Abstract
Neutralization or deletion of tumor necrosis factor (TNF) causes loss of control of intracellular pathogens in mice and humans, but the underlying mechanisms are incompletely understood. Here, we found that TNF antagonized alternative activation of macrophages and dendritic cells by IL-4. TNF inhibited IL-4-induced arginase (Arg) 1 expression by decreasing histone acetylation, without affecting STAT6 phosphorylation and nuclear translocation. In Leishmania major-infected C57BL/6 wild-type mice, type 2 nitric oxide (NO) synthase (NOS2) was detected in inflammatory dendritic cells/macrophages, some of which coexpressed Arg1. In TNF-deficient mice Arg1 was hyperexpressed causing an impaired production of NO in situ. A similar phenotype was seen in L. major-infected BALB/c mice. Arg1 deletion in hematopoietic cells protected these mice from an otherwise lethal disease, although their disease-mediating T cell response (Th2, Treg) was maintained. Thus, deletion or TNF-mediated restriction of Arg1 unleashes the production of NO by NOS2 which is critical for pathogen control.
INTRODUCTION
Myeloid cells such as macrophages play pivotal roles in the immune system. They are essential for the uptake, killing and degradation of pathogens, the processing and presentation of antigens, and the activation of effector T cell populations, but also for the termination of T cell responses, the resolution of inflammatory processes and for tissue homeostasis. The functional diversity of macrophages largely results from exposure to different microenvironmental cues including cytokines and tissue-specific signals (Ginhoux et al., 2015; Lavin et al., 2014). Whereas interferon (IFN)-γ and tumor necrosis factor (TNF) are associated with the induction of ‘classically’ activated or M1-like macrophages expressing antimicrobial effector functions, cytokines such as interleukin (IL)-4, IL-10, IL-13 or transforming growth factor (TGF)-β limit the release of proinflammatory factors by macrophages and promote macrophage phenotypes that suppress T cell responses and/or support tissue repair (Ginhoux et al., 2015). These macrophages, which were operationally dubbed “suppressor” (Kirchner et al., 1975), “deactivated” (Tsunawaki and Nathan, 1986), “alternatively activated” (Stein et al., 1992) or “M2” (Mills et al., 2000) depending on their exact secretory and immunomodulatory function and surface phenotype, are characterized by distinct transcriptional and proteomic profiles (Murray et al., 2014). IL-4, for example, causes upregulation of the mannose receptor 1 (Mrc1; CD206), the chitinase-3-like-protein 3 (Ym1, Chi3l3), the resistin-like molecule alpha (Relma, Retnla; also termed “found in inflammatory zone-1” [Fizz1]), macrophage galactose-type C-type lectin 2 (Mgl2; CD301b), programmed death ligand 2 (Pdl2; CD273) or of arginase 1 (Arg1).
Arg1 is a cytosolic enzyme that hydrolyzes L-arginine into urea and ornithine. It is a constitutive and essential component of the hepatic urea cycle, which explains the postnatal lethality of Arg1−/− mice (Iyer et al., 2002), but inducible in many other cell types, including macrophages, endothelial and epithelial cells (Morris, 2009). In the immune system, arginine metabolism by Arg1 has been conceptually linked to three major processes. First, as ornithine is a precursor of polyamines or proline that are required for cell proliferation or collagen synthesis, respectively, Arg1 expression is characteristic for wound healing and tissue regeneration, whereas excessive Arg1 activity can cause organ fibrosis (Wynn et al., 2013). Second, Arg1 expression by ‘suppressor’ myeloid cells might deprive T cells of arginine and thereby impair their activation and proliferation during anti-infectious or anti-tumor immune responses (Bogdan, 2015; Pesce et al., 2009). Third, Arg1 competes with inducible or type 2 nitric oxide (NO) synthase (iNOS, NOS2), a key antimicrobial and immunoregulatory pathway, for their common substrate L-arginine (Bogdan, 2015; El-Gayar et al., 2003; Rutschman et al., 2001). Accordingly, Arg1 activity correlated with increased pathogen loads in infectious diseases (De Muylder et al., 2013; Iniesta et al., 2005; Kropf et al., 2005). On the other hand, Arg1 was also observed to limit immunopathology and fibrosis (Pesce et al., 2009) and to prevent microbial growth, notably in the absence of NOS2 activity (Duque-Correa et al., 2014).
Due to the diverse and possibly pathological effects of Arg1, its expression requires tight control. In the past, research very much focused on defining factors that upregulate Arg1. Several studies revealed that not only cytokines, but also microbes or microbial products (De Muylder et al., 2013; El Kasmi et al., 2008), hypoxia or lactic acid (Colegio et al., 2014; Louis et al., 1998) induced Arg1. In contrast, the negative regulation of Arg1 in situ during infections has not yet been addressed.
TNF is essential for the defense against intracellular microorganisms and for maintaining life-long control of latent pathogens. Experiments with anti-TNF antibodies, TNF-receptor (R)−/− or Tnf−/− mouse strains as well as treatment of humans suffering from autoimmune diseases with TNF antagonists have demonstrated the protective role of TNF (Allenbach et al., 2008; Flynn et al., 1995; Grivennikov et al., 2005; Marino et al., 1997; Novosad and Winthrop, 2014; Pfeffer et al., 1993; Vassalli, 1992). However, the TNF-mediated molecular mechanisms of pathogen control in vivo are still poorly defined. Tnf−/− C57BL/6 mice locally infected with the protozoan parasite Leishmania (L.) major (FEBNI strain) succumbed to progressive cutaneous and visceral disease (Wilhelm et al., 2001). Unexpectedly, the development of type 1 T helper (Th1) cells and the expression of NOS2, which are strictly required for the resolution of Leishmania infections (Diefenbach et al., 1998; Liew et al., 1990), were completely preserved in Tnf−/− mice (Wilhelm et al., 2001), raising the question of the mechanism underlying the exquisite susceptibility of this mouse strain.
Here, we tested the hypothesis that TNF causes protection by inhibiting the expression of Arg1 and the development of alternatively activated macrophages. We observed a strong negative regulatory effect of TNF on Arg1 expression in macrophages and dendritic cells. In L. major-infected wild-type (WT) mice, Arg1+ myeloid cells represented a subpopulation of monocyte-derived dendritic cells/macrophages that frequently coexpressed NOS2. In Tnf−/− mice, Arg1 was strongly upregulated and impeded the production of NO in situ, a phenotype that was shared by highly susceptible BALB/c mice. Cell type-specific deletion of Arg1 prevented an otherwise lethal course of infection.
RESULTS
TNF inhibits the IL-4-induced expression of Arg1 and other M2 markers
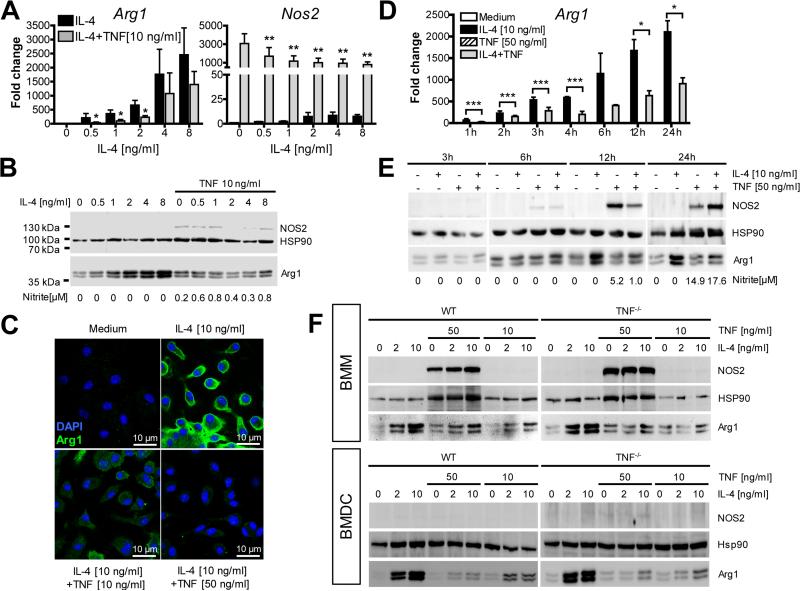
We first tested the effect of TNF on the IL-4-induced Arg1 expression in bone marrow-derived macrophages (BMM) and dendritic cells (BMDC). Simultaneous addition of TNF caused downregulation of Arg1 mRNA and protein in BMM (Fig. 1A and 1C). The suppression was more prominent at higher TNF/IL-4 ratios (Fig. S1A), except for IL-4 concentrations (≤1 ng/ml) that were too low to induce Arg1 protein above control levels (Fig. S1B and 1C). The inhibitory effect of TNF was noticeable once Arg1 mRNA or protein were induced by IL-4 above the level of unstimulated cells (Fig. 1D and 1E). TNF alone induced NOS2 mRNA in BMM and, at high concentrations, also NOS2 protein and activity (Fig. 1A, 1B, 1E, and 1F).
Fig. 1. TNF inhibits the expression of Arg1 in BMM and BMDC.
BMM (A-F) or BMDC (F) of C57BL/6 WT (A-E) or Tnf−/− mice (F) were cultured with or without IL-4 (0.5-10 ng/ml) or TNF (10 or 50 ng/ml) for 1-24 h followed by the determination of NO2− in the supernatants (B, E).
(A, D) RT-qPCR analysis of BMM. Results were calibrated to the medium value and represent means ± SEM of 6-8 (A) or 2-4 (D) experiments. *p < 0.05; **p < 0.01; ***p < 0.005, two-tailed Mann-Whitney U test comparing IL-4 ± TNF.
(B, E, F) Western blot analysis of BMM or BMDC. One of 3 (B), 2 (E) or 4 (F) experiments.
(C) CLSFM of BMM. One of 3 experiments.
See also Fig. S1A-F.
The suppressive activity of TNF on IL-4-triggered Arg1 expression was largely independent of NOS2-derived NO (Fig. S1D), maintained in macrophages that were infected with L. major (Fig. S1E), and also observed with BMDC (Fig. 1F, Fig. S1C) and resident peritoneal macrophages (data not shown). In both BMM and BMDC, the effect was somewhat more pronounced when the cells were derived from Tnf−/− mice (Fig. 1F, Fig. S1C); cultures from Tnf−/− BMDC, however, did not differ from WT BMDC in the total yield of cells, the DC maturation state (expression of CD40, CD80, CD86 and MHC class II) or the expression of TNFR1 (CD120a) and TNFR2 (CD120b) (data not shown).
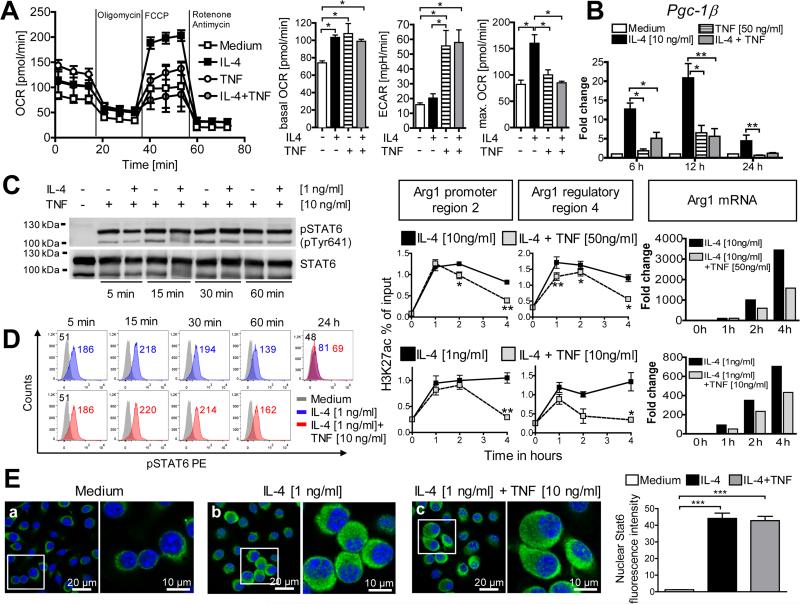
In addition to Arg1, TNF also antagonized the IL-4-mediated mRNA upregulation of the M2 markers Retnla and Ym1, whereas others (Mgl2, Mrc1) were not affected (Fig. S1F). To further corroborate that TNF counteracts the development of M2 macrophages, we analyzed the metabolism of BMM. As expected (Huang et al., 2014; Vats et al., 2006), IL-4 caused a shift towards mitochondrial oxidative phosphorylation via upregulation of the transcriptional PPARγ-coactivator-1β (PGC-1β), leading to a marked rise of the maximal oxygen consumption rate (OCR). TNF-stimulated BMM, in contrast, showed an increased extracellular acidification rate (ECAR) indicating that they rely on aerobic glycolysis. Simultaneous addition of IL-4 and TNF to unbiased BMM prevented the induction of Pgc-1β mRNA by IL-4 and the subsequent metabolic changes (Fig. 2A and 2B).
Fig. 2. TNF suppresses IL-4-induced oxidative phosphorylation and histone acetylation, but does not affect activation and translocation of STAT6.
C57BL/6 BMM were cultured with or without IL-4 ± TNF for 5 min to 24 h.
(A) 24h-cultures of stimulated BMM (10 ng/ml IL-4; 50 ng/ml TNF) were analyzed for basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). In addition, the OCR response to oligomycin (ATP synthase inhibitor), FCCP (uncoupler) or rotenone/antimycin (electron transport inhibitors) was measured. One of 4 experiments (left panel). Bar graph panels show means ± SEM of basal OCR, ECAR and maximal OCR.
(B) RT-qPCR analysis. Means ± SEM calibrated to medium values of 4 experiments.
(C, D) STAT6 phosphorylation monitored by flow cytometry or by STAT6-immunoprecipitation (followed by Western blot of pSTAT6 [pTyr641] and reprobing of total STAT6 protein). In a parallel Western blot, suppression of IL-4-induced Arg1 by TNF was ascertained after 24 h (not shown). One of 5 experiments.
(E) Nuclear translocation of STAT6 protein in BMM after 3h of stimulation, visualized by CLSFM after staining with Alexa Fluor® 488-conjugated anti-STAT6 (green) and DAPI (nuclei, blue). One of 3 experiments. Quantification of nuclear STAT6 fluorescence intensity of 100 cells per condition from 3 experiments (right panel).
(F) H3K27ac-ChIP of fixed BMM. Eluted and quantified DNA was subjected in duplicates to qPCR analysis using primer pairs spanning the Arg1 promoter region 2 or the putative regulatory/enhancer region 4 (see supplemental experimental procedures). Results of 2 experiments are shown as relative enrichment compared to 1% input DNA. Aliquots of the same BMM cultures were analyzed in parallel for Arg1 mRNA by RT-qPCR (right panel).
*p ≤ 0.05; **p ≤ 0.01; ***p < 0.005, ****p < 0.0001, two-tailed Mann-Whitney U test.
see also Fig. 1SG
As the IL-4-induced expression of Arg1 and Pgc-1β strictly depends on Stat6 (Rutschman et al., 2001; Vats et al., 2006), we thought that TNF inhibited the Stat6 pathway. However, TNF neither suppressed the phosphorylation (Fig. 2C and 2D) nor the nuclear translocation of STAT6 (Fig. 2E). Furthermore, TNF did not affect the stability of Arg1 mRNA (data not shown). To explore, whether TNF might act by diminishing the accessibility of transcription factors to the Arg1 promoter and enhancer regions, we performed chromatin immunoprecipitations (ChIP) using an antibody against acetylated lysine 27 of histone H3 (H3K27ac). This histone mark is highly enriched at active cis-regulatory regions (promoters and enhancers) and directly correlates with chromatin accessibility and transcription of the target gene (Creyghton et al., 2010; Rada-Iglesias et al., 2011). In line with previous results (Ostuni et al., 2013), stimulation of BMM with IL-4 triggered a rapid and persistent gain of H3 acetylation at Arg1 promoter and putative enhancer sites (see supplemental information). Co-administration of TNF to IL-4-stimulated cultures markedly reduced the histone acetylation. TNF-mediated inhibition of H3 acetylation was maximal at 4 h and correlated well with the reduction of Arg1 mRNA at the same time-point (Fig. 2F, Fig. S1G).
Thus, TNF counteracts various components of the IL-4-driven activation of myeloid cells. This appears to be at least partly due to an impaired opening of the chromatin as demonstrated for the Arg1 promoter and enhancer elements.
Arg1 and other M2 marker are upregulated in Tnf−/−mice
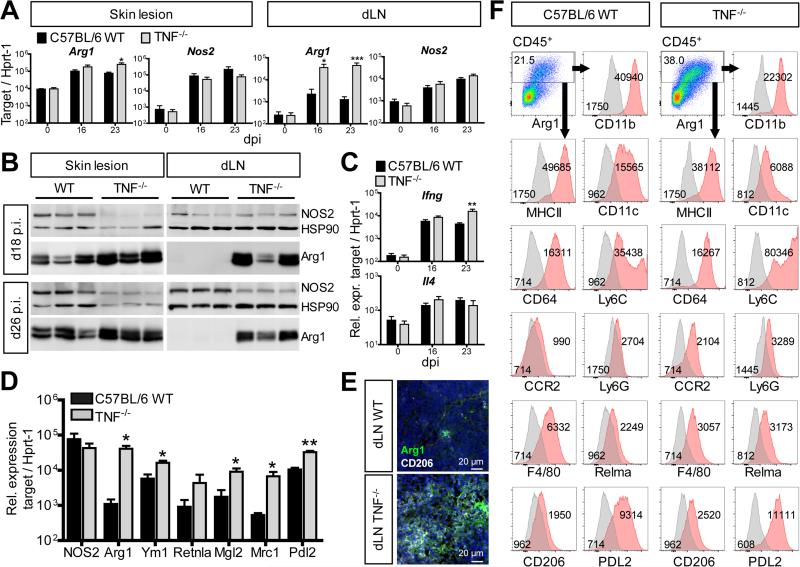
To address whether the TNF-mediated suppression of Arg1 also occurred in vivo, we used the C57BL/6 L. major infection model. The control of L. major FEBNI strain in C57BL/6 mice strictly required NOS2 and TNF (Diefenbach et al., 1998; Wilhelm et al., 2001). Confirming our earlier data (Wilhelm et al., 2001), C57BL/6 Tnf−/− mice infected with L. major showed more severe skin lesions and higher numbers of parasites at the site of infection and in the lymphatic organs compared to C57BL/6 WT controls (Fig. S2A). We observed an upregulation of the mRNA (Fig. 3A), protein and activity of Arg1 (Fig. 3 B; Fig. S2B, S2C, S2D and S2E) in the tissues of L. major-infected Tnf−/− mice. Skin lesions of WT mice contained substantially higher amounts of Arg1 than draining lymph nodes (dLNs). Consequently, the differences in Arg1 expression between Tnf−/− and WT mice were more prominent in dLNs than at the site of infection. Unlike to Arg1, infected WT and Tnf−/− mice had similar amounts of Nos2 mRNA in the skin lesions and dLNs (Fig. 3A), whereas the expression of NOS2 protein in total organ lysates of Tnf−/− mice was tentatively (skin) or significantly (dLN) lower than in WT controls (Fig. 3B, Fig. S2B).
Fig. 3. Arg1 and other M2 markers are upregulated in L. major-infected Tnf−/− mice.
C57BL/6 WT and Tnf−/− mice were infected with L. major.
(A, C) RT-qPCR analysis of skin lesions and dLNs at different days after infection (means ± SEM of 4 experiments with 2-3 mice per group and time point).
(B) Western blot of skin lesions and dLNs. One of 5 experiments with 3-4 mice per group and time point.
(D) RT-qPCR analysis of CD11b+ cells sorted from dLNs (day 21-27 post infection [dpi]) (means ± SEM of 3 experiments with 4 mice per group).
(E) CLSFM of dLN sections (d18 p.i.) stained with anti-Arg1 or anti-CD206 antibodies and DAPI (nucleus, blue). The image is representative of 3-4 mice per group and two experiments.
(F) Flow cytometry of cell suspensions of skin lesions (d23-29 p.i.) from 2-3 mice per group and time point which were pooled prior to staining. Within viable CD45+ cells excluding doublets (see Fig. S3A) Arg1+ cells were gated (red) and analyzed for the expression of surface markers (red: surface marker, grey: isotype or fluorescence minus one [fmo] control; number: mean fluorescence value). One of 4 experiments.
*p < 0.05; **p < 0.01; ***p < 0.005, two-tailed Mann-Whitney U test.
see also Fig. S2
The enhanced expression of Arg1 in L. major-infected Tnf−/− mice is unlikely to result from a cytokine imbalance in this mouse strain, as there was no significant increase of Il4 or a lack of Ifng mRNA in Tnf−/− as compared to WT mice (Fig. 3C). In fact, at later time points the expression of Ifng was even enhanced in infected Tnf−/− mice, demonstrating that Tnf−/− mice were fully capable to mount a Th1 response in response to the increased parasite burden.
In CD11b+ myeloid cells sorted from the dLNs of TNF−/− mice at day 21 to 27 of L. major infection, the expression of Arg1 and of other genes related to alternatively activated macrophages (e.g. Ym1, Mrc1) were significantly upregulated, whereas the amounts of Nos2 mRNA were comparable in Tnf−/− versus WT cells (Fig. 3D). CLSFM analysis confirmed the increased Mrc1 (CD206) expression in Tnf−/− dLNs on the protein level (Fig. 3E).
Together, these data show that TNF, apart from its participation in the induction of NOS2, restricts the in vivo expression of Arg1 and of other IL-4-dependent genes in myeloid cells.
Increased frequency of Arg1+NOS2+ myeloid cells in L. major-infected Tnf−/− mice
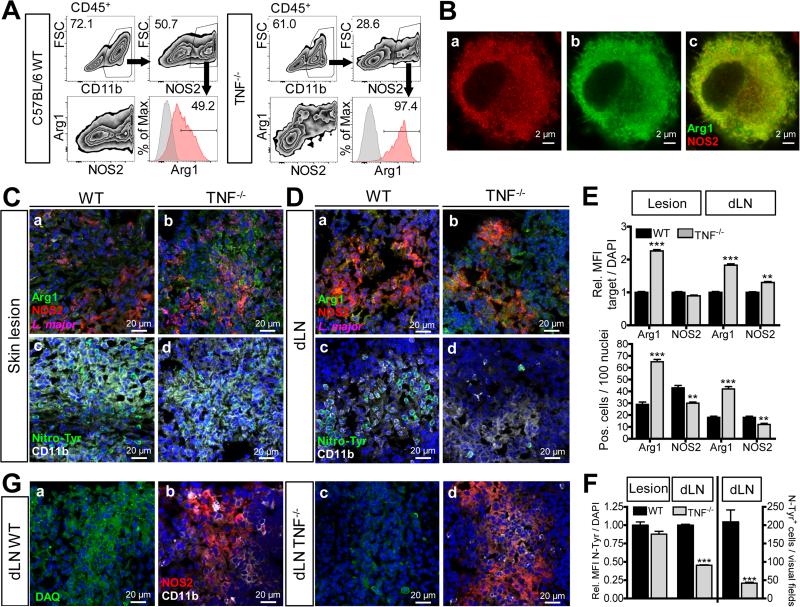
In addition to myeloid cells, Arg1 and NOS2 can also be expressed by endothelial cells and fibroblasts (Morris, 2009; Witte et al., 2002). To define the cell population that accounts for Arg1 expression in C57BL/6 WT and Tnf−/− mice during L. major infection, single cell analyses using multicolour flow cytometry (gating strategy see Fig. S3A) were performed. In the skin lesions of WT mice, Arg1 was detected in CD11b+CD11c+MHCII+ cells that were stained for other surface molecules in different combinations including at least two of the above-mentioned markers. Co-expression was seen for CD64, Ly6C, CCR2, Ly6G, F4/80, Relma, CD206 and PDL2 (Fig. 3F). Importantly, Arg1+ cells were not detected in the CD45− population (Fig. S3B). The specificity of the Arg1 staining was verified by isotype controls and by the absence of Arg1+ cells in Arg1-deficient C57BL/6 mice (Fig. 3F, Fig. S3C). Although we cannot formally exclude the existence of subsets within the Arg1+ cells (as not all the markers were stained in one sample), our data strongly point to one major Arg1+ cell population. The surface phenotype of the Arg1+ cells in the skin lesions identifies them as monocyte-derived inflammatory cells and allows to categorize them either as dendritic cells or as macrophages (Fromm et al., 2012; Gautier et al., 2012; Tamoutounour et al., 2013).
The surface markers of the Arg1+ cells were identical in WT and Tnf−/− mice. However, in line with the mRNA analyses of sorted CD11b+ cells (Fig. 3D), the levels of M2 markers (Relma, CD206, PDL2) were considerably higher in Arg1+ cells from Tnf−/− mice, as was the expression of Ly6C and CCR2, whereas CD11c and F4/80 were reduced on Arg1+ cells from Tnf−/− mice as compared to WT controls (Fig. 3F). Most notably, the overall percentage of Arg1+ cells was 2.14 (± 0.32)-fold greater in Tnf−/− lesions than in WT lesions (mean±SD of 3 experiments) (Fig. 3F), which corroborates our Western blot and CLSFM data. In two Tnf−/− mice we detected the same Arg1+ cells also in dLNs, but the much weaker expression of Arg1 and the limited sensitivity of flow cytometry precluded their routine detection in this organ (data not shown).
Since the size, granularity and surface phenotype of NOS2+ cells (Fig. S3A and S3D) and of Arg1+ cells (Fig. 3F and Fig. S3A) were very similar, we specifically assessed the potential co-expression of Arg1 and NOS2. As depicted in Fig. 4A, the CD11b+ cell population of skin lesions from Tnf−/− mice contained 2.2 (± 0.68) less NOS2+ cells than WT lesions (mean ± SD of 3 experiments). In addition, almost all NOS2+ cells from Tnf−/− mice, but only around 50% from WT mice, co-expressed Arg1, and the mean fluorescence signal for Arg1 was higher in NOS2+ cells from skin lesions of Tnf−/− mice. Overall, the percentage of Arg1+NOS2+ cells was 2.42 (± 0.7)-fold higher in Tnf−/− as compared to WT mice (Fig. 4A). Thus, the preponderance of single-positive NOS2+ cells seen in the myeloid cell population of WT mice is shifted towards NOS2+Arg1high double-positive cells in the absence of TNF.
Fig 4. Increased frequency of Arg1+NOS2+ myeloid cells correlates with decreased detection of nitrotyrosine and NO in Tnf−/− mice.
(A, C-F) C57BL/6 WT and Tnf−/− mice were infected with L. major.
(A) Flow cytometry of skin lesions (d23-29 p.i.). NOS2+ cells within viable CD11b+CD45+ single cells were gated and analyzed for co-expression of Arg1. The percentage of Arg1+NOS2+ cells within the NOS2+ population was determined. One of 3 experiments.
(B) BMM were stimulated with IL-4 (10 ng/ml), IFN-γ (20 ng/ml) plus LPS (200 ng/ml) for 24 h. Co-localization of Arg1 and NOS2 was analyzed by superresolution microscopy. One of 5 experiments.
(C, D) Serial sections from skin lesions (C) or dLNs (D) at d26 p.i. were stained for Arg1 (green), NOS2 (red) and L. major (magenta) (a,b), or nitrotyrosine (green) and CD11b (white) (c,d). Nuclei staining with DAPI (blue). Due to marked differences in signal strength, exposure times had to be reduced for Arg1 (green) in skin sections of Tnf−/− mice. Representative images from one of 4 experiments with 3-4 mice per group and time point (d19-29 p.i.).
(E, F) Quantification of the number and mean fluorescence intensity (MFI) of Arg1- (E), NOS2- (E) or nitrotyrosine (F)-positive cells in tissue sections of WT vs Tnf−/−mice (d19-29 p.i.). The target MFI was normalized to the fluorescence signal of DAPI. The number of positive cells was determined per 100 nuclei. 3-4 visual fields (× 200) per tissue section were evaluated. 17 dLNs and lesions of WT and Tnf−/− mice from 4 experiments were analyzed.
(G) CLSFM of serial sections of dLNs from WT vs. Tnf−/− mice stained for NO using DAQ (green) (a,c) or for NOS2 (red) and CD11b (white) (b,d). Representative images of 4 experiments with 3-4 mice per group and time point (d18-34 p.i.). **p < 0.01; ***p < 0.0001, two-tailed Mann-Whitney U test.
see also Fig. S3 and S4
In activated macrophages NOS2 activity is equally distributed between the cytosol and a vesicular compartment (Bogdan, 2015). We therefore investigated, to which extent NOS2 and Arg1 colocalize when induced in one and the same cell. Superresolution microscopy of NOS2+Arg1+ BMM (stimulated with IL-4 plus IFN-γ/LPS) revealed an overlapping expression of NOS2 and Arg1 in the cytoplasm, whereas the punctate (vesicular) NOS2+ structures remained Arg1-negative (Fig. 4B).
Hyperexpression of Arg1 in Tnf−/− mice impairs the production of NO
Arginine depletion by Arg1 can inhibit NO production (due to substrate competition) or even downregulate NOS2 protein (El-Gayar et al., 2003; Rutschman et al., 2001). As Arg1 and NOS2 were co-expressed within the same cell, we tested whether Arg1 hyperexpression in Tnf−/− mice indeed correlates with a reduced activity of NOS2 in situ.
CLSFM-analysis of skin lesions not only confirmed the presence of NOS2+, Arg1+ or NOS2+Arg1+ myeloid cells, but also the substantially higher amounts of Arg1 (both in terms of cell number and MFI) and the lower expression of NOS2 (cell number) in Tnf−/− mice as compared to WT controls (Fig. 4C, panel a vs. b; Fig. 4E; Fig. S4A, panel a vs. b). Leishmania parasites were preferentially found in Arg1+ foci and much more abundant in Tnf−/− lesions (Fig. 4C, panel a vs. b). dLNs of Tnf−/− mice also contained more Arg1+ cells and parasites than the respective WT organs, whereas the number of NOS2+ cells was slightly reduced in Tnf−/− dLNs (Fig. 4D, panel a vs. b; Fig. 4E; Fig. S4A, panel c vs. d).
NO is a precursor of peroxynitrite that causes tyrosine nitration of proteins and thereby acts as footprint of NOS2 activity in tissues (Bogdan, 2015). Anti-nitrotyrosine staining of skin and dLN sections from L. major-infected WT mice yielded significantly stronger signals (number and MFI of positive cells) than of tissues from Tnf−/− mice (Fig. 4C and 4D, c vs. d; Fig. 4F). Nitrotyrosine+ cells stained positively for CD11b (Fig. 4C and 4D, c vs. d). In serial sections of Tnf−/− dLNs, expression of Arg1 by CD11b+ cells correlated with a lack of nitrotyrosine staining despite the presence of NOS2 protein (Fig. S4B). The nitrotyrosine staining was absent in tissue sections from naive C57BL/6 and infected Nos2−/− mice (Fig. S4C, a vs. b; Fig. S4D) and after blocking of the antiserum with 3-nitro-L-tyrosine (data not shown). To verify that the reduced nitrotyrosine-staining in Tnf−/− mice indeed reflects a lack of locally produced NO, we treated the sections with the NO-reactive compound 1,2-diamino-anthraquinone (DAQ). Clearly, much more NO was detected in infected dLNs of WT mice compared to dLNs of Tnf−/− mice (Fig. 4G; Fig. S4E). Serial sections from infected Nos2−/− mice gave no signal with DAQ despite the presence of CD11b+ cells (Fig. S4C, c vs. d).
Together, these results show that Arg1 hyperexpression in Tnf−/− mice impairs the local production of NO and subsequently the control of Leishmania parasites.
Arg1 expression accounts for the non-healing course of L. major infection
To test whether the increased Arg1 expression in Tnf−/− mice contributed to the detrimental course of L. major infection, we treated L. major-infected C57BL/6 WT or Tnf−/− mice with Nω-hydroxy-nor-L-arginine (nor-NOHA). Nor-NOHA, an Arg inhibitor that is neither tissue- nor isoform-selective (as it also blocks the Arg2 isoform) (Custot et al., 1997), did not impede the arginase of L. major (Kropf et al., 2005) (data not shown), but was reported to ameliorate the severity of L. major skin lesions in BALB/c mice (Iniesta et al., 2005; Kropf et al., 2005). Although in our hands this compound potently blocked Arg1 in macrophages in vitro (El-Gayar et al., 2003), the clinical course of L. major infection and the parasite loads in nor-NOHA-treated C57BL/6 WT or Tnf−/− mice were indistinguishable from the respective controls (data not shown). Furthermore, when testing nor-NOHA in L. major-infected BALB/c mice, neither local nor systemic application led to consistent protective effects (data not shown). We therefore abandoned the pharmacological approach and resorted to a model of cell type-specific deletion of the Arg1 gene using Tie2cre+/−Arg1fl/fl mice (El Kasmi et al., 2008). The Tie2cre deleter is active in hematopoietic and endothelial cells. However, as Arg1 expression in L. major-infected mice was confined to CD45+ cells (Fig. 3SB) identified as myeloid cells (Fig. 3F), the Tie2cre+/−Arg1fl/fl mice are an efficient means of achieving myeloid-specific deletion of Arg1 (Duque-Correa et al., 2014).
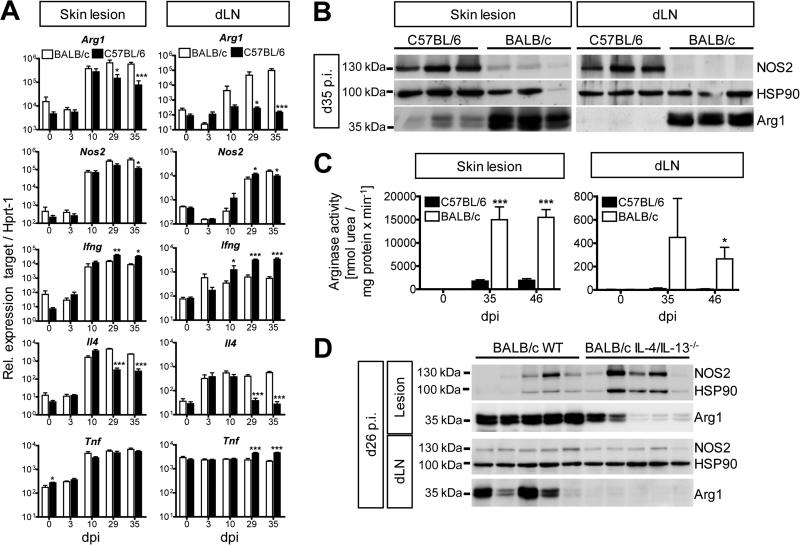
Tie2cre+/−Arg1fl/fl mice were backcrossed to BALB/c. We chose the BALB/c background, because similar to C57BL/6 Tnf−/− mice (Fig. S2A) BALB/c WT mice developed progressive disease after L. major infection (Fig. S5A). The amounts of Arg1 mRNA, protein and activity in the skin lesions (Iniesta et al., 2005; Kropf et al., 2005) and dLNs of L. major-infected BALB/c mice were much higher than in self-healing C57BL/6 mice (Fig. 5A, 5B and 5C; Fig. S5B and S5C). In both strains of mice, Arg1 was more prominent in the skin than in dLNs, which is in line with the differential expression of IL-4 at these two sites (Fig. 5A). Despite comparable quantities of Nos2 mRNA in BALB/c and C57BL/6 mice (Fig. 5A), NOS2 protein was markedly reduced in L. major-infected BALB/c mice (Fig. 5B). The reciprocal expression of Arg1 and NOS2 protein suggests that the translational control of NOS2 by Arg1 activity observed in vitro (El-Gayar et al., 2003) also occurs in vivo.
Fig 5. L. major-infected BALB/c mice express increased levels of Arg1 in the skin lesions and dLNs compared to self-healing C57BL/6 mice.
BALB/c vs. C57BL/6 or BALB/c WT vs Il4/Il13−/− mice were infected with L. major. Skin lesions and dLN tissue were analyzed at the time points indicated.
(A) RT-qPCR analysis (means ± SEM from 2 experiments with a total of 6 mice per group and time point).
(B) Western blot analysis. One of 4 experiments.
(C) Arginase activity in protein lysates of skin lesions and dLNs (means ± SEM from 2-3 experiments with 3 mice per group and time point).
(D) Western blot analysis. One of 2 experiments.
*p < 0.05; **p < 0.01; ***p < 0.005, two-tailed Mann-Whitney U test.
see also Fig. S5 and S6
In BALB/c mice the increased amounts of Arg1 were accompanied by persistent Il4 mRNA and reduced levels of Ifng and Tnf mRNA (Fig. 5A). To test whether the Th2 cytokine milieu and Arg1 hyperexpression were causally linked, we used Il4−/− or IL-4/IL-13-double deficient BALB/c mice. Unlike to BALB/c WT mice, both Il4−/− and Il4/Il13−/− mice contained the infection (Fig. S5D and S5G) and showed a strong reduction (lesions) or abolition (dLNs) of Arg1 mRNA and protein expression (Fig. 5D; Fig. S5E, S5F, S5H, S5I and S5J). The residual amount of Arg1 in the skin lesions of Il4/Il13−/− mice might reflect the activity of other Arg1-inducing cytokines or of microbial ligands. The Arg1+ myeloid cell population in skin lesions of L. major-infected BALB/c WT mice showed the same surface phenotype (i.e. CD11b+CD11c+MHCII+Ly6C+CD64+PDL2+CD207−CCR7−) (Fig. S6) as seen in C57BL/6 WT and Tnf−/− mice.
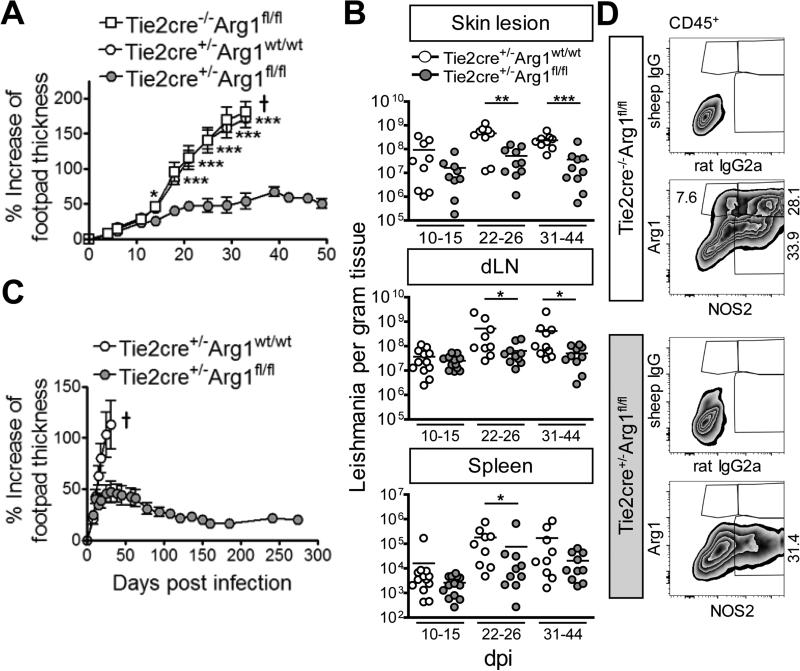
As expected, Arg1-expressing Tie2cre−/−Arg1fl/fl or Tie2cre+/−Arg1wt/wt BALB/c mice developed ulcerative skin lesions and visceral disease following L. major infection. In contrast, the Arg1-deficient Tie2cre+/−Arg1fl/fl littermates presented only small, nonulcerative and non-progressive skin swellings after low (1×104) (data not shown) or high infection inocula (3×106) (Fig. 6A) and showed a significantly reduced parasite burden at the site of infection, in the dLNs and the spleen (Fig. 6B). No spontaneous reactivation of disease in Tie2cre+/−Arg1fl/fl BALB/c mice occurred during the observation period (≤274 days) (Fig. 6C). The efficiency of deletion of Arg1 expression was >99.5% (Fig. 6D; Fig. S7A and S7B).
Fig. 6. Arg1-deficient BALB/c mice are protected from progressive cutaneous leishmaniasis.
Arg1-deficient BALB/c mice (Tie2cre+/−Arg1fl/fl) and control littermates (Tie2cre+/−Arg1wt/wt or Tie2cre−/−Arg1fl/fl) were infected with L. major.
(A, C) Clinical course of infection. 6-9 (A) or 4-6 (C) mice were used per group (means ± SEM). One of 6 (A) or 2 (C) experiments. † Mice had to be euthanized for ethical reasons.
(B) Parasite numbers in the tissues (results represent the mean of 9-12 mice that were analyzed in 3-4 experiments with 3 mice per group and time point).
(D) Flow cytometry of skin lesions (d23 p.i.). Cells were gated on viable single CD45+ cells (see Fig. S3A) and the percentage of Arg1+, NOS2+ or Arg1+NOS2+ cells was determined using the respective isotype controls. One of 2 experiments.
*p < 0.05; **p < 0.01; ***p < 0.005, two-tailed Mann-Whitney U test.
see also Fig. S7A, S7B
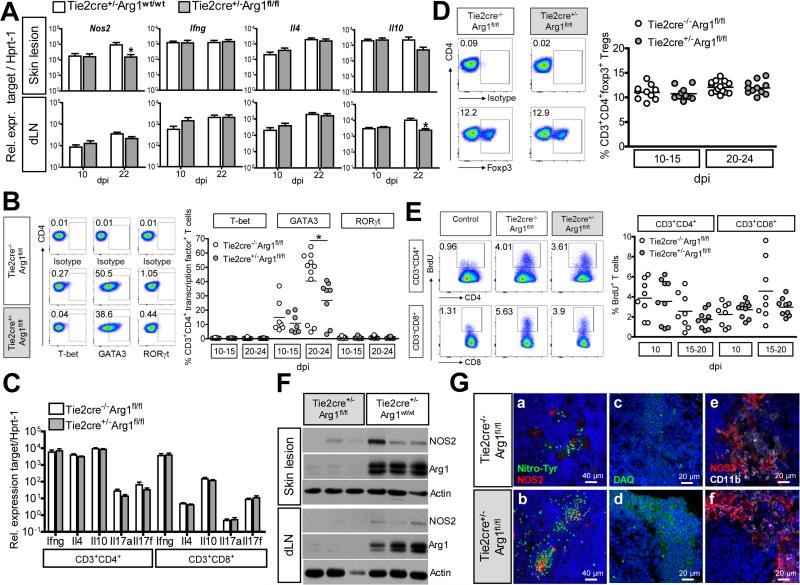
Analyses of infected tissues revealed a slight, but significant reduction of Il10 mRNA in the dLNs from Arg1-deficient Tie2cre+/−Arg1fl/fl BALB/c mice, whereas Ifng and Il4 mRNAs remained unaltered (Fig. 7A). Considering that Arg1 activity can diminish the availability of L-arginine and thereby might affect the T cell differentiation and/or proliferation (Bogdan, 2015; Duque-Correa et al., 2014; Munder et al., 2009; Pesce et al., 2009), we studied the T cell response in L. major-infected Tie2cre−/−Arg1fl/fl BALB/c versus Tie2cre+/−Arg1fl/fl BALB/c mice. First, CD4+CD3+ T cells of dLNs from both mouse strains were analyzed for their expression of the transcription factors T-bet (Th1), GATA3 (Th2) and RORγt (Th17) to evaluate the expansion of Th cell subsets. Whereas T-bet+ or RORγt+ CD4+ T cells were hardly detectable, the high percentage of GATA3+ T cells in both WT and Arg1-deficient dLNs demonstrated a sustained Th2 response (Fig. 7B). Despite a significantly reduced (ca. 30%) frequency of CD4+GATA3+ T cells at later time points of infection in the Arg1-deficient BALB/c mice, the Il4 mRNA expression was neither altered in whole dLNs (Fig. 7A) nor in sorted CD4+CD3+ dLN cells (Fig. 7C). Thus, the net response of Th2 effector cells was not reduced in Arg1-deficient BALB/c mice. Second, the expression of Ifng, Il10 and Il17a or Il17f mRNA was comparable in CD4+ or CD8+ T cells sorted from both groups of mice (Fig. 7C). Third, there was also no difference in the percentage of Foxp3+CD4+ regulatory T cells (Treg) in the dLNs at different time points of infection (Fig. 7D). Finally, when we measured the proliferative capacity of CD4+ or CD8+ T cells in vivo using BrdU incorporation, absence of Arg1 was associated with either an unaltered (two experiments) or a slightly reduced (one experiment), but not with an improved T cell proliferation (Fig. 7E).
Fig. 7. Increased NO production, but unaltered T cell response in Arg1-deficient BALB/c mice.
Arg1-deficient BALB/c mice (Tie2cre+/−Arg1fl/fl) and control littermates (Tie2cre+/−Arg1wt/wt or Tie2cre−/−Arg1fl/fl) were infected with L. major.
(A) RT-qPCR analysis (means ± SEM of 3 experiments with 3-4 mice per group and time point).
(B) dLN cells (stained for CD3, CD4, CD8, and NKp46 and T-bet, GATA3 or RORγt) were gated on viable CD3+CD4+CD8−Nkp46− cells and the percentage of T-bet-, GATA3- or RORγt-positive cells was determined. Representative dot plots and means of 3 experiments with 2-4 mice per group and time point.
(C) RT-qPCR analysis of CD3+CD4+ and CD3+CD8+ T cells sorted from the dLNs of infected mice (3 per group) at d17-22 p.i.. Means ± SEM of 3 experiments.
(D) dLN cells (stained for CD3, CD4, CD8 and Foxp3) were gated on viable CD3+CD4+CD8− T cells and the percentage of Foxp3+ T cells (Tregs) was determined. Representative dot blots and means of 4 experiments with 2-4 mice per group and time point.
(E) dLN cells from BrdU-injected mice were stained for CD3, CD4, CD8 and BrdU. BrdU+ cells (excluding doublets and dead cells) within the CD3+CD4+ or CD3+CD8+ T cell population were gated. Representative dot plots and means of 3 experiments with 3 mice per group and time point. dLN cells from infected WT mice without BrdU treatment were used as controls for the BrdU staining.
(F) Western Blot analysis of skin lesions and dLNs (day 18 p.i.). One of 5 experiments with 3-4 mice per group and time point.
(G) CLSFM analysis of dLNs that were stained for nitrotyrosine (green) and NOS2 (red) (a,b) and in serial sections for NO (c,d; visualized by DAQ [green]) and NOS2 (red) and CD11b (white) (e,f). Nuclei were stained with DAPI (blue). Representative images from 3 experiments with 4 mice per group and time point analyzed at d17-32 p.i.; *p < 0.05; two-tailed Mann-Whitney U test.
see als Fig. S7C and S7D
The deletion of Arg1 did not lead to an upregulation of Nos2 mRNA or protein at the site of infection or in the dLNs; in fact, the RT-qPCR analyses and the Western blot experiments even revealed a slightly diminished Nos2 mRNA and protein expression in Arg1-deficient mice (Fig. 7A and 7F; Fig. S7A). Nevertheless, Arg1-deficiency caused a marked increase of nitrotyrosine-positive cells and of the production of NO (as detected by DAQ) in the skin lesions and dLNs (Fig. 7G; Fig. S7C and D).
Taken together, these data allow to conclude that hyperexpression of Arg1 as seen in non-healing BALB/c or C57BL/6 Tnf−/− mice does not deviate the T cell response against L. major, but markedly impairs the in situ generation of protective NO and thereby interferes with disease control.
DISCUSSION
TNF as regulator of immune responses
TNF is best known for its potent proinflammatory effects resulting from its ability to promote the proliferation or enhance the function of macrophages, dendritic cells, B lymphocytes, T lymphocytes, NK cells and other types of immune cells (Vassalli, 1992). While this immunostimulatory function of TNF confers protection of the host organism against infectious pathogens (Flynn et al., 1995; Grivennikov et al., 2005; Körner et al., 2010; Marino et al., 1997; Novosad and Winthrop, 2014; Pfeffer et al., 1993; Vassalli, 1992), overproduction of TNF is a typical feature of autoimmune and other chronic inflammatory diseases leading to tissue destruction in mice and humans (Atreya et al., 2014; Riminton et al., 1998; Vassalli, 1992). Previous studies showed that TNF can mediate control of intracellular infections by the stimulation of the antimicrobial activity of phagocytes (Bogdan, 2015; Yazdanpanah et al., 2009), the activation of T helper cells or cytotoxic T cells (Vassalli, 1992), the differentiation, functional maturation or survival of myeloid cells (Caux et al., 1992; Fromm et al., 2012; Sade-Feldman et al., 2013), and the generation of non-necrotizing granulomas (Flynn et al., 1995; Marino et al., 1997).
Our present work revealed that TNF downregulated Arg1 in vitro and in vivo and thereby unleashed the enzymatic activity of NOS2 and the production of NO. TNF also antagonized other aspects of macrophage activation by IL-4 including the IL-4-induced oxidative metabolism. TNF did not inhibit the phosphorylation or nuclear translocation of STAT6, but suppressed IL-4-triggered histone acetylation of the Arg1 promoter and putative regulatory/enhancers regions which is likely to impede access or binding of STAT6 to these sites and/or its assembly with other relevant transcription factors (e.g. C/EBPβ) (Pauleau et al., 2004; Sheldon et al., 2013). We currently investigate whether TNF regulates histone acetyl-transferases or deacetylases which are critical for the induction of Arg1 by IL-4 (Serrat et al., 2012). Hyperexpression of Arg1, due to the failure to shut down IL-4 or resulting from the lack of TNF, presumably accounts for the exquisite L. major susceptibility of BALB/c or Tnf−/− mice, respectively, because deletion of Arg1 alone was sufficient to restore local NO production and to prevent non-healing disease in BALB/c mice. Furthermore, we defined inflammatory monocyte-derived dendritic cells/macrophages as the principle source of Arg1 at the infection site. The high rate of intracellular co-expression of Arg1 and NOS2 seen by high resolution microscopy in IL-4/IFN-γ/LPS-stimulated macrophages in vitro and in infected Tnf−/− mice certainly facilitated the ability of Arg1 to effectively impair the production of NO in situ.
Earlier studies reported an upregulation of Arg1 by TNF in endothelial cells in a model of coronary ischemia and reperfusion injury (Gao et al., 2007) or in myeloid suppressor cells during chronic inflammation elicited by repeated s.c. injections of heat-killed Mycobacterium bovis BCG (Sade-Feldman et al., 2013). While these experimental settings were entirely different from the infection model used here, our results do not exclude a divergent role of TNF during infections with other pathogens or in other organs.
Arginase and pathogen control
Arg1 expression might impair antimicrobial defense in vivo by three different mechanisms (Bogdan, 2015): (1) Arg1 can deplete myeloid cells of L-arginine thereby impeding the generation of NO by NOS2; (2) Arg1 of myeloid cells is capable to deprive T lymphocytes of L-arginine, which was reported to decrease their proliferation, survival and/or expression of costimulatory molecules in vitro; and (3) Arg1 activity leads to the generation of ornithine and via the ornithine decarboxylase (ODC) pathway to the synthesis of polyamines, which are essential for the growth of various parasites including Leishmania (Colotti and Ilari, 2010). However, as L. major expresses its own arginase and ODC (Reguera et al., 2009), its survival is unlikely to be affected by exogenous polyamines derived from host cells. With respect to the second possible mechanism of Arg1 action, we avoided the use of in vitro culture systems which are prone to distort the true proliferative capacity of T cells due to the high L-arginine content of the culture media. Instead, we studied the differentiation and proliferation of CD4+ or CD8+ T cells in vivo using Tie2cre−/−Arg1fl/fl or Tie2cre+/−Arg1fl/fl mice. These analyses revealed an unaltered proliferation of T cells in situ without any apparent shift in the T helper cell differentiation, the abundance of Treg or the expression of Th cell cytokines, irrespective of the presence or absence of Arg1. In contrast, the generation of NO by NOS2 was severely impaired when Arg1 was hyperexpressed. Thus, our data clearly show that the counterprotective function Arg1 primarily results from its competition with NOS2.
TNF and leishmaniasis
Previous studies emphasized the role of TNF as a coactivator of macrophages for the expression of NOS2 activity. In vitro, exogenous or endogenous TNF clearly synergized with IFN-γ for the production of NO by inflammatory macrophages (Wilhelm et al., 2001) and for the killing of intracellular Leishmania parasites (Green et al., 1990; Liew et al., 1990). At least in BMM, TNF might induce NOS2 even on its own as shown in the present study (Fig. 1A, F). So far, it has been assumed that the remarkable susceptibility to L. major seen in mice lacking TNF (Allenbach et al., 2008; Titus et al., 1989; Wilhelm et al., 2001) results from an impaired macrophage activation. However, TNFR1/TNFR2 double-deficient mice (Nashleanas et al., 1998) and Tnf−/− mice (Wilhelm et al., 2001) maintained the expression of Nos2 mRNA and protein in the dLNs, and L. major-infected macrophages from these mice were still capable to express NOS2 and to kill intracellular parasites in response to IFN-γ (Nashleanas et al., 1998). These findings already indicated that the synergism with IFN-γ might not be the only or leading mechanism of action of TNF in vivo.
Our study demonstrates that TNF suppresses the IL-4-dependent expression of Arg1 in myeloid cells and thereby generates a micromilieu that allows for enhanced generation of NO by NOS2 in situ and improved parasite control. It will be of interest to assess whether TNF functions in the same way in patients with autoimmune diseases, who have an increased risk to develop severe infections with Leishmania or other intracellular pathogens following anti-TNF treatment (Novosad and Winthrop, 2014; Tung Chen et al., 2014). The recent observations that TNF-signaling antagonized M2 gene expression in spinal cord injury (Kroner et al., 2014) and in solid tumors (Kratochvill et al., 2015) and that two other TNF superfamily members (BAFF and APRIL) also caused downregulation of M2 markers (Allman et al., 2015), suggest that the suppression of Arg1 by TNF and TNF-related molecules forms a general paradigm in immunology. Considering that Arg1-expressing myeloid cells contribute to the pathogenesis of numerous diseases ranging from chronic infections to malignant tumors and progressive organ fibrosis, our findings have implications for novel treatment strategies and call for caution when using TNF-neutralizing agents.
EXPERIMENTAL PROCEDURES
Mouse strains
C57BL/6 or BALB/c WT or transgenic mice (C57BL/6 Tnf−/−, C57BL/6 Nos2−/−, BALB/c Il4−/−, BALB/c Il4/Il13−/−, conditional Arg1-deficient BALB/c or C57BL/6 mice [Tie2Cre+/−Arg1fl/fl vs. Tie2Cre+/−Arg1wt/wt or Tie2Cre−/−Argfl/fl]) were used at 8-14 weeks of age. Infections were performed with female mice and the respective age-matched WT or littermate controls.
Infection and in vivo treatment of mice
Mice were infected bilaterally into the skin of the hind foot with 3×106 stationary-phase L. major promastigotes (strain MHOM/IL/81/FEBNI) in 50μl PBS. The clinical monitoring of the infection by measuring the footpad thickness, the determination of the parasite burden by limiting dilution analyses and the applications of drugs are described in the supplemental experimental procedures.
Cell culture and measurements of cellular activities
BMDC and BMM were generated from bone marrow cells (Prajeeth et al., 2011) and used at day 7 or 8 to 10 of culture, respectively. rmIL-4 (0.5-10 ng/ml) and rmTNF (10 or 50 ng/ml) (R&D Systems) were added as stimulant(s). At the concentrations used, the LPS content of all reagents and the supplemented media was ≤10 pg/ml (colorimetric Limulus amebocyte lysate assay, Whittaker M.A. Bioproducts, Walkersville, MD).
The composition of the culture media and the determination of nitrite, arginase activity and energy metabolism (oxygen consumption rate [OCR], extracellular acidification rate [ECAR]) are described in the supplemental experimental procedures.
Quantitative PCR
Total RNA was extracted from homogenized tissue or cultured cells using the TRIfast reagent (Peqlab). 1-5 μg RNA were reverse transcribed using the High Capacity cDNA Archive Kit (ThermoFisher Scientific). qPCR was performed on ABI7900HT Fast Real Time PCR system (Applied Biosystems™; ThermoFisher Scientific). For the gene-specific assays used see the supplemental experimental procedures. mRNA levels were calculated by the following formula: relative expression = 2-(CT(Target)-CT(Endogenous control)) × f, with f = 104 as an arbitrary factor. In some of the experiments relative expression was calibrated to controls as indicated in the legends.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described (Garber et al., 2012; Ostuni et al., 2013). Briefly, nuclear extracts obtained from 12×106 fixed BMM were sheared by sonication and incubated overnight at 4°C with protein G Dynabeads (Invitrogen) coupled with 2.5 μg of anti-H3K27ac antibody (ab4729, Abcam). After ChIP, beads were magnetically recovered, washed, and DNA was eluted and decrosslinked overnight at 65°C. DNA was then purified with solid-phase reversible immobilization beads (Agencourt AMPure XP, Beckman Coulter) and quantified with PicoGreen (Invitrogen). For ChIP-qPCR experiments, 1 μl of purified DNA (IP and 1% input) was used for amplification on an ABI7500 machine. Primers are listed in the supplemental experimental procedures.
Flow cytometry
Western blot analysis and immunoprecipitations
Immunohistology and microscopy
Statistical analyses
Results are displayed as mean +/− SEM and were statistically analyzed using GraphPad Prism version 5 or 6 as detailed in the figure legends.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Claudia Kurzmann, Rosa Mammato and Heidi Sebald for excellent technical assistance during the starting phase of this project, to Katharina Pracht for participating in pilot experiments, and to Ralf Willebrand, Christian Schwartz and Manfred Lutz for advice. We also thank Tristan Nowak and Ralf Palmisano from the Optimal Imaging Center Erlangen (OICE) of the FAU for help with the STED microscopy, the operators of the Core Facility for Cell-Sorting and Immunomonitoring, and the personnel of the Franz-Penzoldt Animal Center Erlangen of the FAU for animal care. This study was supported by grants to C.B., U.S. and D.D. from the Deutsche Forschungsgemeinschaft (SFB620, A9; GRK1660, A5; SFB643, A6, A7; SFB1181, C4, A7), to C.B. and U.S. from the Interdisciplinary Center for Clinical Research of the FAU (A24, A61), to C.B. from the Emerging Field Initiative of the FAU (consortium “Medicinal Inorganic Redox Chemistry”) and from the Dr. Robert Pfleger-Stiftung, to D.D. from the Bavarian Genome Network (BayGene), to P.M. from the National Institutes of Health (CA189990, Cancer Center Core Grant P30 CA21765) and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital, and to H.K. from the National Health and Medical Research Council of Australia (NHMRC 485807).
Abbreviations
- Arg1
arginase 1
- BMM
bone marrow-derived macrophages
- BMDC
bone marrow-derived dendritic cells
- CLSFM
confocal laser scanning fluorescence microscopy
- dLN
draining lymph node
- dpi
days post infection
- iNOS or NOS2
inducible or type 2 nitric oxide (NO) synthase
- OCR
oxygen consumption rate
- ECAR
extracellular acidification rate
- Treg
regulatory T lymphocytes
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHORS’ CONTRIBUTIONS
Conceived and designed the experiments: U.S., H.K., R.O. and C.B.
Performed the experiments: U.S., K.P., A.D., R.O., S.O., D.M., T.K., J.K., and C.B.
Analyzed the data: U.S., K.P., A.D., T.K., J.K., D.M., H.K. and C.B.
Contributed reagents, materials, mouse strains, technical know-how and ideas: E.R., D.D., P.M.
Wrote the paper: C.B., U.S., K.P., P.M., and H.K.
References
- Allenbach C, Launois P, Mueller C, Tacchini-Cottier F. An essential role for transmembrane TNF in the resolution of the inflammatory lesion induced by Leishmania major infection. Eur. J. Immunol. 2008;38:720–731. doi: 10.1002/eji.200737662. [DOI] [PubMed] [Google Scholar]
- Allman WR, Dey R, Liu L, Siddiqui S, Coleman AS, Bhattacharya P, Yano M, Uslu K, Takeda K, Nakhasi HL, Akkoyunlu M. TACI deficiency leads to alternatively activated macrophage phenotype and susceptibility to Leishmania infection. Proc. Natl .Acad. Sci. U S A. 2015;112:E4094–4103. doi: 10.1073/pnas.1421580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya R, Neumann H, Neufert C, Waldner MJ, Billmeier U, Zopf Y, Willma M, App C, Munster T, Kessler H, et al. In vivo imaging using fluorescent antibodies to tumor necrosis factor predicts therapeutic response in Crohn's disease. Nat. Med. 2014;20:313–318. doi: 10.1038/nm.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotti G, Ilari A. Polyamine metabolism in Leishmania: from arginine to trypanothione. Amino Acids. 2010;40:269–285. doi: 10.1007/s00726-010-0630-3. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custot J, Moali C, Brollo M, Boucher JL, Delaforge M, Mansuy D, Tenu JP, Zimmermann JL. The new α-amino acid Nω-hydroxy-nor-L-arginine: a high-affinity inhibitor of arginase well adapted to bind to its manganese cluster. J. Am. Chem. Soc. 1997;119:4086–4087. [Google Scholar]
- De Muylder G, Daulouede S, Lecordier L, Uzureau P, Morias Y, Van Den Abbeele J, Caljon G, Herin M, Holzmuller P, Semballa S, et al. A Trypanosoma brucei kinesin heavy chain promotes parasite growth by triggering host arginase activity. PLoS Pathog. 2013;9:e1003731. doi: 10.1371/journal.ppat.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A, Schindler H, Donhauser N, Lorenz E, Laskay T, MacMicking J, Röllinghoff M, Gresser I, Bogdan C. Type 1 interferon (IFN-α/β) and type 2 nitric oxide synthase regulate the innate immune response to a protozoan parasite. Immunity. 1998;8:77–87. doi: 10.1016/s1074-7613(00)80460-4. [DOI] [PubMed] [Google Scholar]
- Duque-Correa MA, Kuhl AA, Rodriguez PC, Zedler U, Schommer-Leitner S, Rao M, Weiner J, 3rd, Hurwitz R, Qualls JE, Kosmiadi GA, et al. Macrophage arginase-1 controls bacterial growth and pathology in hypoxic tuberculosis granulomas. Proc. Natl. Acad. Sci. U S A. 2014;111:E4024–4032. doi: 10.1073/pnas.1408839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gayar S, Thüring-Nahler H, Pfeilschifter J, Röllinghoff M, Bogdan C. Translational control of inducible nitric oxide synthase by IL-13 and arginine availability in inflammatory macrophages. J. Immunol. 2003;171:4561–4568. doi: 10.4049/jimmunol.171.9.4561. [DOI] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- Fromm PD, Kling J, Mack M, Sedgwick JD, Korner H. Loss of TNF signaling facilitates the development of a novel Ly-6C(low) macrophage population permissive for Leishmania major infection. J. Immunol. 2012;188:6258–6266. doi: 10.4049/jimmunol.1100977. [DOI] [PubMed] [Google Scholar]
- Gao X, Xu X, Belmadani S, Park Y, Tang Z, Feldman AM, Chilian WM, Zhang C. TNF-alpha contributes to endothelial dysfunction by upregulating arginase in ischemia/reperfusion injury. Arterioscler. Thromb. Vasc. Biol. 2007;27:1269–1275. doi: 10.1161/ATVBAHA.107.142521. [DOI] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2015;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- Green SJ, Crawford RM, Hockmeyer JT, Meltzer MS, Nacy CN. Leishmania major amastigotes initiate the L-arginine-dependent killing mechanism in IFN-γ stimulated macrophages by induction of tumor necrosis factor-α. J. Immunol. 1990;145:4290–4297. [PubMed] [Google Scholar]
- Grivennikov SI, Tumanov AV, Liepinsh DJ, Kruglov AA, Marakusha BI, Shakhov AN, Murakami T, Drutskaya LN, Forster I, Clausen BE, et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity. 2005;22:93–104. doi: 10.1016/j.immuni.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O'Neill CM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta V, Carcelen J, Molano I, Peixoto PMV, Redondo E, Parra P, Mangas M, Monroy I, Campo ML, Nieto CG, Corraliza IM. Arginase I induction during Leishmania major infection mediates the development of disease. Infect. Immun. 2005;73:6085–6090. doi: 10.1128/IAI.73.9.6085-6090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RK, Yoo PK, Kern RM, Rozengurt N, Tsoa R, O'Brien WE, Yu H, Grody WW, Cederbaum SD. Mouse model for human arginase deficiency. Mol. Cell. Biol. 2002;22:4491–4498. doi: 10.1128/MCB.22.13.4491-4498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner H, Holden HT, Herberman Splenic suppressor macrophages induced in mice by injection of Corynebacterium parvum. J. Immunol. 1975;115:1212–1216. [PubMed] [Google Scholar]
- Körner H, McMorran B, Schluter D, Fromm P. The role of TNF in parasitic diseases: still more questions than answers. Int. J. Parasitol. 2010;40:879–888. doi: 10.1016/j.ijpara.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Kratochvill F, Neale G, Haverkamp JM, Van de Velde LA, Smith AM, Kawauchi D, McEvoy J, Roussel MF, Dyer MA, Qualls JE, Murray PJ. TNF Counterbalances the Emergence of M2 Tumor Macrophages. Cell Rep. 2015;12:1902–1914. doi: 10.1016/j.celrep.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner A, Greenhalgh AD, Zarruk JG, Passos Dos Santos R, Gaestel M, David S. TNF and increased intracellular iron alter macrophage polarization to a detrimental M1 phenotype in the injured spinal cord. Neuron. 2014;83:1098–1116. doi: 10.1016/j.neuron.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Kropf P, Fuentes JM, Fahnrich E, Arpa L, Herath S, Weber V, Soler G, Celada A, Modolell M, Müller I. Arginase and polyamine synthesis are key factors in the regulation of experimental leishmaniasis in vivo. FASEB J. 2005;19:1000–1002. doi: 10.1096/fj.04-3416fje. [DOI] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Li Y, Millott S. Tumor necrosis factor-α synergizes with IFN-γ in mediating killing of Leishmania major through the induction of nitric oxide. J. Immunol. 1990;145:4306–4310. [PubMed] [Google Scholar]
- Louis CA, Reichner JS, Henry WL, Jr., Mastrofrancesco B, Gotoh T, Mori M, Albina JE. Distinct arginase isoforms expressed in primary and transformed macrophages: regulation by oxygen tension. Am. J. Physiol. 1998;274:R775–782. doi: 10.1152/ajpregu.1998.274.3.R775. [DOI] [PubMed] [Google Scholar]
- Marino MW, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, et al. Characterization of tumor necrosis factor-deficient mice. Proc. Natl. Acad. Sci. U S A. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- Morris SM., Jr. Recent advances in arginine metabolism: roles and regulation of the arginases. B.r J. Pharmacol. 2009;157:922–930. doi: 10.1111/j.1476-5381.2009.00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder M, Choi BS, Rogers M, Kropf P. L-arginine deprivation impairs Leishmania major-specific T-cell responses. Eur. J. Immunol. 2009;39:2161–2172. doi: 10.1002/eji.200839041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashleanas M, Kanaly S, Scott P. Control of Leishmania major infection in mice lacking TNF receptors. J. Immunol. 1998;160:5506–5513. [PubMed] [Google Scholar]
- Novosad SA, Winthrop KL. Beyond tumor necrosis factor inhibition: the expanding pipeline of biologic therapies for inflammatory diseases and their associated infectious sequelae. Clin. Infect. Dis. 2014;58:1587–1598. doi: 10.1093/cid/ciu104. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ. Enhancer-mediated control of macrophage-specific arginase I expression. J. Immunol. 2004;172:7565–7573. doi: 10.4049/jimmunol.172.12.7565. [DOI] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer K, Matsuyama T, Kundig TM, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi PS, Kronke M, Mak TW. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- Prajeeth CK, Haeberlein S, Sebald H, Schleicher U, Bogdan C. Leishmania-infected macrophages are targets of NK cell-derived cytokines, but not of NK cell cytotoxicity. Infect. Immun. 2011;79:2699–2708. doi: 10.1128/IAI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera RM, Balana-Fouce R, Showalter M, Hickerson S, Beverley SM. Leishmania major lacking arginase (ARG) are auxotrophic for polyamines but retain infectivity to susceptible BALB/c mice. Mol. Biochem. Parasitol. 2009;165:48–56. doi: 10.1016/j.molbiopara.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riminton DS, Korner H, Strickland DH, Lemckert FA, Pollard JD, Sedgwick JD. Challenging cytokine redundancy: inflammatory cell movement and clinical course of experimental autoimmune encephalomyelitis are normal in lymphotoxin-deficient, but not tumor necrosis factor-deficient, mice. J. Exp. Med. 1998;187:1517–1528. doi: 10.1084/jem.187.9.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Stat6-dependent substrate depletion regulates nitric oxide production. J. Immunol. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- Sade-Feldman M, Kanterman J, Ish-Shalom E, Elnekave M, Horwitz E, Baniyash M. Tumor necrosis factor-alpha blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity. 2013;38:541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Serrat N, Pereira-Lopes S, Comalada M, Lloberas J, Celada A. Deacetylation of C/EBPbeta is required for IL-4-induced arginase-1 expression in murine macrophages. Eur. J. Immunol. 2012;42:3028–3037. doi: 10.1002/eji.201242413. [DOI] [PubMed] [Google Scholar]
- Sheldon KE, Shandilya H, Kepka-Lenhart D, Poljakovic M, Ghosh A, Morris SM., Jr. Shaping the murine macrophage phenotype: IL-4 and cyclic AMP synergistically activate the arginase I promoter. J. Immunol. 2013;191:2290–2298. doi: 10.4049/jimmunol.1202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. IL-4 potently enhances murine macrophage receptor activity, a marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Titus RG, Sherry B, Cerami A. Tumor necrosis factor plays a protective role in experimental murine cutaneous leishmaniasis. J. Exp. Med. 1989;170:2097–2104. doi: 10.1084/jem.170.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunawaki S, Nathan CF. Macrophage deactivation. Altered kinetic properties of superoxide-producing enzyme after exposure to tumor cell-conditioned medium. J. Exp. Med. 1986;164:1319–1331. doi: 10.1084/jem.164.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Chen Y, Perales C, Lacruz J, Senent L, Salavert M. Visceral leishmaniasis infection during adalimumab therapy: a case report and literature review. Int. J. Rheum. Dis. 2014;17:822–824. doi: 10.1111/1756-185X.12427. [DOI] [PubMed] [Google Scholar]
- Vassalli P. The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm P, Ritter U, Labbow S, Donhauser N, Röllinghoff M, Bogdan C, Körner H. Rapidly fatal leishmaniasis in resistant C57BL/6 mice lacking tumor necrosis factor. J. Immunol. 2001;166:4012–4019. doi: 10.4049/jimmunol.166.6.4012. [DOI] [PubMed] [Google Scholar]
- Witte MB, Barbul A, Schick MA, Vogt N, Becker HD. Upregulation of arginase expression in wound-derived fibroblasts. J. Surg. Res. 2002;105:35–42. doi: 10.1006/jsre.2002.6443. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanah B, Wiegmann K, Tchikov V, Krut O, Pongratz C, Schramm M, Kleinridders A, Wunderlich T, Kashkar H, Utermohlen O, et al. Riboflavin kinase couples TNF receptor 1 to NADPH oxidase. Nature. 2009;460:1159–1163. doi: 10.1038/nature08206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.