Abstract
Aberrant epigenomic programming is a hallmark of acute myeloid leukemia. This is partially due to somatic mutations that perturb cytosine methylation, histone post-translational modifications and transcription factors. Remarkably, mutations in the IDH1 and IDH2 genes perturb the epigenome through all three of these mechanisms. Mutant IDH enzymes produce high levels of the oncometabolite (R)-2-hydroxyglutarate that competitively inhibits dioxygenase enzymes that modify methylcytosine to hydroxymethylcytosine and histone tail methylation. The development of IDH mutant specific inhibitors may now enable the therapeutic reprogramming of both layers of the epigenome spontaneously to revert the malignant phenotype of these leukemias and improve clinical outcome for acute myeloid leukemia patients with IDH mutations.
Keywords: : AML, DNA methylation, epigenomics, histone tail methylation, hydroxymethylcytosine, IDH, leukemogenesis, mutation, targeted therapy, transcriptional regulation
Acute myeloid leukemia (AML) is a heterogeneous hematologic malignancy characterized by abnormal hematopoiesis and clonal expansion of immature myeloid cells. As compared with other tumor types AML features a paucity of somatic mutations [1]. Conversely, epigenetic dysregulation and epigenomic reprogramming have emerged as a hallmark of AML and as central mechanisms driving leukemogenesis. Perturbation of the epigenome in AML is at least in part due to specific somatic gene mutations, which can provide critical information about disease mechanism and help guide treatment decisions [2]. A frequent category of genes mutant in AML perturb epigenetic programming through effects on cytosine nucleotides (including, but not limited to 5-methylcytosine and hydroxymethylcytosine, 5mC and 5hmC, respectively) and histone tail modifications, including point mutations of the IDH1 and IDH2 genes [1,3]. Understanding how these mutant proteins work could have fundamental implications for understanding epigenetic mechanisms that can contribute to disease pathogenesis. Excluding acute promyelocytic leukemia [4], there have been a limited number of significant advancements in treatments for AML patients (examples include [5,6]). A better understanding of disease pathogenesis is required to expand upon novel and effective therapeutic approaches needed to improve clinical outcomes. The following review aims to offer insight into the molecular effects and biological downstream consequences of IDH mutations, and address potential implications in AML clinical treatment and outcomes.
Role of IDH1 & IDH2 in cellular functions
IDH1 and IDH2 contribute to generating and shuttling cellular pools of NADPH used as reductive potential in a variety of biological processes. While IDH1 is cytosolic, IDH2 is mitochondrial and functions within the context of the tricarboxylic acid (TCA) cycle. These enzymes reversibly catalyze the oxidative decarboxylation of isocitrate while producing α-ketoglutarate (α-KG), NADPH and carbon dioxide in the forward direction (Figure 1; blue box). These reactions not only facilitate the function of α-KG dependent dioxygenases but also supply NADPH necessary for lipid biogenesis and protection from oxidative and radiation-induced damage [7].
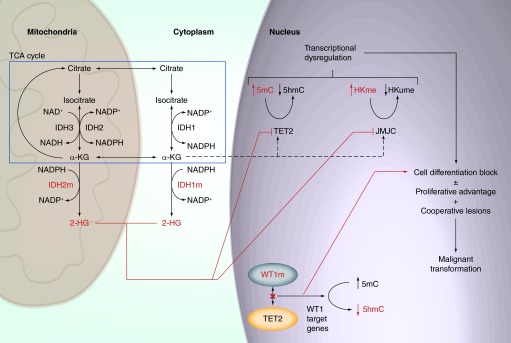
Figure 1. . Overview of the IDH–TET2–WT1 leukemogenic axis.
Mitochondrial and cytosolic IDH enzymes as well as a subset of normal enzymatic steps from the TCA cycle are represented (blue box). In IDH mutant cells, IDH1 and IDH2 neomorphic enzymes (IDH1m and IDH2m) produce the oncometabolite 2-HG at high levels. 2-HG can inhibit the function of dioxygenase enzymes, including epigenetic modifiers (TET2, JMJC). TET2 and JMJC inhibition results in elevated levels of 5mC and histone lysine methylation respectively. These changes result in transcriptional dysregulation, which facilitates the acquisition of proliferative advantage and/or cell differentiation blockade. Malignant transformation can occur in IDHm cells in the presence of cooperative mutations. Mutations in WT1 (WT1m) that disrupt TET2 recruitment to WT1-target genes result in an alternative mechanism for transcriptional dysregulation and cell differentiation blockade.
α-KG: α-ketoglutarate; 2-HG: (R)-enantiomer of 2-hydroxyglutarate; HKme: Methylated histones; hmC: Hydroxymethylcytosine; HMume: Unmethylated histones; IDHm: Mutant IDH enzymes; mC: Methyl-cytosine; TCA: Tricarboxylic acid.
Concurrent with its metabolic role in the TCA cycle, α-KG functions as a central intermediate in glutamine metabolism. Glutamine metabolism can supply a carbon source for cells and facilitate the use of biosynthetic intermediates derived from glucose and the TCA cycle. Through the process of glutaminolysis, glutamine-derived α-KG can be oxidatively metabolized via the TCA cycle into lactate [8]. Alternatively, cells can implement reductive carboxylation in which glutamine-derived α-KG can be converted into citrate. This is in part mediated through reversible IDH1 enzymatic activity in the cytoplasm [9].
Mutant isocitrate dehydrogenase enzymes in malignant disorders
Acquired mutations in IDH genes in malignant disorders were originally reported in glioblastoma multiforme [10]. In AML, somatic mutations of IDH1 were first reported in a normal karyotype AML patient [3]. Studies profiling AML genetics have determined that mutations in IDH1 and IDH2 are highly recurrent. For example the overall incidence of mutations in IDH1 and IDH2 (IDH1/2) in the TCGA cohort was 9.5 and 10%, respectively [1]. IDH1/2 mutations are almost exclusively heterozygous and occur more frequently in AML patients with normal cytogenetics [1,11–13].
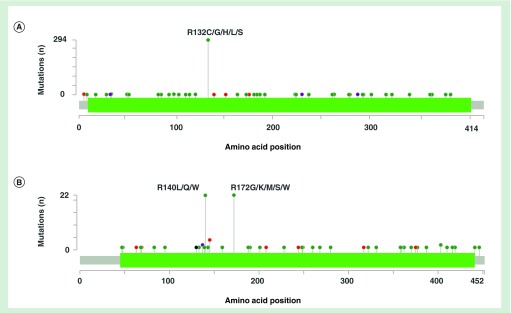
The most frequently detected mutations of IDH enzymes in AML include mutations in DNA codons for Arg132 in IDH1 (IDH1m) and Arg140 or Arg172 in IDH2 (IDH2m) residues (Figure 2A & B). These affect substrate-binding arginine residues within the enzyme catalytic domain [14]. Subsequent studies have identified additional mutations (Figure 2A & B) at codons encoding residues in or near the enzymes’ active site and at other locations [15], however, their functional outcomes are yet to be fully defined. While IDH1m and IDH2m mutations impair the enzymes’ forward catalytic activity by reducing the affinity for isocitrate, they do not cripple enzymatic capacity completely. In fact, these mutations enhance the enzymes’ capacity to catalyze the conversion of α-KG to the metabolite (R)-2-hydroxyglutarate (2-HG) while oxidizing NADPH to NADP+ (Figure 1) [16,17]. This chemical reaction occurs at low levels under normal conditions with wild-type IDH enzymes but is greatly enhanced in cells with mutant forms of IDH1 and IDH2. This is biologically relevant since high serum 2-HG is present in both mouse models of IDHm enzymes [18–20] and in IDH1m and IDH2m AML patients [21].
Figure 2. . IDH1 and IDH2 mutations in malignant disorders.
cBioPortal [22,23] frequency plots of amino acid substitutions resulting from mutations in IDH1 (A) and IDH2 (B) in malignant disorders (all cancer studies included). Green = missense mutations; red = inframe mutations (inframe deletion or insertion); black = truncating mutations (nonsense, nonstop, frameshift deletion, frameshift insertion, splice site); purple = other mutations (all other mutation types).
IDH1 & IDH2 mutations in AML are associated with a dysregulation of DNA cytosine modifications
To better understand the possible role epigenetic dysregulation has in AML, cytosine methylation patterning has been studied in large cohorts of AML patients [1,24–25]. These studies established that aberrant DNA cytosine methylation (DNA methylation) is a hallmark of AML. In a cohort of 344 newly diagnosed AML patients cytosine methylation was explored using HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) microarrays. DNA methylation patterning allowed AML to be classified into epigenetically defined disease subtypes, each of which features specific DNA methylation signatures and distinct clinical outcomes. Some of these epigenetically defined AML subtypes were characterized by dominant hyper- or hypo-methylation signatures while others displayed more balanced changes in DNA methylation levels. IDH1 and IDH2 were not initially sequenced in this cohort [24]. DNA methylation profiling using HELP in a second cohort of 398 newly diagnosed AML patients revealed similar clustering and included sequencing of IDH1 and IDH2 genes. In this cohort patients with IDH1m and IDH2m tended to cluster together based on cytosine methylation patterns. Compared to normal bone marrow controls and AML patients without IDH1m and IDH2m, these groups featured dominant cytosine hypermethylation patterns [25]. The hypermethylation signature was confirmed in studies of AML patients using Illlumina arrays [1] as well as enhanced reduced representation bisulfite sequencing (ERRBS) [26,27]. Mouse models expressing IDH1m recapitulated the hypermethylation signatures. Importantly, the mouse and human signatures overlapped at genes involved in hematopoietic cell proliferation and differentiation, leukemogenesis and leukemic stem cell maintenance [18].
5hmC, originally described as an intermediate during 5mC demethylation [28], is now a recognized epigenetic modification with its own independent effects. 5hmC has been identified as a mark within intergenic regions, including distal regulatory elements (enhancers, CTCF binding sites, DNase hypersensitivity sites), near transcription factor bindings sites, as well as within genic regions [29–36]. Rampal et al. [26] reported the first high-resolution genome-wide mapping of 5hmC patterns in AML patients. Genome-wide 5hmC profiling in IDHm AML patient samples revealed a dominant pattern of focal and regional 5hmC loss compared with nonmutated AMLs, although there was also a smaller subset of focal regions with increased 5hmC compared with normal bone marrow controls.
Based on these data, IDH1/2 mutations likely define a unique subset of AML patients [25,37]. IDH1 and IDH2 mutations were mutually exclusive, and their associated cytosine hypermethylation and hydroxymethylation signatures were highly overlapping [11,25–26], suggesting a common downstream mechanism of action in AML. Likewise, differentially expressed genes in IDH1m and IDH2m patients compared with normal bone marrow controls also showed a significant overlap of gene sets involved in cell cycle, DNA repair processes as well as hematopoiesis [25] suggesting a role in leukemia pathogenesis.
Functional links between IDH1/2 & other mutant genes in AML
TET2
The link between mutations in the genes encoding the metabolic IDH1 and IDH2 enzymes located in the mitochondria and cytoplasm, with cytosine hypermethylation and changes in hydroxymethylcytosine patterning occurring in the cell nucleus, was initially a puzzle. However, the clue to solving this puzzle was the observation that IDH1 and IDH2 mutations are almost mutually exclusive with loss-of-function mutations in TET2 (TET2m) [25,38]. TET2 is a member of the ten-eleven translocation DNA hydroxylase family of enzymes (TET1–3). These enzymes are dependent on α-KG and Fe(II) and catalyze the conversion of 5mC to 5hmC and other intermediate products in the process of DNA demethylation [28,39]. TET2 mutations in AML are almost exclusively heterozygous and occur in 7.6% of patients [38]. AML patients with TET2 mutations tended to cluster with IDH1 and IDH2 mutant cases in DNA methylation profiling suggesting common targets of aberrant cytosine hypermethylation [25]. Furthermore, TET2m AMLs exhibited a dominance of regional 5hmC loss and aberrant gene expression signatures compared with normal bone marrow controls that highly overlapped with those of IDH1 and IDH2 mutant cases [25,26]. Given that TET2 is dependent on α-KG, and that 2-HG produced by IDH1m and IDH2m enzymes is structurally similar to α-KG [17], it was theorized that 2-HG might competitively inhibit TET2. In this way, loss of TET2 might constitute a key target pathway downstream of IDH1 and IDH2 mutations. To determine if TET2 could be affected by this mechanism, 5mC and 5hmC levels were assessed in 293T cells expressing wild-type TET2 and IDH1m. IDH1m expression blocked TET2 from increasing cellular 5hmC levels, resulting as well in accumulation of global 5mC levels [25]. This block was then confirmed to be mediated by 2-HG functioning as a competitive inhibitor [40]. Hence, hypermethylation signatures in IDH1 and IDH2 mutant AMLs are driven at least in part by 2-HG inhibition of TET2 function (Figure 1). However, since the magnitude of hypermethylation in IDHm cases is greater than TET2m AML it cannot be ruled out that part of the IDHm effect could be linked to inhibiting TET1 and TET3.
WT1
Further evaluation of the mutational landscape in AML revealed that mutations in the gene encoding the transcription factor WT1 are also almost mutually exclusive with mutations in IDH1, IDH2 and anticorrelated with mutations in TET2 [11,25–26,41]. DNA methylation profiling in WT1 mutant (WT1m) AML patient samples revealed a cytosine methylation pattern that featured hypermethylation compared with normal bone marrow controls. There was a significant overlap between CpGs affected by aberrant cytosine methylation in WT1m patients with patients harboring IDH1m and IDH2m as well as TET2m. Concordantly, 5hmC profiling in WT1m patients revealed a loss of the mark at a subset of regions identified in IDHm cases that significantly overlapped with those lost in TET2m patients [26].
Given that TET2 and WT1 mutations are nearly mutually exclusive in AML and associate with similar 5mC and 5hmC profiles, a functional interaction was proposed. Targeting WT1 using small hairpin RNAs resulted in a loss of 5hmC in primary murine BM cells. WT1 did not alter 5hmC levels by changing the expression level of TET2. Rather, WT1 could directly interact with TET2, suggesting that these two factors cooperate to regulate transcription [26]. TET2 loss of function resulted in perturbed expression of WT1 target genes [26,41] further supporting the hypothesis for a functional link between these proteins. Indeed TET2 was found to not only be recruited to the target genes by WT1, but to also facilitate enrichment for 5hmC and associate with H3K4me3 activating marks at those loci [41]. Mutant forms of WT1 disrupted the function of TET2 and resulted in reduced 5hmC levels at these loci (Figure 1) [26].
Collectively, these findings suggest that AMLs with IDH1 or IDH2 mutations and loss-of-function WT1 and TET2 mutations form a sub-type of disease. An IDH-TET2-WT1 leukemogenic axis in which either production of 2-HG by IDH1m or IDH2m, TET2 loss of function or WT1 mutations result in disruption of a common set of genes through loss of 5hmC and gain of 5mC (Figure 1). In spite of these clear functional overlaps, it still must be underlined that the epigenomic and biological impact of these somatic mutations are not exactly equivalent to each other. For example, unlike IDH1 and IDH2 mutations, neither TET2 nor WT1 mutations are expected to perturb global histone modification patterns (see next section). It will be critical to perform detailed analyses of methyl-histone distributions in IDH1m and IDH2m cases and their interrelationship with 5mC and 5hmC patterning to fully resolve 2-HG's full epigenetic effects.
IDH1m & IDH2m are associated with changes in histone methylation patterning
Histone tail methylation is associated with both activation and repression of gene transcription depending on the identity of the residue modified and the chromatin context of the modification [42,43]. Jumonji C domain-containing (JMJC) proteins are histone demethylases and similar to TET proteins, JMJC proteins are α-KG-dependent dioxygenases. In vitro enzymatic and cell-based assays established that 2-HG inhibits the function of KDM7A (Histone [H] 3 lysine [K] 9 and H3K27 demethylase), KDM2A (H3K36 demethylase) and KDM5B (H3K4 demethylase) (Figure 1) [40,44]. While AML patient samples have not been interrogated extensively for the effects of IDH1 and IDH2 mutations on histone methylation patterning, IDH2m expression in the cytokine-dependent human erythroleukemia TF-1 cell line, was associated with an increase in H3K4, H3K9, H3K27 and H3K36 methylation [45]. This is in agreement with reports of IDH1m and IDH2m expression in other cell based systems [46,47] and suggests that histone methylation patterning may also be contributing to gene expression aberrations in IDH1/2 mutant AMLs.
Potential mechanisms of aberrant gene expression in IDH1 & IDH2 mutant AML cells
2-HG production in IDH1 and IDH2 mutant AMLs can potentially affect gene expression through several mechanisms. The first is altered patterning of cytosine modifications:
DNA hypermethylation within genes and their associated regulatory elements. While a gain in gene body methylation could facilitate transcriptional activation, increased 5mC at gene promoters could result in repression of gene transcription. Indeed, there is a general inverse correlation between cytosine methylation of gene promoters and gene expression in IDHm patient samples [25]. Changes in 5mC can also impact transcription via the induction of altered occupancy at distal regulatory elements, including enhancers [48]. One example is the DNA methylation dependent chromatin architectural protein CCCTC-binding factor (CTCF). CTCF can function as a transcriptional regulator as well as an insulator between topologically associated chromatin domains [49]. In IDHm glioma cells, CTCF displacement from its insulator binding site upstream of an oncogene (PDGFRA) results in disruption of a topological domain. The resulting juxtaposition of a constitutively active enhancer leads to over expression of PDGFRA which might contribute to the malignant transformation [50]. Another mechanism through which 5mC could impact transcriptional regulation is hypermethylation at enhancers. TET2 loss of function was found to associate with a gain of DNA methylation within enhancers in both an AML mouse model (AML1–ETO expression with TET2 loss of function) as well as within human TET2m AML patient samples. The acquisition of DNA methylation was associated with lower expression of nearby genes [51];
DNA methylation effects on gene splicing. Gene splicing is a central mechanism by which cells can diversify their transcriptional landscape. 5mC levels can regulate binding of CpG methylation specific factors (CTCF and MeCP2) and result in alternative exon inclusion via modulation of RNA-polymerase II elongation rates [52];
Cytosine hydroxymethylation within genes and associated regulatory elements. 5hmC is proposed to contribute to gene expression regulation [33,36]. In AML, 5hmC distribution is perturbed in IDHm, TET2m and WT1m cases compared with nonmutated cases and changes in 5hmC levels at gene body and distal regulatory regions had a positive correlation with gene expression [26]. Hence, in IDHm AML cells, transcriptional repression may be mediated via lower levels of 5hmC.
The second mechanism by which IDHm could impact transcription is via effects on histone modifications [53]. Inhibition of histone lysine demethylases by 2-HG in IDHm cells can result in the accumulation of a number of methylated histone marks with functional consequences. Transcriptional activation could be mediated through H3K4me3 accumulation at gene promoters and bodies. Conversely, transcriptional repression could be mediated through H3K27me3 and/or H3K9me3 accumulation [54,55]. Additionally, H3K36me3 can result not only in transcriptional activation, but also in alternative exon splicing [54,56]. However, a cautionary note about the influence of individual methylated lysines is warranted since the context of other histone modifications present can influence the transcriptional activity (e.g., bivalent domains) [55]. The full functional consequences of histone lysine methylation are yet to be determined in IDHm AML cells, however, accumulation of these marks might have profound effects on transcriptional regulation.
Transcriptional dysregulation could be targeted via interactions between TET2 and specific transcription factors. CCAAT/enhancer binding protein alpha (C/EBPa) and PU.1 are key transcription factors that regulate genes important in the myeloid lineage cell differentiation [57]. C/EBPa is a transcriptional activator of TET2, and transcriptional activation is associated with the acquisition of 5hmC and loss of 5mC at TET2 protein target gene promoters [58]. TET2 can directly interact with the transcription factors PU.1 [59] and WT1 [26,41] and likely cooperates to regulate their target genes. Thus, 2-HG inhibition of TET2 activity could result in aberrant gain of 5mC at both PU.1 and WT1 target gene promoters resulting in gene transcription repression. Interestingly, 5mC patterning in IDHm AML specimens was enriched for disruption of PU.1 binding sites, as well as other hematopoietic regulators such as GATA1 and GATA2 perhaps indicating an additional mechanism through which IDH1 and IDH2 mutations could antagonize transcription factors [25–27].
An alternative mechanism by which 2-HG may affect transcriptional regulation is independent of epigenetic marks. Hypoxia inducible factor (HIF) transcription factors mediate responses to hypoxic stress in normal and malignant tissues. Both HIF1α and HIF1β have been implicated in oncogenesis. EglN family hypoxia-inducible factors are α-KG-dependent dioxygenases, which regulate the HIF transcription factor(s) through targeting for polyubiquitination and subsequent proteosomal degradation. Under normal oxygen conditions, HIF activity and transcriptional activity regulated by HIFs is low, however, under hypoxic conditions which malignant tumors are subjected to, HIF target genes involved in angiogenesis, cell proliferation, and glucose metabolism can be upregulated [60]. Conflicting reports have been published about 2-HG effects on EglN function, some supporting a role for the (S)-2-HG enantiomer in facilitating the competitive inhibition in this case [14,44,61–62]. Further studies are indicated to decipher the full functional impact this mechanism may play in IDHm AML cells.
Are there combinatorial effects of cytosine modifications in IDHm leukemias?
Little is known about the potential for cooperative or opposing effects that 5mC and 5hmC can have at the level of individual genes. In the setting of mutations that can affect the balance of 5mC and 5hmC within specific regulatory elements or genic regions, there is potential for combinatorial regulation of transcriptional activity by these marks. In IDHm AMLs 5mC levels within gene promoters inversely correlate with changes in gene transcription [25,26]. Strikingly, there was a stronger positive correlation between changes in gene expression and 5hmC levels regardless of the genomic region assessed. While there was some overlap between regions with 5mC gain and 5hmC loss in IDHm AML cases, many of the changes localized proximal to or more distal from transcription start sites respectively [26]. While intra-tumor cell heterogeneity may underlie these differences, the patterns detected may truly represent a mixed epigenetic landscape within the same cells. This raises the possibility that 5hmC might have an effect that is both distinct of and potentially cooperative with 5mC depending on the genomic location. The available comparisons between 5hmC and 5mC were made using ERRBS [26], which provides quantitative 5mC levels at a base-pair resolution, but is still a regional approach that does not provide full genomic coverage. Furthermore, the 5hmC profiling was performed using a genome wide enrichment method [26] and quantitative measurements at base pair resolution [30] are still not available for these patients. Hence, additional studies are needed to determine if the two marks can cooperatively regulate specific gene loci.
There are, however, some indications for ways in which 5mC and 5hmC may interact from the functional standpoint. For example, Methyl-CpG binding protein 2 (MeCP2) was originally identified as a 5mC binding protein. It functions in the regulation of gene transcription, chromatin topology and splicing [63,64]. In nerve cells, MeCP2 has been identified as both a 5mC and a 5hmC binding factor [65]. The functional implications for this dual binding capacity in IDH1m and IDH2m AML cells harboring an aberrant balance of the two cytosine modifications are unknown. An altered balance of 5mC and 5hmC might alter the binding kinetics of MeCP2 or its ability to form chromatin modifying complexes and/or topological domains and result in an altered transcriptional landscape. Finally, it is unknown if CTCF occupancy is influenced by 5hmC as opposed to solely being regulated by 5mC at its binding sites. 5hmC was found to be enriched in regions flanking CTCF binding sites [30]. Future studies will determine if an altered balance between 5mC and 5hmC in IDHm cells might also impact CTCF's insulator function.
IDH1/2 mutation role in leukemogenesis
The epigenomic effects of IDH1 and IDH2 mutations appear to play an important role in leukemogenesis. Expression of IDH2 R140Q in primary murine bone marrow cells resulted in inhibition of myeloid differentiation and an increase in the proportion of immature myeloid progenitor cells [25]. Concordantly, expression of IDH2 R140Q or IDH1 R132H in the cytokine-dependent TF-1 human erythroleukemia cell line induced cytokine independent growth and a block in cell differentiation [45,62,66]. In the IDH1 R132H model the phenotype required several passages to manifest, was inducible upon exposure to a cell permeable 2-HG and was reversible upon 2-HG withdrawal. Whether 5mC and/or histone methylation changes were the driver in facilitating the cellular phenotypes was not assessed. However, in a differentiation model of adipogenesis (3T3-L1 cell line), expression of IDH2 R172K resulted in overproduction of 2-HG and increased histone methylation (H3K9me3 and H3K27Me3) prior to the gain of 5mC at gene promoters of transcription factors essential to adipogenesis (Cebpa and Pparg). Furthermore, in immortalized human astrocytes, retroviral transduction with IDH1 R132H resulted in a step-wise gain of histone methylation (12 passages: H3K9me3 and later at 17 passages: H3K27me3 and H3K79Me2) preceding a global gain in 5mC (passage 22) [47]. Collectively these data support the hypothesis that epigenetic mechanisms may contribute to IDHm induced phenotypes, perhaps in a stepwise manner.
A more detailed assessment of phenotype effects of these mutations has emerged from the study of mice engineered to express mutant IDH1 or IDH2. Expression of IDH1 R132H or IDH2 R140Q in hematopoietic cells results in extramedullary hematopoiesis and splenomegaly suggestive of a myeloproliferative disease [18,20]. By 12 months of age, the IDH1m conditional knock-in mice developed reduced bone marrow cellularity. These mice had elevated proportions of hematopoietic stem and myeloid lineage-specific progenitor cells (Lin-/Sca1+/cKit+ cells; LSK cells) in the spleen and bone marrow. Bisulfite sequencing of DNA isolated from bone marrow derived macrophage cells revealed a gain of 5mC. Furthermore, cell lysate immunoblots revealed increased methylation of multiple H3 lysine residues (including but not limited to H3K4me3, H3K79me2 and H3K36me3) in LSK cells as well. The relevance of this model was suggested by the fact that similar sets of genes involved in hematopoietic cell functions (WNT, NOTCH and TGF-β) are hypermethylated in IDHm human AML patients [18]. Similarly, expression of IDH2 R140Q (in tetracycline-inducible transgenic mice) induced expansion of LSK cells, in vitro enhancement of self-renewal capacity and block of erythroid differentiation potentially linked to repression of GATA1 transcriptional activity. The cellular phenotype was reversible upon treatment with AGI-6780, an IDH2 R140Q specific inhibitor, with restoration of more limited self-renewal capacity, and decreased intracellular 2-HG production [20].
In other experiments, overexpression of IDH2 R140Q and IDH2 R172K was achieved using retroviral transfection of cKit+ hematopoietic stem cells. Competitive mouse transplantations using these cells resulted in a differentiation block with decreased myeloid progenitor cells and reciprocal accumulation of immature cells (LSK). While this was not associated with global changes in 5mC or histone methylation, IDH2m overexpression resulted in elevated levels of 2-HG production and a global loss of 5hmC. These mice developed a myeloid disorder, however, with some dysplastic features [67]. Interestingly, transplantation of LSK cells overexpressing IDH2 R140Q using a different retroviral vector (MSCV) resulted in a more heterogeneous set of phenotypes. In this case some mice had no hematological abnormalities, others developed either a myeloproliferative disorder, a more myelodysplastic phenotype, or a B-cell (B220+) or T-cell (CD3+) lymphoma [68]. Indeed IDH mutations have also been reported to occur in humans with certain kinds of lymphomas [69], suggesting that perhaps the levels or timing of IDH mutations might yield perturbation of different hematopoietic lineages.
Collectively, these data suggest a role for IDHm in preleukemic progenitor cell expansion due to enhanced self-renewal and/or differentiation block. Furthermore they show that 2-HG functions as an oncometabolite in the hematopoietic system. However, the range of 5mC, 5hmC and histone methylation changes detected upon IDHm expression in cells raises questions about which epigenetic mark is necessary and/or what order of change in the marks is required for the cellular phenotype changes observed.
IDHm cooperate with other oncogenes to induce leukemogenic transformation
Despite the epigenetic reprogramming and hematopoietic phenotype effects of IDHm, expression of IDH mutant enzymes alone does not cause leukemic transformation in mice. IDHm tend to occur in combination with other mutations in human AMLs [1,11,25]. This suggests that IDH1 and IDH2 mutations must cooperate with other lesions to drive leukemogenesis. Along these lines, IDHm enzymes have been tested for cooperative effects with other leukemia oncogenes. HoxA9/Meis1a over-expressing mice develop a growth factor-dependent oligoclonal acute myeloid leukemia within three months [70]. Secondary transplants of cells overexpressing HoxA9, Meis1a and tetracycline-inducible IDH2 R140Q developed AML within two months. The malignant phenotype was reversible in the majority of mice upon withdrawal of doxycycline, as manifested by elimination of leukemia blast cells and normal hematopoiesis restoration. Similarly, while Hoxa9 expression alone resulted in a latent myeloproliferative disorder without leukemic transformation in mice [70], co-expression of IDH1 R132C in Hoxa9 immortalized mouse bone marrow cells resulted in a short-latency myeloproliferative disorder [19]. Flt3-ITD transgenic mice also develop a myeloproliferative neoplasm but not acute leukemia [71]. Conversely, a compound transgenic mouse of Flt3-ITD and IDH2m R140Q developed both myeloid and lymphoid acute leukemias [20]. Finally, NrasG12D expression in hematopoietic cells results in a latent myeloproliferative disorder [72]; however, co-expression of IDH2 R140Q or R172K resulted in an acute leukemia [67]. Collectively, these data suggest that the cellular and phenotypic aberrancies resulting from IDHm enzymes are insufficient for oncogenic transformation. However, IDHm can cooperate with additional mutations to accelerate disease onset and/or facilitate leukemogenesis.
Therapeutic targeting of IDH mutant enzymes
Direct IDHm inhibitors
Wang et al. [66] and Rohle et al. [73] reported small molecules (AGI-5198 and AGI-6780, respectively) that specifically inhibit two of the most common mutant IDH isotypes (R132H mutant IDH1 and R140Q mutant IDH2, respectively). These were shown to reverse putative IDHm transforming effects, and promote cellular differentiation in leukemia [45,66] and glioma [73] model systems. The IDH1m inhibitor AGI-5198 was able to significantly decrease intracellular 2-HG in glioblastoma cells [73] and intra- and extra-cellular 2-HG levels in in vitro models of chondrosarcoma [74]. Since these initial reports, additional inhibitors have been developed. A distinct IDH1 R132C chemical inhibitor (2-[2-[3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1, 4-dimethylpiperazine) impairs cellular proliferation and induces leukemic cell differentiation in IDH1 mutated cells [75]. Furthermore, a selective IDH1 R132 mutant inhibitor (GSK321) also reduced intracellular 2-HG, was associated with significant reversal of DNA cytosine hypermethylation, and myeloid cell differentiation in human IDH1m AML cells in ex vivo culture conditions. Importantly, GSK321 treatment of IDH1m human AML cells mice xenotransplants resulted in a reduction in leukemia blasts and a relative increase in differentiated cells [76].
Clinical trials of some direct inhibitors are underway for AML patients with IDH1 and IDH2 mutations. Early results from a Phase I clinical trial using a selective IDH2m inhibitor in patients with relapsed AML (AG-221; NCT01915498) showed responses in 41% of patients, including a subset that achieved complete remission. Interestingly, rapid rises in neutrophil cell counts after treatment initiation were detected, suggesting induction of differentiation [77]. An IDH1 selective inhibitor is also in a Phase I clinical trial (AG-120; NCT02074839). Finally, 2HG is being evaluated for biomarker potential (NCT01385150) [78,79].
Indirect targeting of IDHm in AML
Data suggest that IDHm AML cases can also be effectively targeted using indirect therapeutic approaches. BET protein bromodomain inhibitors [80] are one example. The BRD4 inhibitor JQ-1 can suppress the growth of AML cells [81]. JQ-1 is especially effective in IDH mutant AMLs. In the case of IDH2 mutant cells isolated from the NrasG12D/IDH2R172K murine AML model, JQ-1 was effective in facilitating rapid differentiation of AML cells in vitro and in extending median survival of transplanted mice. While Myc activity was inhibited upon JQ-1 treatment, there was no significant change in 2-HG levels suggesting an effect independent to the direct inhibition of the neomorphic enzyme activity [67].
A synthetic lethality screen was an alternative approach utilized to discover potential therapeutic targets in IDHm cells. Using this approach, BCL2 was identified as a top target. Treatment with a BCL2-inhibitor in IDHm cell lines and ex vivo culture of IDHm primary patient AML samples resulted in higher cell death rates compared with nonmutated controls. Furthermore, BCL2-inhibitor administration in mouse xenotransplants of human IDHm AML cells resulted in reduced serial transplantation capacity consistent with a reduced leukemia stem cell activity. These results suggest that BCL2 is a downstream effector of mutant IDH enzymes and a potential therapeutic target in mutated cases [82].
IDH's central role in cell metabolism raises the possibility that the oncogenic effects in IDHm cells may not all result from epigenetic effects. Since IDHm cells have reduced α-KG, they are potentially addicted to glutamine-based metabolism. Indeed, exposure of primary IDH-mutated AML patient samples in vitro to a selective allosteric inhibitor of glutaminase resulted in a significant induction of cell death in IDHm patient samples compared with controls, without significant changes in 2-HG levels [83].
Together, these newly identified inhibitory approaches may offer treatment opportunities in the event that resistance develops to the direct mutant IDH enzyme inhibitors. Further testing is needed to determine if alternative therapeutic targeting approaches will have efficacious impact on 2-HG levels in a de novo AML model and serve as a viable alternative to direct IDHm targeting. Regardless, since IDH2 R172K has been a challenging target to date, these approaches open possibilities for effective treatments in such cases.
Concluding remarks & future perspective
Somatic mutations in IDH1 and IDH2 are recurrent features in AML. While mutations in other enzymes in the TCA cycle have been identified in malignant disorders [84], IDH1m and IDH2m represent the first identified in AML. They results in intra- and extra-cellular elevations of 2-HG and changes in 5mC, 5hmC and histone methylation patterning. Hence, they represent the first somatic mutations that link metabolism and epigenetics in AML. The altered epigenetic landscape likely helps facilitate transcriptional reprogramming in the disease cells through direct effects at gene regulatory elements and via indirect effects on transcriptional factors. Despite the accumulating evidence of 2-HG effects on epigenetic modifiers upon expression of IDH1m or IDH2m enzymes, the primary epigenetic change that drives cellular effects in AML remains largely unknown. Furthermore, how the changes in the different epigenetic marks cooperate to regulate disease-related gene sets in AML is yet to be fully determined. Finally, while 2-HG production has been a focus of study in recent years, the presence of IDH1m/IDH2m results in the depletion and production of additional metabolites [85], whose cellular effects in AML are yet to be characterized. Future studies will determine the full range of cellular mechanisms that facilitate the cellular transformation facilitated by IDH1m/IDH2m.
Despite the gap in knowledge above-noted, in the short time span since the discovery of malignant mutations in IDH1 and IDH2 genes, great progress has been made in developing potential therapeutic agents. Current clinical trials are helping to pave the way to potential effective treatment options in IDH mutant myeloid malignancies. However, data from in vitro models of AML suggest that the acquisition of a pathogenic mutation in IDH1 or IDH2 is insufficient to drive leukemogenesis independently. Furthermore, IDH1 and IDH2 mutations in human AML patients frequently co-occur with mutations in other genes. Together, these findings suggest that it is likely that combination therapies will be required to successfully target and eliminate IDHm disease clones. Regardless, targeting IDH1 and IDH2 gene mutant enzymes offers a welcome opportunity for epigenetic therapy in AML.
Executive summary.
Main characteristics of IDH1 & 2 mutant enzymes
IDH1 and IDH2 are recurrently mutated in acute myeloid leukemia.
Mutations occur in the catalytic domain and result in enhanced production of (R)-2-hydroxyglutarate (2-HG).
IDH1 and IDH2 mutations are associated with a gain of cytosine and histone tail methylation and loss of cytosine hydroxymethylation.
2-HG competitively inhibits dioxygenase enzymes, including Ten-eleven translocation DNA hydroxylase and Jumonji C domain-containing proteins resulting in impaired DNA and histone demethylation respectively.
Role of IDH1 & IDH2 mutant enzymes in leukemogenesis
IDH1 and IDH2 mutant enzyme expression in in vitro and in vivo models results in the overproduction of 2-HG; 2-HG can function as an oncometabolite.
IDH1 and IDH2 mutant enzyme expression results in a block of hematopoietic cell differentiation and expansion of immature cell populations.
Genes targeted by aberrant epigenetic patterning and differential expression compared with controls in IDH1 and IDH2 mutant cells are functional in malignant transformation and hematopoietic cell differentiation.
IDH1 and IDH2 mutant enzymes alone are insufficient to induce a malignant leukemia phenotype; cooperative oncogenes are needed.
Selective IDH1 & IDH2 mutant enzyme inhibitors are under development
Selective inhibitors can reverse cellular phenotypes and 2-HG production in IDH1 and IDH2 mutant cells and in vivo models of acute myeloid leukemia.
AG-221 and AG-220 are direct IDH1 and IDH2 mutant inhibitors that are currently in clinical trials to determine treatment efficacy.
Indirect inhibitors of IDH1 and IDH2 mutant enzymes are being developed and tested for efficacy.
Footnotes
Financial & competing interests disclosure
The authors wish to thank the following sources of financial support: NCI R01CA198089 to AM Melnick, NCI K08CA169055 and funding from the American Society of Hematology (ASHAMFDP-20121) under the ASH-AMFDP partnership with The Robert Wood Johnson Foundation to FEG Bakelman. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides evidence to support that IDH1 and IDH2 genes are recurrently mutated in acute myeloid leukemia (AML).
- 2.Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15(9):e382–e394. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reports first finding of IDH gene mutations in AML.
- 4.Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111(5):2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 5.Stone RM, Mandrekar S, Sanford BL, et al. The multi-kinase inhibitor midostaurin (M) prolongs survival compared with placebo (P) in combination with daunorubicin (D)/cytarabine (C) induction (ind), high-dose C consolidation (consol), and as maintenance (maint) therapy in newly diagnosed acute myeloid leukemia (AML) patients (pts) age 18–60 with FLT3 mutations (muts): an international prospective randomized (rand) P-controlled double-blind trial (CALGB 10603/RATIFY [Alliance]) Blood (ASH Annual Meeting Abstracts) 2015;126(23):6. [Google Scholar]
- 6.Kantarjian HM, Thomas XG, Dmoszynska A, et al. Multicenter, randomized, open-label, Phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J. Clin. Oncol. 2012;30(21):2670–2677. doi: 10.1200/JCO.2011.38.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J. Natl Cancer Inst. 2010;102(13):932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deberardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides evidence suggesting that IDH mutations are recurrent in AML and can be informative in risk classification criteria.
- 12.Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116(15):2779–2782. doi: 10.1182/blood-2010-02-270926. [DOI] [PubMed] [Google Scholar]
- 13.Green CL, Evans CM, Zhao L, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118(2):409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 14.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]; •• Illustrates that IDH mutations are associated with high production of 2-hydroxyglutarate.
- 16.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaturvedi A, Araujo Cruz MM, Jyotsana N, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122(16):2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 20.Kats LM, Reschke M, Taulli R, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14(3):329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides evidence that aberrant DNA methylation is a hallmark of AML.
- 25.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides evidence linking IDH mutations with a hypermethylated DNA phenotype in AML.
- 26.Rampal R, Alkalin A, Madzo J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9(5):1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Illustrates that IDH mutations in AML are associated with changes in hydroxymethylation patterning.
- 27.Akalin A, Garrett-Bakelman FE, Kormaksson M, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 32.Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szulwach KE, Li X, Li Y, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7(6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H, D'Alessio AC, Ito S, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Wu F, Tan L, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2010;28(14):2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaidzik VI, Paschka P, Spath D, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J. Clin. Oncol. 2012;30(12):1350–1357. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 39.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides evidence that 2-hydroxyglutarate is a competitive inhibitor of dioxygenase enzymes.
- 41.Wang Y, Xiao M, Chen X, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell. 2015;57(4):662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 43.Chi P, Allis CD, Wang GG. Covalent histone modifications-miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kernytsky A, Wang F, Hansen E, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125(2):296–303. doi: 10.1182/blood-2013-10-533604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 49.Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen KD, Jia G, Johansen JV, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29(9):910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31(5):274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 56.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 58.Kallin EM, Rodriguez-Ubreva J, Christensen J, et al. Tet2 facilitates the derepression of myeloid target genes during CEBPalpha-induced transdifferentiation of pre-B cells. Mol. Cell. 2012;48(2):266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De La Rica L, Rodriguez-Ubreva J, Garcia M, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14(9):R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adkins NL, Georgel PT. MeCP2: structure and function. Biochem. Cell Biol. 2011;89(1):1–11. doi: 10.1139/O10-112. [DOI] [PubMed] [Google Scholar]
- 64.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Travins J, Delabarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 67.Chen C, Liu Y, Lu C, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27(18):1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mylonas E, Janin M, Bawa O, et al. Isocitrate dehydrogenase (IDH)2 R140Q mutation induces myeloid and lymphoid neoplasms in mice. Leukemia. 2014;28(6):1343–1346. doi: 10.1038/leu.2014.18. [DOI] [PubMed] [Google Scholar]
- 69.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Piloto O, Nguyen HB, et al. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111(7):3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Liu Y, Li Z, et al. Endogenous oncogenic Nras mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood. 2010;116(26):5991–6002. doi: 10.1182/blood-2010-04-281527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, Paz AC, Wilky BA, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS ONE. 2015;10(9):e0133813. doi: 10.1371/journal.pone.0133813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaturvedi A, Cruz MMA, Goparaju R, et al. A novel inhibitor of mutant IDH1 induces differentiation in vivo and prolongs survival in a mouse model of leukemia. Blood. 2014;124(21):3598–3598. [Google Scholar]
- 76.Okoye-Okafor UC, Bartholdy B, Cartier J, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat. Chem. Biol. 2015;11(11):878–886. doi: 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein EM, Dinardo C, Altman JK, et al. Safety and efficacy of AG-221, a potent inhibitor of mutant IDH2 that promotes differentiation of myeloid cells in patients with advanced hematologic malignancies: results of a Phase 1/2 trial. Blood. 2015;123(23):323. [Google Scholar]
- 78.Fathi AT, Sadrzadeh H, Borger DR, et al. Prospective serial evaluation of 2-hydroxyglutarate, during treatment of newly diagnosed acute myeloid leukemia, to assess disease activity and therapeutic response. Blood. 2012;120(23):4649–4652. doi: 10.1182/blood-2012-06-438267. [DOI] [PubMed] [Google Scholar]
- 79.Dinardo CD, Propert KJ, Loren AW, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121(24):4917–4924. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014;13(5):337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 81.Herrmann H, Blatt K, Shi J, et al. Small-molecule inhibition of BRD4 as a new potent approach to eliminate leukemic stem- and progenitor cells in acute myeloid leukemia AML. Oncotarget. 2012;3(12):1588–1599. doi: 10.18632/oncotarget.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan SM, Medeiros BC, Majeti R. BCL-2 inhibition as a synthetic lethal approach to target isocitrate dehydrogenase mutations in acute myeloid leukemia stem cells. Blood (ASH Annual Meeting Abstracts) 2013;122(21):885. [Google Scholar]
- 83.Emadi A, Jun SA, Tsukamoto T, Fathi AT, Minden MD, Dang CV. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp. Hematol. 2014;42(4):247–251. doi: 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 84.Cardaci S, Ciriolo MR. TCA cycle defects and cancer: when metabolism tunes redox state. Int. J. Cell Biol. 2012;2012:161837. doi: 10.1155/2012/161837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl Acad. Sci. USA. 2011;108(8):3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]