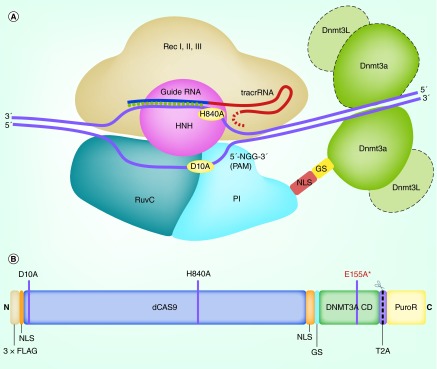
Figure 1. . Anatomy of example CRISPR/dCas9-based EpiEffector: dCas9–DNMT3A.
(A) The dCas9–DNMT3A fusion protein complexes with the sgRNA (composed of the fused crRNA or guide RNA and tracrRNA) to localize the DNMT3A effector domain to the target region. The dCas9 segment is composed of a recognition lobe (Rec I, II and III) and an inactivated nuclease lobe (HNH, RuvC and PI domains). The DNMT3A effector is fused to the PI domain on the nuclease lobe by an NLS and a GS peptide linker. The DNMT3A catalytic domain recruits partners for dimerization to carry out targeted methylation. (B) Linear order of domains on the dCas9–DNMT3A fusion protein. The N-terminal begins with the 3× FLAG epitope tag and the NLS, followed by the nuclease-inactivated dCas9 domain (inactivating mutations D10A and H840A are indicated). dCas9 is followed by a second NLS, and a GS peptide linker which fuse it to the catalytic domain of human DNMT3A. In this domain, E155A indicates the DNMT3A inactivating mutation used as a negative control. The mRNA for this fusion protein also contains a puromycin resistance gene transcript (protein domain-PuroR) or EGFP gene (not shown) for selection of successfully transfected cells. During translation, this selector separates from the EpiEffector when the T2A self-cleaving peptide detaches the fusion protein's C terminal end.
GS: Gly4Ser; HNH: Nuclease lobe cleaving the target strand; NLS: Nuclear localization signal; PI: PAM-interacting.