Abstract
KIR3DL1 is a Natural Killer (NK) cell receptor that recognizes the Bw4 epitope of HLA class I molecules. Following hematopoietic stem cell transplantation for patients lacking Bw4, KIR3DL1-expressing NK cells from Bw4-positive donors can be alloreactive and eliminate tumour cells. However, KIR3DL1 alleles having T instead of C at nucleotide 320 (encoding leucine 86 instead of serine 86) are not expressed on the cell-surface. Thus, not all individuals testing positive for KIR3DL1 are optimal donors for Bw4-negative recipients. Therefore we developed a method for genotyping codon 86, which was validated by its perfect correlation with NK cell phenotype for 100 donors of diverse KIR3DL1/S1 genotype. We typed 600 donors and found that ~12.2% had the KIR3DL1 gene, but did not express cell-surface KIR3DL1. By contrast, high-expressing allotypes were identified when haplotypes from four families with duplicated KIR3DL1/S1 genes were characterized at high resolution. Identifying donors who have KIR3DL1 but lack cell-surface KIR3DL1 would refine donor selection. With this technique, the number of individuals identified who may not be optimal donors for Bw4 negative patients increases by three-fold, when compared with standard methods. Taken together, we propose that allele typing of KIR polymorphisms should become a standard practice when selecting donors.
Keywords: Killer cell Ig-like receptor (KIR), Natural killer (NK) cell, HLA haploidentical hematopoietic stem cell transplantation, Donor selection, Next-generation sequencing
Graphical Abstract
Killer immunoglobulin-like receptors (KIR) genotyping identifies hematopoietic stem cell donors having NK cells potentially alloreactive towards patient leukemia. KIR3DL1 binds HLA molecules expressing the Bw4 ligand, but some KIR3DL1 alleles are not expressed. Donor selection using PCR and sequencing identifies individuals who could become optimum donors for Bw4 negative patients.
INTRODUCTION
Hematopoietic stem cell transplantation (HSCT) is a very successful strategy for the treatment of hematological malignancies [1, 2]. Donors are commonly matched for only one of the patient’s HLA haplotypes (Haplo-HSCT), which creates the potential for alloreactivity. In this context, successful clinical outcome is correlated with both the generation and persistence of donor-derived alloreactive natural killer (NK) cells during post-transplant hematopoietic reconstitution [2]. This is because donor NK cells can eliminate residual leukaemia cells, and thus reduce the incidence of tumour relapse. Donor NK cells also remove host dendritic cells (DCs) and T cells, reducing graft-versus-host disease and graft rejection, respectively [3].
NK cells are cytotoxic lymphocytes that can kill cancer and infected cells and also release cytokines that promote inflammation. NK cell activity is regulated through the integration of signals from multiple cell-surface receptors, which promote both self-tolerance and cytotoxicity against diseased cells [4]. The most polymorphic NK receptor family in humans is the Killer Ig-like Receptor (KIR) family, comprising activating and inhibitory receptors [5]. Inhibitory KIRs are expressed during NK cell maturation and their interaction with HLA class I is crucial for NK cell education (also described as licensing). This process leads to the generation of fully competent, self-tolerant NK cells that express at least one inhibitory receptor for HLA class I molecules [6, 7]. Inhibitory KIRs are clonally distributed and the result is that NK cell subsets can differentially recognize and kill autologous cells that selectively down-regulate expression of HLA class I, through a mechanism of “missing self recognition” [8, 9]. KIR3DL1 is a receptor specific for HLA-A and HLA-B allotypes expressing the Bw4 epitope [10, 11]. KIR3DL1 has three Ig-like domains, all involved directly in the interaction with peptide-presenting HLA class I molecules [12], and a long cytoplasmic tail bearing two Immunoreceptor Tyrosine-based Inhibitory Motifs (ITIMs) [4].
Following Haplo-HSCT, a KIR-ligand mismatch in the graft-versus-host direction occurs when donor-derived NK cells express inhibitory KIRs that recognise ligands present in the donor but not the recipient [13, 14]. Grafts that have been enriched for CD34+ hematopoietic precursor cells prior to transplantation retain this activity [2]. This is likely because NK cells can become licensed in the recipient through interaction with HLA class I expressing cells originating from the donor [15, 16]. Nevertheless, the existence and the size of alloreactive NK cell subsets in the cells transplanted from donor to recipient are important criteria in the selection of optimal Haplo-HSCT donors [1]. For Bw4-negative patients, alloreactive donors can be selected based on the presence of educated NK cells that express KIR3DL1 [13, 17]. Because KIR3DL1 alleles have different levels of cell-surface expression [18, 19], phenotype analysis by flow cytometry has also been used to identify suitable donors for Bw4-negative patients [17]. However, in some cases (e.g. cord blood transplantation) there is insufficient material for an assay of NK cell activity or KIR phenotype and the donor selection must be performed based solely on the results of KIR genotyping. It therefore becomes imperative to develop genetic tests for accurate donor selection.
The KIR region is located on chr19q13.4 and varies in both gene and allele content [20, 21]. Inhibitory KIR3DL1 and its activating counterpart KIR3DS1 segregate as alleles of the same gene (KIR3DL1/S1), which has three phylogenetic clades of alleles [22]. These clades encode three types of receptor, the inhibitory KIR3DL1*005 and KIR3DL1*015 (with low and high expression levels, respectively) and activating KIR3DS1. KIR3DS1 has little polymorphism, with KIR3DS1*01301 being the dominant allele in all populations [22]. By contrast, the two KIR3DL1 clades are highly polymorphic, having been subjected to many events of point mutation and recombination [23].
Of particular interest here are eight KIR3DL1 alleles (KIR3DL1*004, *019, *021, *036, *037, *039, *056 and *072). These alleles are part of the KIR3DL1*005 clade and all encode KIR3DL1 allotypes that have a leucine residue at position 86 in the D0 domain [19, 24]. Other KIR3DL1 alleles (the majority) encode a serine residue at position 86. The substitution of leucine for serine is due to substitution of T for C at nucleotide 320 in exon 3 encoding the D0 domain, and is frequent in European populations. Leucine 86 disrupts a WSAPS motif that ensures the correct folding of Ig-like domains. Consequently, KIR3DL1 allotypes with leucine 86 are retained in the endoplasmic reticulum and do not reach the plasma membrane [19]. This means that NK cells transcribing only KIR3DL1 alleles encoding leucine 86 do not display Bw4 alloreactivity having the same phenotype as NK cells lacking a KIR3DL1 allele.
In this study, we developed a strategy to analyse and distinguish KIR3DL1 polymorphism at codon 86, thereby predicting the presence or absence of surface KIR3DL1 on the NK cells of prospective transplant donors. This method improves the identification of HSCT donors who have the potential for alloreactivity in Bw4 negative recipients.
RESULTS AND DISCUSSION
Codon 86 genotype predicts surface expression of KIR3DL1 by NK cells
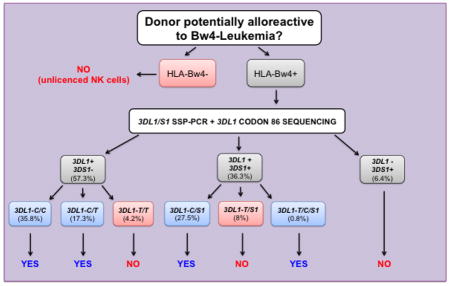
Using our current donor selection strategy, donors, with the potential for alloreactive NK cells that attack Bw4-negative leukemic cells, are selected using three sets of PCR primers that identify KIR3DL1 (3DL1), KIR3DS1 (3DS1) and KIR3DL1 alleles with thymidine at position 320 (3DL1-T), respectively. The 3DL1-T alleles encode leucine at residue 86, which prevents proper folding and cell-surface expression of the KIR [19]. Due to the structural complexity of the KIR locus, a specific PCR for the functional 3DL1 alleles that have cytosine (C) at position 320 (3DL1-C) has not been achieved. Consequently, 3DL1 homozygous donors testing positive for 3DL1-T can be either homozygous for 3DL1-T or heterozygous for 3DL1-T and 3DL1-C. Importantly, 3DL1-T/3DL1-C heterozygotes have potential for NK cell mediated alloreactivity towards Bw4-negative patients, whereas the homozygous 3DL1-T donors do not.
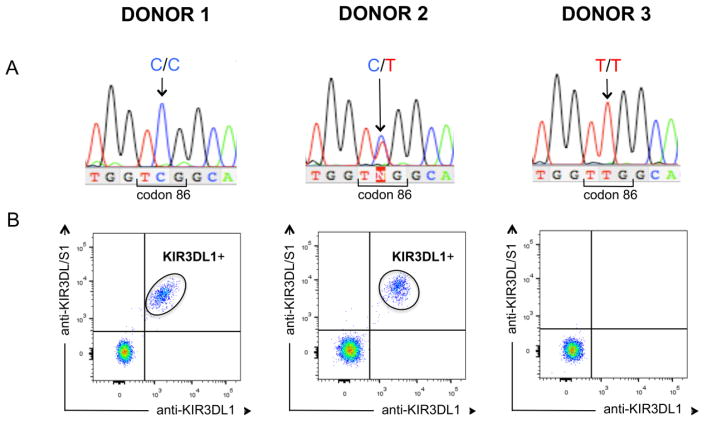
To eliminate this ambiguity, we developed a robust method of genotyping codon 86 based on nucleotide sequencing exon 3 of 3DL1. To validate the method we selected 20 individuals with each of the following genotypes: 3DL1-C/C, 3DL1-T/T, 3DL1-C/T, 3DL1-C/3DS1 and 3DL1-T/3DS1. NK cells from these 100 donors were then analysed for their cell-surface expression of 3DL1 and 3DS1. As shown in Figure 1 and Supporting Information Figure 1, there was clear correspondence between codon 86 genotype and 3DL1 expression, as observed previously [17, 19]. Thus, our sequencing method can correctly identify all prospective transplant donors who express only 3DL1 allotypes with leucine 86 and who are not optimal donors for Bw4 negative recipients.
Figure 1.
Sequencing codon 86 of KIR3DL1 discriminates allotypes that are expressed at the NK cell surface from those that are not.
Shown are three individuals who were (A) genotyped by nucleotide sequencing codon 86 of KIR3DL1 and then (B) had their KIR3DL1 phenotype determined using flow cytometry. (B) Two-colour flow cytometry was performed using KIR3DL1 (DX9) and KIR3DL1/S1 (Z27) specific mAb double staining of donor NK cells. Donors 1 and 2 have KIR3DL1 alleles that are expressed (3DL1 120-C) and Donor 3 is homozygous for an allele that is not expressed (3DL1 120-T) We selected 20 individuals each from the three genotypes, C/C, C/T and T/T (60 individuals total). Each individual was phenotyped once, with similar results obtained for each respective genotype. Each data point shown is thus representative of 20 independent experiments which are described in Supporting information Figure 1.
About one in five potential transplant donors lack functional KIR3DL1
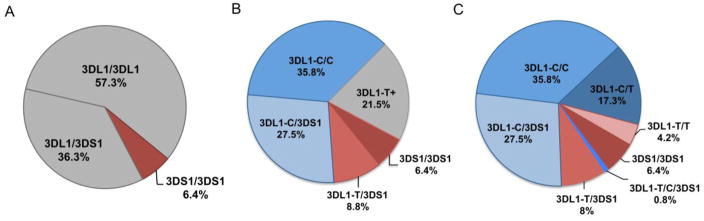
Our PCR and sequencing method was used to determine frequencies for 3DL1-T and 3DL1-C in a cohort of 600 unrelated individuals in the donor pool. From these results we determined the proportion of donors who lack cell-surface expression of 3DL1. The PCR assay identified 38 donors (~6.4%) homozygous for 3DS1, 344 donors (~57.3%) homozygous for 3DL1 and 218 (~36.3%) heterozygous donors (Figure 2A). The observed frequencies of 0.75 3DL1 and 0.25 3DS1 were in Hardy-Weinberg equilibrium (data not shown). These results are in agreement with previous studies of European populations [22]. Among 218 3DL1/3DS1 donors 53 were characterized by 3DL1-T, and they correspond to ~8.8% of the cohort. Moreover, of 344 3DL1 homozygotes, 25 were 3DL1-T homozygotes, and they represent ~4.2% of the potential donor pool. With addition of 3DL1-T/3DS1 heterozygotes (~8.8%) and 3DL1-T homozygotes (~4.2%) to the cohort of 3DS1/S1 homozygote donors, the percentage of donors shown to lack functional 3DL1 expression at cell surfaces increased almost three fold from 6.4% (Figure 2A) to 19.4% (Figure 2B–C). Thus 13% of donors that would have been identified as potentially alloreactive are likely tolerant towards Bw4-negative cells because their NK cells do not express 3DL1.
Figure 2.
Donor selection is refined by genotyping codon 86 of KIR3DL1.
Shown are 600 donors who had their KIR3DL1/S1 genotypes determined first by (A) gene content analysis, and then selectively by (B) KIR3DL1-T specific PCR (3DL1 positive donors) and (C) codon 86 sequencing (3DL1-T positive donors). The KIR3DL1 expression status for each genotype (Figure 1) is shown by colour; blue shades indicate genotypes known to produce KIR3DL1 positive NK cells, red shades indicate genotypes with no expressed KIR3DL1 alleles and grey indicates an unknown phenotype. Each individual genotype was determined once. Data shown are pooled from the 600 donors.
One in ten donors has the 3DL1-T/3DL1-C/3DS1 duplicated KIR haplotype
Several different KIR haplotypes formed by en-bloc gene duplications have been described [23, 25–27]. Among these, a tandem duplication of the KIR3DP1, KIR2DL4 and KIR3DL1/S1 genes is the most frequent [25]. This haplotype likely originated with an event of unequal crossing-over, mediated by sequence homology in the 5′-regions of KIR3DP1 and KIR2DL5A [26]. Individuals having this duplicated haplotype could be good transplant donors for Bw4-negative recipients, when the haplotype contains a 3DL1-C allele that encodes a functional 3DL1 allotype. So far, such individuals were excluded from the donor pool if they typed by PCR as 3DL1-T/3DS1. To study their KIR3DL1/S1 genotypes, we sequenced 3DL1 exon 3 from the 53 3DL1-T/3DS1 heterozygous donors (~8.8% of the donor cohort). Five of them also have 3DL1-C (Figure 2C). These results were confirmed using quantitative PCR, which showed that each of these donors had two copies of 3DL1. Thus, ~9.4% of the 3DL1-T/3DS1 individuals have the 3DL1-T/3DL1-C/3DS1 genotype. Such individuals can now be considered good candidate donors of haplo-HSCT Bw4-negative patients, and therefore the percentage of donors lacking functional 3DL1 has been adjusted from 19.4% to 18.6 % (Figure 2C).
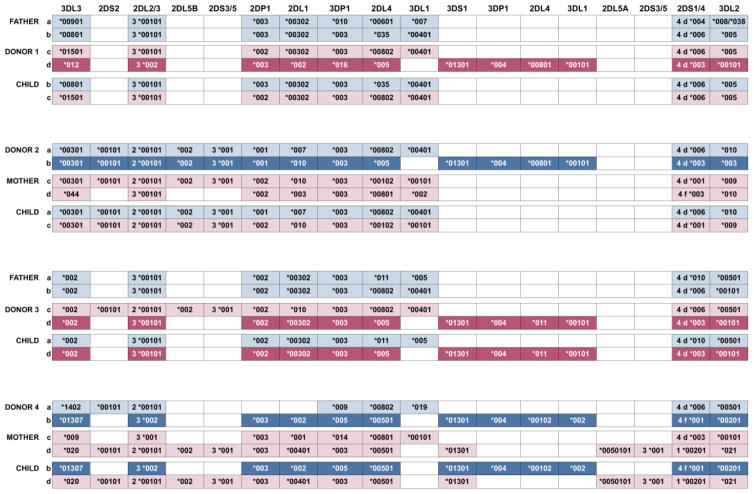
Allele-level characterization of KIR3DL1/S1 duplicated haplotypes
KIR3DL1 allotypes exhibit a clear hierarchy in their effects on NK cell activity, with 3DL1*007 being the weakest and 3DL1*001 the strongest inhibitor [18]. To determine the alleles of KIR3DL1/S1 present, we analysed four donors carrying the gene duplication and their immediate family members. We used a next-generation sequencing method that identifies both gene copy number and alleles to define the haplotypes at high resolution (Supplementary Figure 2 and Figure 3). In this manner we demonstrated that all the duplicated haplotypes carry 3DL1-C and 3DS1 (Supplementary Figure 3). Of note, none of the duplicated KIR haplotypes carry 3DL1-T, thus both 3DL1 and 3DS1 are expressed from each of these haplotypes [23]. The duplicated haplotypes of donors 1, 2 and 3 carry KIR3DL1*001, whereas the duplicated haplotype of donor 4 carries KIR3DL1*002. Both KIR3DL1*001 and KIR3DL1*002 are common alleles of European populations [22], and they are both also highly expressed and strong inhibitors [18]. Thus all of the duplicated haplotypes have potential for facilitating therapeutic alloreactivity towards Bw4-negative leukemia cells.
Figure 3.
High resolution analysis of KIR duplicated haplotypes.
Shown are the KIR haplotypes of 12 individuals belonging to four families (3 individuals/family) who have segregating duplications of the KIR3DL1/S1 gene. All individuals shown were analysed by targeted sequencing of the full KIR genomic region, which determines both the KIR gene and allele content at high resolution. Within each family the four segregating KIR haplotypes are denoted by the letters a, b, c and d. Those haplotypes derived from each parent are shaded with a distinct colour, and duplication haplotypes are shown as darkened colours with white text. The alleles determined for each KIR gene are indicated as text in the coloured boxes, and a clear box indicates the gene is absent. KIR2DS1 and KIR2DS4, which are mutually exclusive, are shown at the same site (indicated as 1 and 4, respectively). Full-length alleles of KIR2DS4 are designated 4f and alleles having the 22 nucleotide frame-shifting deletion are designated 4d. Data shown are derived from a single experiment performed for each individual, each producing full (100%) and high depth (min 100X) coverage of the KIR genomic region.
The duplicated haplotypes described here all contain KIR3DP1*004, the allele previously associated with the 3DP1-2DL4-3DL1/S1 gene duplication [28] (Figure 3). This allele is a hybrid formed by the recombination event that created the duplicated haplotype [28]. In addition to the duplication, haplotypes from Donors 1, 3 and 4 have identical gene content, but differ in their alleles, and the haplotype from Donor 2 differs also in gene content (Figure 3). These observations suggest that one crossover event created a duplication haplotype, which was then diversified by further recombination. The high-resolution analysis also showed that KIR3DL1*00401 is the 3DL1-T allele in donors 1, 2 and 3, whereas in donor 4 the 3DL1-T allele is KIR3DL1*019. The KIR3DL1*00401 allele is a common variant in European populations, whereas KIR3DL1*019 is rare [22]. On the whole, these segregation patterns are consistent with 3DL1-T having been selected because it causes loss of 3DL1 function, whereas the duplicated haplotypes have been selected for gain of 3DL1 function.
CONCLUDING REMARKS
Tens of thousands of hemato-onocological patients are treated with Haplo-HSCT every year in Europe and the USA. For Haplo-HSCT, donor selection is based on the donor and patient HLA types, as well as the donor’s KIR gene content. We have shown that KIR gene content alone is an insufficient criterion for donor selection based on NK alloreactivity, because the allelic polymorphism of the KIR genes can also have major functional effects. This is vividly illustrated by the effect of a dimorphism at position 86 in 3DL1 that divides the allotypes into two functional groups. Allotypes with serine 86, which are expressed at the NK cell surface have potential to mediate graft-versus leukemia alloreactions when transplanted into Bw4-negative recipients. In contrast, allotypes with leucine 86 are not expressed at the cell surface and cannot mediate alloreactivity when transplanted into Bw4-negative recipients. Here we have described a combined PCR and sequencing method to discriminate the two types of alleles and thus subdivide prospective donors into those whose 3DL1 allotypes could mediate antitumour alloreactivity and those whose 3DL1 allotypes cannot. A PCR-based method for genotyping KIR3DL1 alleles was described recently [29]. The method identifies 3DL1-null alleles by targeting adenine (A) at position 193, which is linked to the thymidine (T) at position 320 in some alleles. However, not all alleles with A193 also possess T320 [24]. Our method, providing the precise nucleotide at position 320 by sequencing, therefore represents an improvement over the previous method. Using our method in a cohort of 600 mostly European donors we show ~6.4% lacked 3DL1, ~12.2% had only non-functional 3DL1 allotypes that cannot mediate alloreactivity, and ~81.4% have functional 3DL1 allotypes that have potential to mediate therapeutic alloreactivity. Distinguishing these three groups represents significant progress towards refining donor selection for patients given haplo-HSCT.
The critical role of donor NK cell alloreactivity in eliminating residual leukemia cells was recognized using protocols that enriched the graft tissue for CD34+ hematopoietic precursor cells, but depleted the T cells [1, 2]. Multiple further methods of graft manipulation have been developed recently, including those that retain T cells (T cell replete) and those that remove specific lymphocyte subsets (eg. αβ+ T/CD19+ B cell deplete) [15, 30, 31]. The role of donor NK cell alloreactivity following these graft manipulation protocols remains to be investigated. The genotyping method we describe will be valuable for this purpose, and other studies that correlate NK alloreactivity with clinical outcome.
MATERIAL AND METHODS
Samples for analysis
We analysed 600 samples obtained from Ospedale Bambino Gesù (Rome, Italy), IRCSS AOU San Martino (Genoa, Italy) and IRCCS Ospedale San Matteo (Pavia, Italy). The samples were obtained upon informed consent.
KIR gene content analysis
Genomic DNA was isolated from blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and KIR gene presence was assessed using Olerup SSP KIR genotyping kit (GenoVision, Saltsjoebaden, Sweden), following the respective manufacturer’s instructions. Included in the kit are the three sets of primers we used to genotype KIR3DL1/S1. We developed an additional series of primers to amplify and sequence codon 86.
KIR3DL1 codon 86 sequencing
Exons 3, 4 and 5 of KIR3DL1 were amplified using the primer set: ALL3DL1FOR (TCCCATCTTCCATGGCAGA) and reverse primer 3DL1REV (TAGGTCCCTGCAAGGGCAA). Cycles were performed as follows: 2 min at 94°C; 30 cycles of 94°C for 30s, 60°C for 30s, 72°C for 30s; and a final extension step of 10 min at 72°C. The PCR product (1487bp) was purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). DNA sequencing was performed using d- Rhodamine Terminator Cycle Sequencing kit, All3DL1FOR primer, and a 3100 ABI automatic sequencer (PerkinElmer, Wellesley, MA). The polymorphism at position 86 involves a serine codon (TCG) and a leucine codon (TTG). The nucleotide polymorphism is at position 320 in the full-length CDS, corresponding to codon 107 in full-length polypeptide and residue 86 in mature protein. Eight out of 129 3DL1-T+ donors were excluded from this analysis because of material restrictions.
Analysis of KIR3DL1 gene copy number
The 3DL1 gene copy number assay was performed according to the previously published protocol for determining genomic copy number for all KIR genes [25].
Cytofluorimetric analysis
Peripheral blood mononuclear cells were stained using anti-CD3-PECF594 from BD (Biosciences, Bedford, MA), anti-CD56-PC7 and Z27-PE (anti-KIR3DL1/S1) from Beckman Coulter (Marseille, France) and DX9-FITC (anti-KIR3DL1) from Miltenyi Biotech (Bergisch Gladbach, Germany). Polyclonal-activated NK cells, obtained as previously described [13], were also analyzed for KIR3DL1 expression. Data were acquired on a MACSQuant Analyzer (Miltenyi Biotec) and analyzed with FlowJo version 10.7 software.
High-resolution sequence analysis of the KIR region using next-generation sequencing
Oligonucleotide probes were used to capture the KIR genomic regions from libraries of sheared genomic DNA. The enriched DNA was sequenced using a MiSeq instrument (version 3; 2×300 read pairs: Illumina Inc, San Diego. CA), and the data analysed using a custom bioinformatics pipeline [Norman et al., submitted manuscript; see https://github.com/wesleymarin/]. In the duplicated haplotypes the order of the KIR genes was assigned according to that described previously [26]. In the course of this analysis we identified a novel allele of KIR2DL4, named 2DL4*035 [32].
KIR3DL1 and KIR3DS1 allele typing
Total RNA was extracted from NK cell bulk populations using RNeasy micro kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The cDNA synthesis was performed on ~1 μg RNA using oligo(dT) oligonucleotides. The sequences of the used primers are: KIRIg3up: CATGTYGCTCAYKGTCGTC and KIRIg3dw: GGTTTTGAGACAGGGCTG. The amplification products were cloned into pcDNA3.1/V5/His TOPO vector using the Eukaryotic TOPO TA Cloning kit (Invitrogen, Carlsbad, CA) and sequenced.
Supplementary Material
Acknowledgments
(PJN and PP) U.S. National Institutes of Health grants U01 AI090905 and R01 GM109030
(AM, CB) Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.) Investigator Grant 15704 and (CA, DP, RM, AB, FL, AM, CB, MF) special project 5×1000 9962
Ricerca Finalizzata Locatelli 2010 RF-2010-2316606 (FL, AB, LM, MF, DP)
Abbreviations
- KIR
Killer cell Ig-like receptor
- Haplo-HSCT
HLA haplotype-identical (haploidentical) hematopoietic stem cell transplantation
Footnotes
CONFLICT OF INTEREST DISCLOSURE
A. Moretta is founder and shareholder of Innate Pharma (Marseille, France). The remaining authors declare no financial or commercial conflict of interest.
References
- 1.Locatelli F, Pende D, Maccario R, Mingari MC, Moretta A, Moretta L. Haploidentical hemopoietic stem cell transplantation for the treatment of high-risk leukemias: how NK cells make the difference. Clin Immunol. 2009;133:171–178. doi: 10.1016/j.clim.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 3.Moretta A, Pende D, Locatelli F, Moretta L. Activating and inhibitory killer immunoglobulin-like receptors (KIR) in haploidentical haemopoietic stem cell transplantation to cure high-risk leukaemias. Clin Exp Immunol. 2009;157:325–331. doi: 10.1111/j.1365-2249.2009.03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 2011;32:364–372. doi: 10.1016/j.it.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Ciccone E, Pende D, Vitale M, Nanni L, Di Donato C, Bottino C, Morelli L, Viale O, Amoroso A, Moretta A, et al. Self class I molecules protect normal cells from lysis mediated by autologous natural killer cells. Eur J Immunol. 1994;24:1003–1006. doi: 10.1002/eji.1830240434. [DOI] [PubMed] [Google Scholar]
- 9.Karre K. How to recognize a foreign submarine. Immunol Rev. 1997;155:5–9. doi: 10.1111/j.1600-065x.1997.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 10.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FT, Witt CS. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 12.Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, Gras S, Saunders PM, Olshina MA, Widjaja JM, Harpur CM, Lin J, Maloveste SM, Price DA, Lafont BA, McVicar DW, Clements CS, Brooks AG, Rossjohn J. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, Romeo E, Cognet C, Martinetti M, Maccario R, Mingari MC, Vivier E, Moretta L, Locatelli F, Moretta A. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009;113:3119–3129. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 14.Zhao XY, Chang YJ, Zhao XS, Xu LP, Zhang XH, Liu KY, Li D, Huang XJ. Recipient expression of ligands for donor inhibitory KIRs enhances NK-cell function to control leukemic relapse after haploidentical transplantation. Eur J Immunol. 2015;45:2396–2408. doi: 10.1002/eji.201445057. [DOI] [PubMed] [Google Scholar]
- 15.Foley B, Felices M, Cichocki F, Cooley S, Verneris MR, Miller JS. The biology of NK cells and their receptors affects clinical outcomes after hematopoietic cell transplantation (HCT) Immunol Rev. 2014;258:45–63. doi: 10.1111/imr.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas P, Loiseau P, Tamouza R, Cayuela JM, Moins-Teisserenc H, Busson M, Henry G, Falk CS, Charron D, Socie G, Toubert A, Dulphy N. NK-cell education is shaped by donor HLA genotype after unrelated allogeneic hematopoietic stem cell transplantation. Blood. 2011;117:1021–1029. doi: 10.1182/blood-2010-02-269381. [DOI] [PubMed] [Google Scholar]
- 17.Leung W, Iyengar R, Triplett B, Turner V, Behm FG, Holladay MS, Houston J, Handgretinger R. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- 18.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The Protein Made from a Common Allele of KIR3DL1 (3DL1*004) Is Poorly Expressed at Cell Surfaces due to Substitution at Positions 86 in Ig Domain 0 and 182 in Ig Domain 1. The Journal of Immunology. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 20.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 21.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 22.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 23.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, Parham P. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, Cookson WO, Trowsdale J, Traherne JA. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting Edge: Expansion of the KIR Locus by Unequal Crossing Over. The Journal of Immunology. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 27.Williams F, Maxwell LD, Halfpenny IA, Meenagh A, Sleator C, Curran MD, Middleton D. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum Immunol. 2003;64:729–732. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, Vilches C. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. 2005;35:16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- 29.Boudreau JE, Le Luduec JB, Hsu KC. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS One. 2014;9:e99543. doi: 10.1371/journal.pone.0099543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashey A, Solomon SR. T-cell replete haploidentical donor transplantation using post-transplant CY: an emerging standard-of-care option for patients who lack an HLA-identical sibling donor. Bone Marrow Transplant. 2014;49:999–1008. doi: 10.1038/bmt.2014.62. [DOI] [PubMed] [Google Scholar]
- 31.Bertaina A, Merli P, Rutella S, Pagliara D, Bernardo ME, Masetti R, Pende D, Falco M, Handgretinger R, Moretta F, Lucarelli B, Brescia LP, Li Pira G, Testi M, Cancrini C, Kabbara N, Carsetti R, Finocchi A, Moretta A, Moretta L, Locatelli F. HLA-haploidentical stem cell transplantation after removal of alphabeta+ T and B cells in children with nonmalignant disorders. Blood. 2014;124:822–826. doi: 10.1182/blood-2014-03-563817. [DOI] [PubMed] [Google Scholar]
- 32.Alicata C, Bottino C, Guethlein LA, Parham P, Norman PJ. Description of the novel KIR2DL4*035 allele identified using high-throughput sequencing. HLA. 2016 doi: 10.1111/tan.12761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.