Abstract
The curious patterns of imprinted gene expression draw interest from several scientific disciplines to the functional consequences of genomic imprinting. Methods of probing the function of imprinting itself have largely been indirect and correlational, relying heavily on conventional transgenics. Recently, the burgeoning field of epigenome editing has provided new tools and suggested strategies for asking causal questions with site specificity. This perspective article aims to outline how these new methods may be applied to questions of functional imprinting and, with this aim in mind, to suggest new dimensions for the expansion of these epigenome-editing tools.
Keywords: : CRISPR-Cas9, epigenome editing, EpiEffectors, functional consequences, imprinting
Background
Investigation into the functional consequences of various epigenetic marks requires epigenome-editing tools, with the capacity for specific targeting and mitotically heritable modification. Important regulatory targets for epigenome editing include marks such as methylation at CpG islands, methylation at differentially methylated regions (DMRs – these exhibit DNA methylation asymmetry between parental copies) and modifications to histone tails (including activating and repressive methylation and activating acetylation at regulatory chromatin regions) [1]. Targets also include more complex configurations such as bivalent chromatin domains and changes in higher chromatin structure [1,2]. While we have some knowledge of the effect of various marks (for instance that histone 3, lysine 27 trimethylation [H3K27me3] is repressive), they often exist within a more complicated architecture with conflicts and redundancies. Exploring functional consequences of these marks at specific loci requires fine dissection strategies.
We restrict our discussion here to methods applicable to imprinted genes, which are defined by their parent of origin monoallelic expression [3]. Imprinting occurs when one parental allele is epigenetically marked during embryogenesis, creating a basis for allele-specific regulatory differences. Germline DMRs on the parental genomes are established at fertilization and are protected from genome-wide demethylation during embryogenesis [1]. These differences are reiterated with further modifications in later developmental stages to elaborate and maintain the imprinting mark [4]. The minimal region, as defined by targeted deletions, which regulates an imprinted locus is termed an ‘imprinting center’ or ‘imprinting control region’ (ICR) [4]. While some ICRs regulate imprinted domains extending over large regions of DNA and require complex and extensive epigenetic architecture to regulate a cluster of genes, others regulate a single imprinted protein-coding gene [5,6]. Furthermore, imprinting architecture can vary between tissues and developmental stages, resulting in differing expression patterns in abundance and splice variants. This variation is the result of tissue- and stage-specific epigenetic modification at the locus and is subsequently maintained through several rounds of DNA replication [4]. The long-term maintenance of these marks is an important element for qualifying imprinting architecture as epigenetic rather than simply regulatory and will be important in the assessment of epigenome editing tools we consider for functional research.
There are approximately 120–180 imprinted genes and noncoding RNAs identified in the mouse, and around 100 in humans [7]. While these genes do not appear to associate within any particular biochemical pathway, thematic phenotypic and physiological consequences do emerge. Of these, the most prominent are functions within the embryo, extra-embryonic tissue and the adult CNS [7,8]. Imprinted genes have a role in placental size and function, in utero growth, nutrient transport and placental signaling [9–11]. In metabolic systems, imprinted genes have a role in prenatal growth, appetite, fat and lean mass deposition, energy homeostasis, and insulin sensitivity and production [9]. Imprinted gene expression is particularly prominent in the CNS and in addition to diverse behavioral associations – including maternal care and mother-infant interactions [7,12–14], social dominance [15,16] and exploration [15,17] – there is growing association with neuropsychiatric and neurodevelopmental disorders [3,18–19].
Conventional transgenic strategies
Thus far, we have determined the functional consequence of imprinting without control of specific, localized marks. In fact, most studies examine the degree of association between imprinted regions and disease or disorder [3,19] and/or the function of the genes within imprinted regions, rather than the imprint itself.
Knockout models are a classic starting point used in many functional studies of imprinting. These models often delete only one allele, making use of the monoallelic expression patterns to demonstrate the function of both the gene and its requisite origin. For example, loss of function of the maternal Phlda2 allele resulted in an expanded placental spongiotrophoblast compartment and dysregulated placental hormones [20]. While these approaches are useful in distinguishing the function of allele origin, they often result in broad loss of gene function, rather than providing insight into the inner mechanisms of imprinting or the fine control it exerts in an endogenous setting [21].
Loss of imprinting (LOI) KO models may reveal different and somewhat more specific functional effects by interruption or deletion. Precise placement of these interruptions and deletions can reveal more detailed knowledge of imprint function. For instance, interruption of the maternally inherited Gtl2 promoter by insertion of a ßgeo cassette upstream created a partial LOI which shifted transcript dosage in the chromosome 12 imprinted gene cluster from maternal expression toward paternal-origin expression patterns [22]. LOI models may also distinguish the function of the somatic (established during development and cellular differentiation) or germline (established prior to fertilization) ICRs. Whereas deleting a germline ICR may eliminate the imprinting mark altogether, deletion of a somatic ICR can reveal the tissue-specific roles of certain epigenetic architectures regulating gene dosage at critical stages [4].
Duplication of imprinted regions provides another method for functional investigation, mimicking the effects of LOI at one, or a cluster of imprinted genes. Uniparental disomies and duplications of the imprinted chromosomal region are particularly useful in characterizing the phenotypes of increased dosage [21]. Transgenes are useful tools for introducing duplication. For instance, the addition of an extra copy of maternally expressed Phlda2 carried by a bacterial artificial chromosome significantly reduced placental weight, demonstrating a purpose in growth restriction [23]. Additionally, it is possible to normalize expression levels and thereby rescue these models, by crossing with a targeted deletion model [24]. Various types of transgenes can also be used to define the regional specificity of ICRs and to dissect the functional consequences of imprinted regions regulating multiple genes [21]. These duplication strategies still rely on the endogenous organization of ICRs and the genomic sequences they regulate. However, they still duplicate the coding genome sequences themselves. One way to separate the function of the ICR from the genes it regulates is the insertion of a reporter gene near the ICR. Exogenous sequences inserted within known imprinted regions have been shown to acquire functionally relevant epigenetic imprints following the imprinting pattern of the targeted locus [25,26]. Inserted elements can also interact with long range imprinting signals on their own to generate a new imprinted locus with tissue-specific imprinted expression patterns [27]. These create a useful model for studies of the evolutionary origin and function of imprinted gene clusters. An example is the novel transgenic mouse line Tel7KI (Instm1Lef), which expresses the fluorescent reporter EGFP from an inserted transgene located on chromosome 7 between the imprinting center 1 (IC1) and imprinting center 2 (IC2) regions [27]. This insert gained maternal allele-specific expression specifically in embryonic tissues while remaining biallelic in placenta. The authors suggest this as a model for how novel genes become imprinted during evolution. The tool also provides a strategy for dissecting epigenetic differences between tissue lineages and for interpreting the functional consequences of imprinting signals, independent of endogenous genes.
A drawback, and potentially confounding factor in all these models, is the direct manipulation of the genetic material near the ICR. Strategies circumventing this disrupt the endogenous epigenetic mark or imprinting mechanism. For instance, deletion of essential endogenous effectors such as Dnmt1 (not in itself imprinted) prevents the establishment or maintenance of the imprinting mark [28]. However, this approach is unspecific to the locus of interest and may create widespread changes across the genome, dysregulating many genes and obscuring the resulting LOI phenotype [21]. Equally, pharmacological removal of a mark using agents such as 5-azacytidine (demethylation) has widespread rather than specific effects, obscuring functional consequences of disruption of imprinting [29,30]. Deletion of epigenetic marks by these methods can show their necessity for normal imprinting, but are likely to obscure the specific functional consequence of such a mark. While association, knockout and pharmacological studies are very helpful for discerning the broader impact of imprinting on biological systems, these strategies fall short of the high resolution needed to probe the function of imprinting marks independent of the genetic sequence beneath them.
Tools for targeted epigenome editing
Tools for targeted epigenome editing could allow researchers to manipulate regulatory marks themselves, without changing genomic content. This may enable us to ask more specific and causal questions about the function of imprinting. Epigenome engineering can break down and reconstruct heritable regulatory architecture and build it ectopically to discern both the role of individual parts and the synergistic functional consequences of imprinting structures.
The basic structure of an EpiEffector, or epigenome engineering tool, is a programmable DNA binding domain or mechanism coupled to an epigenetic effector domain. Popular binding domains include ZFNs, TALE nucleases, and the CRISPR/dCas9 system. Laufer catalogues a selection of EpiEffector components in a figure comparing common programmable DNA binding domains and an extensive table of employable epigenetic effector domains [31]. An elegant diagram of a complete EpiEffector example can be found in Figure 1 [32]. This report focuses on CRISPR-based tools because of the ease and multiplexing capacity of targeting by synthetic guide RNAs (sgRNAs) compared with the more difficult design process of ZFNs and TALE. However, all types of EpiEffector are applicable. CRISPR-based tools specifically mentioned in this report are summarized in Table 1.
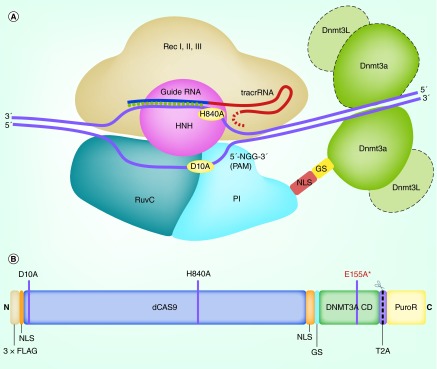
Figure 1. . Anatomy of example CRISPR/dCas9-based EpiEffector: dCas9–DNMT3A.
(A) The dCas9–DNMT3A fusion protein complexes with the sgRNA (composed of the fused crRNA or guide RNA and tracrRNA) to localize the DNMT3A effector domain to the target region. The dCas9 segment is composed of a recognition lobe (Rec I, II and III) and an inactivated nuclease lobe (HNH, RuvC and PI domains). The DNMT3A effector is fused to the PI domain on the nuclease lobe by an NLS and a GS peptide linker. The DNMT3A catalytic domain recruits partners for dimerization to carry out targeted methylation. (B) Linear order of domains on the dCas9–DNMT3A fusion protein. The N-terminal begins with the 3× FLAG epitope tag and the NLS, followed by the nuclease-inactivated dCas9 domain (inactivating mutations D10A and H840A are indicated). dCas9 is followed by a second NLS, and a GS peptide linker which fuse it to the catalytic domain of human DNMT3A. In this domain, E155A indicates the DNMT3A inactivating mutation used as a negative control. The mRNA for this fusion protein also contains a puromycin resistance gene transcript (protein domain-PuroR) or EGFP gene (not shown) for selection of successfully transfected cells. During translation, this selector separates from the EpiEffector when the T2A self-cleaving peptide detaches the fusion protein's C terminal end.
GS: Gly4Ser; HNH: Nuclease lobe cleaving the target strand; NLS: Nuclear localization signal; PI: PAM-interacting.
Table 1. . CRISPR-based tools in this article.
| Study (year) | CRISPR-based tool | Description | Ref. |
|---|---|---|---|
| Perez-Pinera et al. (2013) |
dCas9-VP64 |
Activates transcription by recruiting a transcription complex to the target |
[33] |
| Hilton et al. (2015) |
dCas9-p300(core) |
Acetylates H3K27 and activates transcription by targeting human E1A-associated protein p300 acetytransferase activity |
[34] |
| Vojta et al. (2016) |
dCas9-DNMT3A |
Adds DNA methylation to CpG sites at the target by directing DNMT3A activity |
[32] |
| Polstein et al. (2015) | CIBN-dCas9-CIBN | Heterodimerizes with CRY2-effector fusions (e.g., CRY2FL–VP64) to enable optogenetic control of targeted effector action | [35] |
Two recently published, simple CRISPR-based EpiEffectors direct the addition of fundamental histone marks. The dCas9-p300(core) tool from Hilton permits the targeted addition of the activating histone 3, lysine 27 acetylation (H3K27ac) mark [34]. H3K27ac was used here as a relative measurement of broad p300 acetyltransferase activity and a widely documented indicator of enhancer activity. This tool causally linked target site enrichment of H3K27ac with highly specific target gene activation at levels greater than those induced by the conventional engineered transactivator dCas9-VP64 [33]. When targeted to DNase I hypersensitive site 2 (the HS2 enhancer) on the mammalian β-globin locus control region (not imprinted) [36], this construct was also more effective than dCas9-VP64 at enriching H3K27ac at distal promoters regulated by the HS2 enhancer [34]. This construct provides causal evidence that the acetylation it effects is sufficient to induce enhancer activity. Hilton et al. use the dCas9-p300(core) tool to systematically dissect the histone regulatory modifications necessary for expression at targeted regulatory regions, but tools such as this could be similarly employed to demonstrate the histone marks sufficient for inducing the expression patterns of imprinting.
The CRISPR/dCas9 tool from Vojta uses the catalytic domain of DNMT3A to add repressive DNA methylation marks [32]. This achieved targeted CpG methylation in an approximately 35 base pair wide region, and multiplexing sgRNAs expanded the region of de novo methylation. The study targeted the CpG islands within the promoter regions of IL6ST and BACH2 (neither imprinted), genes relevant for N-glycosylation of IgG and associated with some autoimmune diseases [36,37]. Expression changes from these genes following targeted CpG methylation provide proof of concept for targeted dosage control by epigenome editing. IL6ST in particular showed more than twofold decrease in transcript level. Moreover, changes in methylation were heritable across mitotic divisions up to 42 days after transfection. Such heritability of regulatory modifications created by EpiEffectors is crucial to explorations of the functional consequences of imprinting. To properly mimic true epigenetic change and reveal long-term developmental consequences, the mark must be long lasting and maintainable. In theory, application of the dCas9–DNMT3A tool to ICRs could engineer epigenetically driven dosage control that avoids changes to endogenous genomic sequence or the introduction of transgenes.
Use of a catalytically inactive DNMT3A domain in the Vojta et al. construct still created low levels of methylation activity at the target [32]. This suggests the EpiEffector recruits cell-endogenous interaction partners with effector activity. EpiEffectors need not act exclusively to add and remove regulatory marks from the loci of interest, but attribution of any functional consequences must acknowledge the full contextual change induced by targeting the EpiEffector to the site. For example, changes in regulatory imprinting marks at the Prader–Willi syndrome ICR could dysregulate the noncoding C/D box snoRNA clusters (SNORDs) within region 15q11.2-q13 [38]. One of these clusters, encoding SNORD116, changes the expression of over 200 genes when overexpressed in HEK293T culture. Changing marks regulating the imprinted expression pattern of these snoRNAs could create a cascading effect on gene expression that conceals the immediate effect of the experimental manipulation. However, while recruitment of endogenous mechanisms might obscure the effect of individual marks, especially if additional marks are altered or other regulatory agents are affected, it is likely these joint changes will better mimic imprinting patterns than simple changes added one by one.
However, in the absence of a single, cascading change establishing full imprinting functions, researchers must consider the use of multiple EpiEffectors to break down and construct complex regulatory architecture. The systematic breakdown of epigenetic architectures can demonstrate the minimum points at which regulatory marks attract these secondary modification and maintenance enzymes. Coordinated changes may be necessary to discern the synergistic function of imprinting structures.
Precise control of EpiEffector activity
We can make further use of the genome editing toolbox by taking advantage of systems for precise control of timing. Adding tissue or stage specific promoters can trigger EpiEffector expression within specific cellular contexts, while cre-loxP recombination systems can eliminate stably integrated EpiEffector expression after specific time points, as in traditional strategies [39]. For more direct access, engineered mechanisms can use optogenetics to drive EpiEffector activity with high spatial and temporal specificity. An example of such an optogenetic actuator is the light-inducible transcriptional effectors (LITEs) employed in Konermann to drive transcriptional effectors [40]. Konermann et al. suggested using this LITE system with a range of successful TALE-histone epigenetic effector fusions (epiTALES), constructed for repression of Grm2 (not imprinted) and a neural lineage-specifying transcription factor, Neurog2 (not imprinted), in primary neurons. Chromatin immunoprecipitation confirmed epiTALE-mediated modification of histone marks including H3K9me1, H4K20me3, H3K27me3, H2K9ac and H4K8ac. LITE mediated Neurog2 expression rose 30 minutes after initial optogenetic stimulation. Modification of histone marks could occur more quickly than this transcriptional output. Additionally, the reversibility of the conformational changes that activate the LITE system enables precise control of editing functions within a narrow temporal window.
Optogenetics has also been employed in light-activated CRISPR-Cas9 effector (LACE) systems to create dynamic and spatially specific transcriptional effectors (or transactivators) [35]. Polstein and Gersbach describe the fusion of parts of the light-inducible heterodimerizing proteins CRY2 (full length: CRY2FL) and CIB1 (N-terminal fragment: CIBN) to transactivation domain VP64 (CRY2FL-VP64) and dCas9 (CIBN-dCas9-CIBN) respectively. This LACE system produced a high level of transcriptional activation in some cases comparable to that observed with constitutively active dCas9-VP64. Under this system, gene expression response levels were sensitive to sequential delayed illumination, light removal and reillumination. Expression from an eGFP plasmid reflected arbitrary illumination patterns projected onto cell culture through a photomask, demonstrating a high degree of spatial precision. LACE systems demonstrate the retargetable flexibility of the CRISPR/dCas9 system is compatible with light-induced recruitment of effector domains. This system could be tested with epigenetic effector domains to achieve fine control over imprinting architectures throughout developmental stages in model systems.
Conclusion: potential epigenome engineering strategies for probing the functional consequences of imprinting
As with genome editing approaches, epigenome engineering strategies can use the ‘necessary and sufficient’ principle to investigate the functional consequences of imprinting architecture. The precise addition or removal of a regulatory mark can reveal consequences for transcription. Changes in gene dosage as a whole, by allele- and tissue-specific transcriptional variants, and by protein production reveal the function of imprinting in maintaining differential usage patterns. Ectopic expression at the removal of a mark can demonstrate a role in tissue- or stage-specific gene expression, and resulting phenotypes provide insight into the importance of the targeted imprinting mark in development or homeostasis.
Ectopic duplication of an epigenetic mark or even a full imprinting architecture within a different tissue or developmental stage can demonstrate the sufficiency of that mark or architecture for reproducing precise expression patterns. Reproduction of this structure on a new genetic locus could also demonstrate whether the function of the imprint is independent from the surrounding genomic sequence or requires complementary genetic sequences to promote or suppress gene expression.
The timing and duration of our modifications will also affect resultant phenotypes. Mitotically, if not meiotically, heritable marks added by EpiEffectors, as in Vojta, allow us to investigate downstream developmental effects and are especially important for questions concerning germline DMRs and imprinting marks that diverge between developing tissues. We can use the addition of a stable mark and its temporally precise removal to define a clear developmental window in which specific imprinting marks are necessary for normal development. Tissue- or stage-specific promoters or optogenetic drivers of EpiEffector expression can provide this precise control of timing both in vitro and in vivo [40].
Conversely, we can create or observe an epigenetic mark or imprinting deficiency and attempt to rescue the model by temporally precise, targeted epigenetic re-engineering at the ICR. If we can restore normal expression by recreating the series of modifications that establish and maintain an imprinting mark, we can demonstrate which marks are sufficient to establish characteristic expression patterns. These modifications require temporal precision to identify their necessary order and duration of relevance during the establishment and maintenance of imprinting. Such reconstructions may also reveal functional redundancies of several marks in the normal imprint architecture.
Future perspective: potential tools
One of the issues of current methods of epigenome editing for imprinted genes is the lack of allele specific activity. Global demethylation by pharmaceutical means or by deletion of, for instance, a DNMT, removes the DMR methylation on one allele that distinguishes between them. While allele-specific EpiEffector targeting has been suggested through SNP differences in the sgRNA targeting mechanism of the CRISPR/dCas9 system, this relies on sufficient differences between alleles and binding fidelity [41]. Nevertheless, successful allele-specific genome editing using the CRISPR-Cas system in vivo suggests this strategy could also be viable for epigenome editing [42]. In Cas9/gRNA-injected F344xDA hybrid embryos heterozygous for coat color alleles of the Tyr gene (not imprinted), SNP-specific gRNAs targeted induced mutations only within their respective alleles. Targeting the recessive allele resulted in six mutations in 21 pups born while targeting the dominant allele resulted in seven mutations in 23 pups born and thereby changed the dominant coat-color phenotype of these mutated pups.
Current strategies of epigenome engineering by EpiEffectors have not yet addressed the possibility of building upon the distinguishing methylation mark to achieve allele-specific regulatory changes (see Box 1). Methylation-sensitive EpiEffector binding could be achieved by adding a methylation-sensitive domain with a conformational inhibition of dCas9 binding or of effector domain activity. An application for such an EpiEffector is suggested in Figure 2.
Box 1. . Model epigenome engineering of the Grb10 CGI2 bivalent chromatin domain.
Epigenome engineering strategies building on existing allelic differences present more options for addressing complex architectures such as bivalent chromatin domains. The manipulation of histone modifications in an allele-specific manner could be used, for example, in a non-neuronal tissue to ectopically resolve the Grb10 bivalent chromatin domain regulating a neuron-specific transcript [2]. This resolution could provide causal demonstration of the function of such marks in regulating transcript expression and help identify functional and phenotypic changes resulting from ectopic resolution of this imprinting mark.
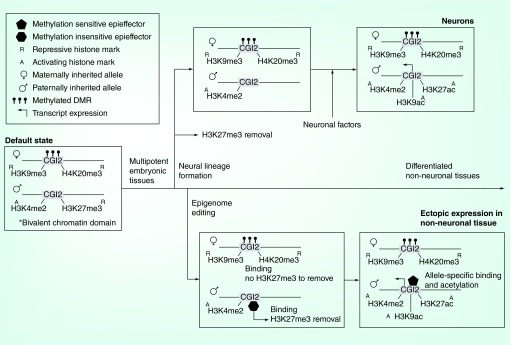
From early developmental stages onward through adult non-neuronal tissues, Grb10 promoters at the imprinted CpG Island 2 (CGI2) differentially methylated region are silent on both alleles [2]. A bivalent chromatin domain featuring both permissive and repressive histone modifications characterizes the paternal allele in these tissues. However, upon commitment to a neuronal lineage, the paternal enrichment for the repressive H3K27me3 mark is lost. The bivalent chromatin domain is resolved, leaving the activating H3K4me2 at the site. This removal of H3K27me3 is unlikely to trigger expression from CGI2 on its own. Tissue-specific promoter reactivation may rely on neuron-specific factors to induce the observed increase of H3K9 and H3K27 acetylation at the site upon differentiation.
Current evidence for the model relies on correlations between histone mark enrichment and repression or expression of the paternal-specific transcripts originating from CGI2. Epigenome engineering strategies could provide additional causal evidence for this model of Grb10 expression by ectopically replicating this sequence of histone modifications (Figure 2). Removal of H3K27me3 would require only a standard EpiEffector (using, for instance, a dCas9 targeting system and the active domain of JMJD3, an H3K27 demethylase [31,43]) localized to the CGI2 locus sequence; binding to the paternal allele achieves the desired effect and binding to the maternal allele does nothing, as there is no H3K27me3 to remove. Any interference with or recruitment of endogenous proteins, which may induce an unexpected change at the maternal site, should be ruled out by testing a control EpiEffector with a catalytically inactive effector domain. Successful epigenome engineering here homogenizes the status of the H3K27 mark on both alleles. Addition of the acetyl groups, on the other hand, requires parent-of-origin specific modification. A methylation-sensitive EpiEffector could bind exclusively to the unmethylated paternal CGI2 without relying on distinguishing SNPs to add H3K9ac and H3K27ac monoallelically. Any resulting reactivation of the CGI2 differentially methylated region promoters would demonstrate the sufficiency of these combined histone modifications for inducing tissue- and parent-of-origin specific expression.
Figure 2. . Model of ectopic resolution of the Grb10 imprinting architecture by epigenome engineering.
The default state of the imprinted Grb10 CpG Island 2 locus is characterized by repressive DNA methylation and histone marks on the maternally inherited allele and a bivalent chromatin domain on the paternally inherited allele. The bivalent chromatin domain is resolved when the repressive H3K27me3 mark is lost during normal neural differentiation. Further on in differentiation, and likely as a result of neuron-specific factors, the site acquires increased H3K9 and H3K27 acetylation – both activating marks. Epigenome editing could be employed to attempt ectopic resolution of the bivalent chromatin domain and to mimic the construction of the activating architecture. In this model, a methylation insensitive EpiEffector with H3K27me3 demethylation activity targeted to the site could resolve the bivalent domain, without effect on the maternal allele. This is followed by treatment with methylation-sensitive EpiEffectors possessing H3K9 and H3K27 acetylation activity. These bind in an allele-specific manner to the unmethylated paternal allele to reconstruct a neuron-specific histone profile in non-neuronal tissues.
DMR: Differentially methylated region.
Modifications to existing EpiEffector components could improve multiplexing capacity as well as temporal and spatial specificity. CRISPR/Cas9 systems have demonstrated synergistic effects through multiplexing, as in Vojta [32] and Perez-Pinera [33]. This capacity can be further expanded by simultaneously using systems, such as TALEs and ZFNs, with entirely different targeting strategies. This relieves the basic guide system from the difficulties of carrying multiple bulky domains and introduces increased specificity by requiring two separate systems to localize to the correct locus for complex editing. The use of multiple systems also allows separate EpiEffector domains to localize to an ICR, yet remain autonomous, without risk of cross targeting by interactions with the other guide system. For instance, Cpf1, a CRISPR-associated two-component RNA-programmable DNA nuclease, provides a smaller, simpler system that could replace or complement Cas9 [44,45]. In particular, differences between the shorter Cpf1 guide CRISPR RNAs and the analogous Cas9 sgRNAs that guide binding could enable different EpiEffectors, separately guided by either Cpf1 or Cas9 systems, to multiplex in close proximity. Finally, arrays of EpiEffectors triggered by optogenetic drivers stimulated by different wavelengths of light further increase the multiplexing capacity of epigenome engineering experiments. The spatial and temporal specificity afforded by optogenetics can be used to direct sequential construction of complex regulatory architectures in a short window.
While genetic models have provided useful insights into the function of imprinting centers and the genes they regulate, a true exploration of the consequences of these epigenetic architectures requires tools targeted toward epigenetic modifications themselves. EpiEffectors provide a method of targeted epigenome editing, and several effective editors have already been published. These have so far focused on simple, fundamental and useful modifications, but there are still several more configurations of components already at hand which may improve our capacity to construct and deconstruct more complex arrangements. Experimental designs using these tools can follow similar principles to genome engineering experiments, but must also acknowledge epigenome-specific requirements: functional consequences may be inferred from epigenome editing only if this editing is heritable across mitotic divisions and is independent from changes to the genome sequence.
Executive summary.
Conventional transgenic strategies
Conventional genetic manipulation is indirect and confounds understanding of the epigenetic mark independent of genomic sequence.
Global manipulation of regulatory DNA methylation and histone marks by pharmacology or genetic knockout obscures functional consequences of specific, localized imprinting architecture.
Tools for targeted epigenome editing
An EpiEffector, or epigenome engineering tool, is a programmable DNA binding domain or mechanism coupled to an epigenetic effector domain.
CRISPR-based tools are appealing because of the ease and multiplexing capacity of targeting by synthetic guide RNAs.
Simple marks such as somatically heritable repressive DNA methylation and histone 3, lysine 27 acetylation are already feasible.
Precise control of EpiEffector activity
Tissue- or stage-specific promoters, cre-lox systems and optogenetic actuators can add an additional layer of spatio-temporal precision to EpiEffector activity.
Potential strategies
Ectopic duplication of imprinting architecture.
Defining critical windows for changes in imprinting marks.
Rescue of imprinting deficiency.
Potential tools
Methylation-sensitive EpiEffectors could make allele-specific regulatory changes based on existing germline imprinting control regions.
Using multiple systems, such as CRISPR/Cas9, Cpf1, transcription activator-like effectors and zinc finger nucleotides could increase multiplexing capacity and targeting specificity for constructing complex architecture.
Footnotes
Financial & competing interests disclosure
KDA Rienecker is supported by a Wellcome Trust 4 Year PhD Studentship (reference 105218/Z/14/A). MJ Hill was supported by Neuroscience and Mental Health Research Institute Fellowship. AR Isles has received research funding from GW Pharmaceuticals in the past. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414(6859):122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- 2.Sanz LA, Chamberlain S, Sabourin JC, et al. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008;27(19):2523–2532. doi: 10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies W, Isles AR, Humby T, Wilkinson LS. What are imprinted genes doing in the brain? Epigenetics. 2007;2(4):201–206. doi: 10.4161/epi.2.4.5379. [DOI] [PubMed] [Google Scholar]
- 4.John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81(5):270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2008;647(1):77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2(11):e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins JF, Ubeda F, Van Cleve J. The evolving landscape of imprinted genes in humans and mice: conflict among alleles, genes, tissues, and kin. Bioessays. 2016;38(5):482–489. doi: 10.1002/bies.201500198. [DOI] [PubMed] [Google Scholar]
- 8.Keverne EB. Genomic imprinting in the brain. Curr. Opin. Neurobiol. 1997;7(4):463–468. doi: 10.1016/s0959-4388(97)80023-2. [DOI] [PubMed] [Google Scholar]
- 9.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr. Opin. Endocrinol. Diabetes Obes. 2007;14(1):3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 10.Costa LM, Yuan J, Rouster J, Paul W, Dickinson H, Gutierrez-Marcos JF. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 2012;22(2):160–165. doi: 10.1016/j.cub.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 11.Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction. 2013;145(5):R117–R137. doi: 10.1530/REP-12-0511. [DOI] [PubMed] [Google Scholar]
- 12.Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci. Biobehav. Rev. 2009;33(4):593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest . Nat. Genet. 1998;20(2):163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 14.Isles AR, Holland AJ. Imprinted genes and mother–offspring interactions. Early Hum. Dev. 2005;81(1):73–77. doi: 10.1016/j.earlhumdev.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Dent CL, Isles AR. Brain-expressed imprinted genes and adult behaviour: the example of Nesp and Grb10 . Mamm. Genome. 2014;25(1–2):87–93. doi: 10.1007/s00335-013-9472-0. [DOI] [PubMed] [Google Scholar]
- 16.Garfield AS, Cowley M, Smith FM, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469(7331):534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plagge A, Isles AR, Gordon E, et al. Imprinted Nesp55 influences behavioral reactivity to novel environments. Mol. Cell. Biol. 2005;25(8):3019–3026. doi: 10.1128/MCB.25.8.3019-3026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNamara GI, Isles AR. Dosage-sensitivity of imprinted genes expressed in the brain: 15q11-q13 and neuropsychiatric illness. Biochem. Soc. Trans. 2013;41(3):721–726. doi: 10.1042/BST20130008. [DOI] [PubMed] [Google Scholar]
- 19.Isles AR, Ingason A, Lowther C, et al. Parental origin of interstitial duplications at 15q11.2-q13.3 in schizophrenia and neurodevelopmental disorders. PLoS Genet. 2016;12(5):e1005993. doi: 10.1371/journal.pgen.1005993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tunster SJ, Creeth HDJ, John RM. The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources. Dev. Biol. 2016;409(1):251–260. doi: 10.1016/j.ydbio.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John RM. Engineering mouse models to investigate the function of imprinting. Brief. Funct. Genomics. 2010;9(4):294–303. doi: 10.1093/bfgp/elq010. [DOI] [PubMed] [Google Scholar]
- 22.Charalambous M, Ferron SR, da Rocha ST, et al. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15(2):209–221. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunster SJ, Van De Pette M, John RM. Isolating the role of elevated Phlda2 in asymmetric late fetal growth restriction in mice. Dis. Model. Mech. 2014;7(10):1185–1191. doi: 10.1242/dmm.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L, Szabó PE, Mann JR. Postnatal survival of mice with maternal duplication of distal chromosome 7 induced by a Igf2/H19 imprinting control region lacking insulator function. PLoS Genet. 2010;6(1):e1000803. doi: 10.1371/journal.pgen.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ainscough JF, Koide T, Tada M, Barton S, Surani MA. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124(18):3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- 26.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 27.Jones MJ, Lefebvre L. An imprinted GFP insertion reveals long-range epigenetic regulation in embryonic lineages. Dev. Biol. 2009;336(1):42–52. doi: 10.1016/j.ydbio.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14(16):1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 29.Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S. Use of epigenetic drugs in disease: an overview. Genet. Epigenet. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 31.Laufer BI, Singh SM. Strategies for precision modulation of gene expression by epigenome editing: an overview. Epigenetics Chromatin. 2015;8(1):34–45. doi: 10.1186/s13072-015-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides a comprehensive catalogue of effector domains with potential for sourcing new epigenome editing tools from zinc finger, transcription-activator-like effector and CRISPR/Cas9 systems.
- 32.Vojta A, Dobrinić P, Tadić V, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44(12):5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Elegant example of an EpiEffector capable of targeting somatically heritable DNA methylation.
- 33.Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilton IB, D'Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Important example of an EpiEffector capable of activating transcription of endogenous genes by the enrichment of a representative marker of acetylation (H3K27ac).
- 35.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11(3):198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An example of optogenetic controlled, retargetable CRISPR/dCas9 transactivator that effects sensitive and temporally precise changes in gene transcription.
- 36.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Bio. 2005;350(5):883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 37.Lauc G, Huffman JE, Pučić M, et al. Loci associated with N-Glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 2013;9(1):e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falaleeva M, Surface J, Shen M, de la Grange P, Stamm S. SNORD116 and SNORD115 change expression of multiple genes and modify each other's activity. Gene. 2015;572(2):266–273. doi: 10.1016/j.gene.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang S, Pfiefer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19 . Genes Dev. 2000;14(10):1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 40.Konermann S, Brigham MD, Trevino A, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Novel demonstration of in vivo and in vitro optogenetic control of epigenetic editing which enables causal testing of epigenetic regulatory mechanisms.
- 41.Bashtrykov P, Kungulovski G, Jeltsch A. Correction of aberrant imprinting by allele-specific epigenome editing. Clin. Pharmacol Ther. 2015;99(5):482–484. doi: 10.1002/cpt.295. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun. 2014;5:4240. doi: 10.1038/ncomms5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131(1):29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 44.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. CpF1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fagerlund RD, Staals RH, Fineran PC. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol. 2015;16(1):251. doi: 10.1186/s13059-015-0824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]