811. [DOI] [PubMed] [Google Scholar]
- Wang W, et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci USA. 2014;111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Bibb M (1997) bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol 179:627–633. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC178740/. Accessed 5 July 2016 [DOI] [PMC free article] [PubMed]
- Yang YH, et al. Rapid functional screening of Streptomyces coelicolor regulators by use of a pH indicator and application to the MarR-like regulator AbsC. Appl Environ Microbiol. 2010;76:3645–3656. doi: 10.1128/AEM.02617-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Winestrand S, Guo X, Chen L, Hong F, Jonsson LJ. Effects of aromatic compounds on the production of bacterial nanocellulose by Gluconacetobacter xylinus. Microb Cell Fact. 2014;13:1475–2859. doi: 10.1186/1475-2859-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.