Abstract
MicroRNAs (miRNAs) are ∼22 nucleotide non-coding RNAs that regulate gene expression by targeting mRNAs for degradation or inhibiting protein translation. To investigate whether miRNAs regulate the pathogenesis in necrotrophic fungus Rhizoctonia solani AG1 IA, which causes significant yield loss in main economically important crops, and to determine the regulatory mechanism occurring during pathogenesis, we constructed hyphal small RNA libraries from six different infection periods of the rice leaf. Through sequencing and analysis, 177 miRNA-like small RNAs (milRNAs) were identified, including 15 candidate pathogenic novel milRNAs predicted by functional annotations of their target mRNAs and expression patterns of milRNAs and mRNAs during infection. Reverse transcription-quantitative polymerase chain reaction results for randomly selected milRNAs demonstrated that our novel comprehensive predictions had a high level of accuracy. In our predicted pathogenic protein-protein interaction network of R. solani, we added the related regulatory milRNAs of these core coding genes into the network, and could understand the relationships among these regulatory factors more clearly at the systems level. Furthermore, the putative pathogenic Rhi-milR-16, which negatively regulates target gene expression, was experimentally validated to have regulatory functions by a dual-luciferase reporter assay. Additionally, 23 candidate rice miRNAs that may involve in plant immunity against R. solani were discovered. This first study on novel pathogenic milRNAs of R. solani AG1 IA and the recognition of target genes involved in pathogenicity, as well as rice miRNAs, participated in defence against R. solani could provide new insights into revealing the pathogenic mechanisms of the severe rice sheath blight disease.
Keywords: rice sheath blight pathogen, microRNA, milRNA-mRNA interaction, gene expression, pathogenic mechanisms
1. Introduction
Small RNAs (sRNAs), which are derived from double-stranded RNA or hairpin-structured RNA, are non-coding RNAs of 19–30 nt in length.1 Over the last decade, several key studies have demonstrated that sRNAs participate in various cellular processes in many organisms, including DNA damage response, the maintenance of genome integrity, and the regulation of developmental processes.2–4 MicroRNAs (miRNAs) are one of three major classes of sRNAs.4,5 And they are commonly 22 nt in length and play important roles in the post-transcriptional regulation of gene expression in plants and animals.2 Mature miRNAs negatively regulate gene expression via complementary binding to the open reading frame or untranslated region (UTR) of specific target genes. The sRNA pathway has been characterized in several filamentous fungi,6,7 but the miRNA pathway remains poorly understood in most fungal species. Recently, it was reported that at least four types of miRNAs have been identified in Neurospora crassa,3 including a Dicer-dependent miRNA pathway. The production of these miRNA-like small RNAs (milRNAs) does not require quelling deficient-1 (QDE-1, a RNA-dependent RNA polymerase) or QDE-3 (a RecQ DNA helicase) but is dependent on Dicer, QDE-2 (an Argonaute protein), the exonuclease QDE-2 interacting protein, and mitochondrial ribosomal protein L3 (a RNase III domain-containing protein). These differences may indicate that fungal miRNA production evolved independently from that in plants and animals. Recently, deep sequencing technology was applied to investigate milRNA in nine filamentous fungi.8–16 However, neither miRNA nor milRNA has been reported in the plant pathogen Rhizoctonia solani, raising the question of whether miRNAs exist in R. solani.
Rhizoctonia solani, a basidiomycete necrotrophic fungal pathogen, is widely distributed and causes the loss of many economically important plants.17 In addition to rice, the fungus can infect the crops of approximately 50 species, including maize, barley, lettuce, sorghum, and tomato.18–20 As the agent of rice sheath blight disease, R. solani anastomosis group AG1 IA infection results in the most serious economic agricultural losses. Currently, studies of R. solani AG1 IA group classification,17 infection process,21 transcriptome, and genome22 have been reported; however, studies describing the milRNA of R. solani AG1 IA are lacking. Given its economic importance, we examined the regulatory role of miRNAs in host–pathogen interactions. We applied Illumina sequencing to investigate milRNA over six phases of R. solani infection process in rice. The main objective of our study is to explore the potential of miRNAs as pathogenetic factors and to elucidate the possible molecular basis of the host–pathogen interactions of the rice-infecting pathogen to improve the yield of food crops. In addition, R. solani AG1 IA milRNA was also the first miRNA reported in the Rhizoctonia genus, and it may serve as a model for studying pathogenic mechanisms.
2. Materials and methods
2.1. Ethics statement
The human cell lines provided by Shanghai Ying Biotechnology Co., Ltd, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Protocols in this study were approved by the Cell Bank of the Chinese Academy of Sciences. All experiments were performed in accordance with the approved guidelines.
2.2. Fungal strains
The R. solani AG1 IA strain was selected from a heavily infected rice plant at South China Agricultural University and was designated the national standard isolate. The fungal strains were germinated and mycelia cultured on potato dextrose agar (PDA; 200 g of potato, 20 g of dextrose, and 20 g of agar) at 28 °C for 2 days. The mycelia were used for infection and RNA extraction.
2.3. RNA extraction and construction of cDNA libraries
To detect the interactions of rice and R. solani AG1, we collected strains at seven infection stages. The seven stages included mycelium cultured on PDA without plant; mycelium incubated on rice for 10-h; infection cushion incubated on rice for 18-h; after mycelium had invaded the host, with light necrosis appearing after 24-h; heavy necrosis after a 32-h incubation; heavy necrosis after a 48-h incubation and light grey sclerotia on infected plant tissue after a 72-h incubation. We extracted total RNA from these samples using a Fungal RNA Kit (Omega, USA). Next, cDNA libraries were constructed using a TransScript First-Stand cDNA Synthesis SuperMix (TransGen, Beijing, China). sRNA was then extracted using a mirVana™ miRNA Isolation Kit (Ambion, USA), and small cDNA libraries were constructed using a One Step PrimerScript miRNA cDNA Synthesis Kit (TransGen).
2.4. Cloning and sequencing of R. solani AG1 sRNAs
Fractions of sRNA between 15 and 30 nt were extracted from the seven-staged samples on a 15% denaturing polyacrylamide gel. Next, a 5' adaptor and a 3' adaptor were ligated sequentially to 15 µg of sRNA. Then, the short RNAs were converted to DNA by reverse transcription-polymerase chain reaction (RT-PCR) and sequenced on an Illumina GA II machine (BGI, Shenzhen, China). The sequenced short read data were deposited in Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) under the accession number GSE68236.
2.5. Identification of miRNAs
After removing adaptor contaminants and low-quality tags from the raw reads, the remaining sRNA sequences were utilized in further analysis. We compared sRNAs against known non-coding RNAs (i.e. rRNA, tRNA, snRNA, and snoRNA) deposited in the Rfam database23 (version: 10.1) to identify other ncRNAs using BLAST (version 2.2.26). sRNAs belonging to the ‘rRNA, etc.’ class were removed. The prediction of R. solani AG1 IA milRNAs was performed using MIREAP (version 0.2; http://sourceforge.net/projects/mireap) and miRDeep2 software. Both methods identified known miRNAs and novel candidates with a canonical hairpin structure. Some changes in the published workflow were adopted to predict milRNAs. Tags that appeared in at least four libraries were selected. The abundance of each tag in each library was normalized to transcripts per million (TPM), and tags with an abundance >2.5 TPM were used to identify candidate milRNAs. Based on the normalized data, we calculated the Pearson correlation coefficient between milRNAs and performed hierarchical cluster analysis using the hclust function in R based on the average agglomeration method, which was used to analyse the expression pattern of the milRNAs. These candidate milRNAs were also aligned to the R. solani AG1 IA genome sequence (accession AFRT00000000.1).24
2.6. Target gene prediction
Miranda v3.3a25 software was used to predict targets of candidate milRNAs with the following parameters: gap open penalty (−8), a gap extend penalty (−2), score threshold (90), energy threshold (−23) kcal/mol, and scaling parameter (2).
2.7. Identification of rice miRNA and their target genes
During the milRNA prediction as shown above, we had obtained the clean reads without other ncRNAs. These reads were aligned to the hairpin sequences of rice miRNAs in the miRBase database26 using BLASTN. Then, we analysed the reads that were perfectly matched to rice miRNA sequences. And reads in at least three libraries (10-h, 18-h, 24-h, 32-h, 48-h, and 72-h) at more than 2.5 TPM were considered as rice miRNA sequence. We further confirmed that all these reads were not matched to R. solani AG1 IA genome sequences by performing BLASTN alignment. As several miRNA sequences in miRBase were actually the same to each other (such as the same sequences found for osa-miR159a.1 and osa-miR159b), they were considered as one miRNA here. Finally, 23 rice miRNAs were discovered. The target genes of these miRNAs had been reported in the miRBase database and one study by Zhu et al.,27 and these resources were used for analysis in our study.
2.8. Transcriptome
The R. solani AG1 IA transcriptome was detailed in our previous study.22,24 Six transcriptome libraries for plant infection at 10-, 18-, 24-, 32-, 48-, and 72-h were prepared and sequenced. The expected fragments per kilobase of transcript per million fragments (FPKM) values were calculated. The FPKM values of milRNA targets were used to calculate the Pearson correlation coefficient and to perform hierarchical cluster analysis (the hclust function in R).
2.9. Construction of the protein-protein interaction network
The protein–protein interactions (PPIs) were obtained in our previous study28 that identify interactions based on the interolog approach and domain–domain interactions and assess the interactions by gene expression profile, gene ontology (GO) annotation, and the results of yeast two-hybrid assay.
2.10. Analysis of miRNA expression using RT-qPCR
Experimental validation of miRNA-like RNAs was performed using the miRNA Purification Kit (Cwbio, CW0627), miRNA cDNA Kit (Cwbio, CW2141), and miRNA Real-Time PCR Assay Kit (Cwbio, CW2142). Briefly, a poly-A tail was added to the 3' end of total RNA. Then, the RNA was reverse-transcribed with an oligo-dT adaptor. Quantitative PCR (qPCR) was then performed using Synergy Brands green detection with a forward primer for mature milRNA sequences and a universal adaptor reverse primer.29 To calculate the expression levels of the target gene, the 2−△△CT relative quantification method was employed.30 All reactions were repeated three times with three biological samples and were detected by 0.8% (w/v) agarose gel electrophoresis.
2.11. Dual-luciferase reporter assay
This assay system was provided by Shanghai Ying Biotechnology Co., Ltd. We used the psiCHECK-2 dual-luciferase reporter vector, which contained both the synthetic Renilla luciferase gene and the synthetic firefly luciferase gene, each with its own promoter and poly (A)-addition sites.31 The target gene of Rhi-milR-16 was synthesized using XhoI and NotI sites at 5' and 3' ends, respectively, and was inserted between the XhoI and NotI restriction sites in the multiple cloning regions of the Renilla Luciferase 3'-UTR. As a negative control, we designed the following mutated Rhi-milR-16 targeting oligonucleotide: 5'-CGCTAGTTCAGAGTGGACGGTCT-3'. For luciferase assays, HEK293T cells were co-transfected with 100 ng of the reporter plasmid (psiCHECK2-AG1IA_02173) and 100 nm of the mimic. Cells were grown at 37 °C in 5% CO2 for 48 h. Renilla and firefly luciferase activities were measured using the Dual-Glo™ Luciferase Assay System Kit (Promega).
3. Results
3.1. Identification of milRNAs in infection stages
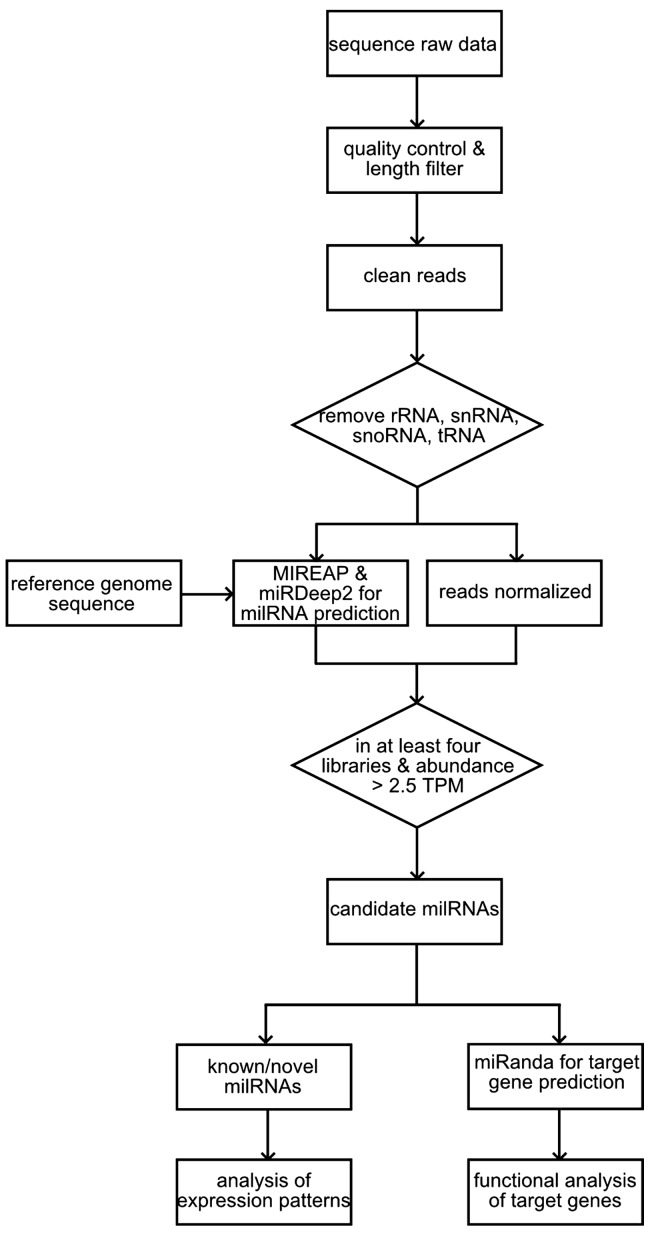
To identify milRNAs in R. solani AG1 IA, we analysed sequencing data collected pre-infection and at six different stages of infection, including 10 h (10-h), 18-h, 24-h, 32-h, 48-h, and 72-h after infection. A total of 77,029,030 clean reads were sequenced. For each library, greater than 1 million unique reads were obtained, at least 67% of which were singletons (Table 1). The lengths of all sequenced sRNAs range from 18 to 30 nt, with the majority distribution on 19–25 nt (Supplementary Fig. S1), which is similar to the standard distribution of sRNAs in plants and animals.2 The composition of the deep sequenced sRNA dataset is very complex and includes a large number of other non-coding RNA species, such as rRNA, tRNA, snRNA, and snoRNA (Supplementary Table S1). Given this complexity and the disproportionate number of singletons in our data that possibly originate from sequencing error, we utilized a method (Fig. 1) originally developed for fungal milRNA prediction with some alterations to the rice miRNA detection workflow.32 In our analysis, reads matched to other non-coding RNA types were removed, and the R. solani AG1 IA genome sequence22 was used as the reference for milRNA detection. Previous research on fungal milRNA detection has indicated that few milRNAs are conserved compared with other eukaryotes.8–15. Two popular and powerful tools, MIREAP and miRDeep2,33,34 can identify fungal milRNAs; however, fewer than 45 miRNAs were found for each of four reported fungi,8,9,13,15 which may be due to the limitation of analytical workflows. However, each method also has its limits of prediction in fungi. Here, we used both MIREAP and miRDeep2 to comprehensively analyse our sequencing data, avoiding their shortages, and identified a total of 177 candidate milRNAs, each identified in at least four libraries at more than 2.5 TPM (Supplementary Table S2). Each of the 177 candidate milRNAs, of which 17 milRNAs were overlapped predicted by MIREAP and miRDeep2, was supported by at least 11 reads in one of the libraries (Supplementary Table S2). Three milRNAs were chosen as examples and are presented in Supplementary Figs S2 and S3.
Table 1.
Summary of R. solani AG1 IA microRNA-like RNA sequencing
| IA | 1-1 | 1-2 | 1-3 | 1-4 | 1-5 | 1-6 | |
|---|---|---|---|---|---|---|---|
| Clean data | 11,141,234 | 11,888,887 | 11,146,287 | 11,196,662 | 10,174,721 | 10,566,997 | 10,914,242 |
| Unique reads | 2,273,938 | 2,020,357 | 1,852,268 | 1,825,295 | 1,083,067 | 1,768,179 | 2,229,097 |
| Singletons | 1,540,073 | 1,478,502 | 1,333,911 | 1,307,798 | 796,228 | 1,240,766 | 1,606,571 |
| Reads length (nt) | 18∼30 | 18∼30 | 18∼30 | 18∼30 | 18∼30 | 18∼30 | 18∼30 |
| Mapped reads | 1,715,684 | 1,542,499 | 1,413,949 | 1,383,724 | 651,796 | 1,057,442 | 1,303,246 |
| Mapped per (%) | 75.45 | 76.35 | 76.34 | 75.81 | 60.18 | 59.80 | 58.47 |
Mapped reads: a count of the unique reads that matched to R. solani AG1 IA genome sequence. IA: library without plant infection; 1-1, 1-2, 1-3, 1-4, 1-5, and 1-6: libraries after infection at 10-h, 18-h, 24-h, 32-h, 48-h, and 72-h, respectively.
Figure 1.
Workflow for the prediction of candidate miRNAs.
3.2. Conserved and novel milRNAs
A comparison of sequences between candidate milRNAs in R. solani AG1 IA and published miRNAs in miRBase,26 detailing milRNAs in other fungi,8–15 revealed that 109 milRNAs exhibited partial sequence conservation (sequence coverage > 50%, identity ≥ 0.88) (Supplementary Table S3). Fourteen milRNAs in R. solani AG1 IA matched 14 miRNA mature sequences belonging to 14 families in miRBase with alignment coverage greater than 70% (greater for Rhi-milR-94; Supplementary Tables S3 and S4). Among the remaining 95 semi-conserved milRNAs, 24 (25.26%) had expression greater than 10 TPM during the infection stages. In addition, 68 novel milRNAs were not conserved among fungi and were exclusively found in R. solani AG1 IA. At least 15 novel milRNAs exhibited expression levels greater than 20 TPM during the infection stages, suggesting their involvement in the pathogenic mechanism of Rhizoctonia. Further study of these milRNAs is needed to improve the understanding of their involvement in the pathogenesis of filamentous fungi.
3.3. Target prediction of milRNAs
Based on the genome sequence of R. solani AG1 IA,22 the 3'-UTRs of 653 genes were predicted to be target regions for 157 milRNAs using Miranda25 (Supplementary Table S5). Targets were not identified for 20 milRNAs, which could be explained by several reasons: they may lack a target, their target may not be annotated in the genome, there may be mismatches between genome and milRNA35 or their targets may be outside of the 3'-UTR.36
Pleiotropic miRNA may target several mRNAs in a given signalling pathway or different mRNAs in converging pathways to exert a large effect on a cell.37 Many of the milRNAs examined in this work regulated multiple mRNA targets. In fact, only 30 (19.11%) milRNAs regulated a single target (Supplementary Table S5). The largest number of targets that we identified for one milRNA (Rhi-milR-135) is 50. Fifteen milRNAs had more than 10 targets, and 103 genes were predicted to be regulated by at least two milRNAs (Supplementary Table S5). Of the 653 targeted genes, 342 (52.37%) have been annotated,22 including transcription factors (TFs), glycoside hydrolases (GHs), haloacid dehalogenase-like hydrolase, ABC transporters, cytochrome P450, E3 ubiquitin-protein ligase, heterotrimeric G proteins, and mitotic checkpoint proteins (Supplementary Table S6). Among them, some of the predicted target factors could be involved in pathogenicity, including cell wall-degrading enzymes and virulence-associated factors in the transduction signal pathway in the Pathogen–Host Interaction (PHI) database.38 Target genes with possible functions in pathogenesis were discovered using GO terms and the EuKaryotic Orthologous Groups, which included pathogenic-related GO classifications (such as hydrolase activity annotated in GH10), large numbers of signal transduction genes, and genes involved in secretion (Supplementary Tables S7 and S8; Supplementary Fig. S4). During infection stages, secreted proteins are typically important factors, which may indicate their involvement in pathogenesis. For example, the expression of AG1IA_00621 (a putative secreted protein) was up-regulated during the infection stages from 10-h to 32-h, while the expression of its milRNA regulator Rhi-milR-91 decreased during these stages (Supplementary Tables S2 and S6).
3.4. Pathogenic protein-protein network regulation
The PPI network of R. solani AG1 IA28 contains the interacting partners of 120 milRNA targets, including 14 PHI genes and 6 secreted proteins (Supplementary Table S9). Among them, some of the genes involved in the cell cycle are contained in one subnetwork (Supplementary Fig. S5), including AG1IA_05392 (a mitotic checkpoint protein), AG1IA_06586 (a g2/mitotic-specific cyclin), AG1IA_03893 (a cell division regulator), AG1IA_00552 (a cyclin-dependent kinase regulatory subunit), and AG1IA_08213 (an exocyst complex protein). AG1IA_08213 regulated by Rhi-milR-48 is involved in the exocyst complex, which plays roles in exocytosis, cell migration, and growth.39 The network interaction between AG1IA_08213 and AG1IA_05392 is regulated by Rhi-milR-115, supporting that AG1IA_08213 is involved in the cell cycle. Two PHI genes—AG1IA_03893 (cell division control) that showed a peak in transcript level at 10-h stage and AG1IA_06586 (a g2/mitotic specific cyclin) that was abundantly expressed during infection process, regulated by Rhi-milR-95 and Rhi-milR-85, respectively—interact with one another, possibly indicating an important role for milRNAs in pathogenesis. In one subnetwork, an ABC-A transporter (AG1IA_05406) interacts with AG1IA_03552 (peptidase), which is subsequently regulated by Rhi-milR-75. Because the AG1IA_05406 homologue (MGG_00937) in Magnaporthe oryzae is essential for formation of the appressoria, a specialized cell of fungal pathogens that is used to infect plants,40 it is possible that AG1IA_05406 plays a similar role in R. solani AG1 IA. This notion is also supported by its expression pattern, which peaks 24-h after infection.22 However, to confirm the function of this gene, further experimental approaches are needed.
AG1IA_05961 is a candidate G protein beta subunit (Gβ), which is an important basic unit of the heterotrimeric G protein-mediated signalling pathway. Heterotrimeric G protein-mediated signalling plays a central role in the virulence, mycelial growth, and development of filamentous fungi.41 In a yeast two-hybrid assay, we explored the protein interactions of a hub (i.e. connected proteins with central roles in network architecture) containing AG1IA_05961 and its nine partners.28 We identified three key milRNA regulators of this hub (Rhi-milR-44, Rhi-milR-120, and Rhi-milR-77) in the transduction signal pathway (Supplementary Fig. S6).
In the ubiquitin ligase (AG1IA_01341) hub, 13 partners, including transporters, actin, ubiquitin, and uncharacterized proteins, were predicted as PPIs. This ubiquitin ligase was predicted to be a potential effector of R. solani AG1 IA during infection stages,22 and Rhi-milR-83 might regulate a key factor in this subnetwork, supporting by more than 2-fold up-regulation of Rhi-milR-83 during 0-h and 18-h stages and the decreased transcription of AG1IA_01341 during infection. In total, functional annotation of milRNA target mRNAs reveals that milRNAs have complex interactions in PPI networks involved in pathogenesis, transcription, signal transduction, and cell cycle regulation.
3.5. milRNA expression patterns during rice infection
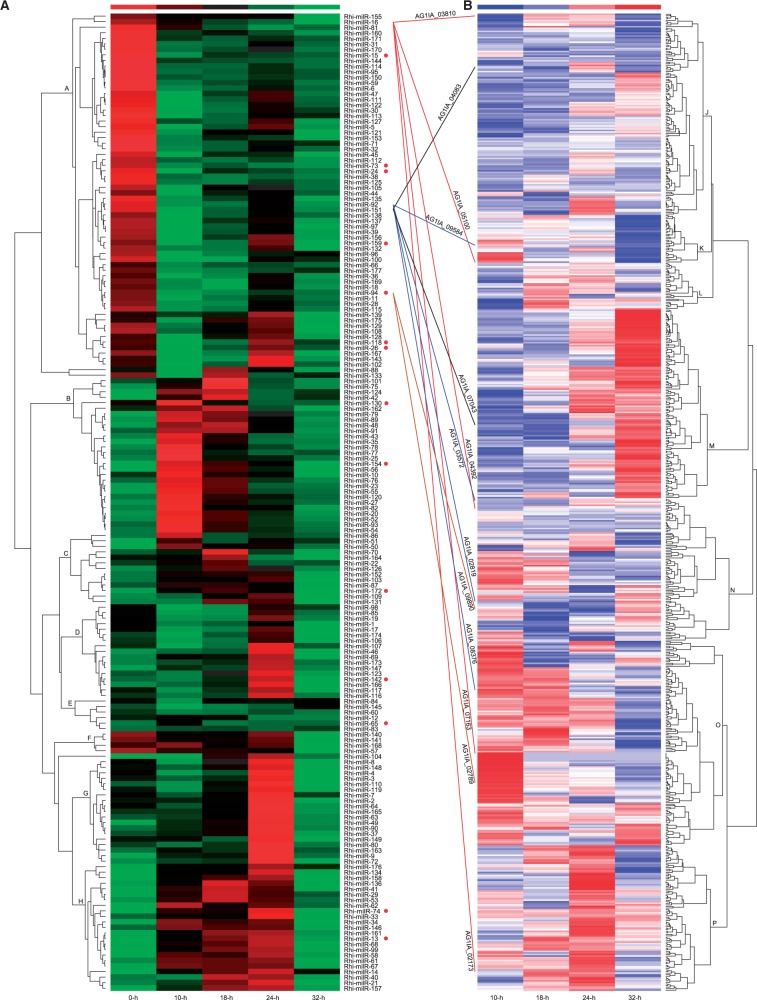
Based on the normalized expression of milRNAs during infection stages, hierarchical clustering (see Materials and methods section) identifies eight major clusters (Fig. 2A). In clade A, expression peaks appear without fungal infection (0-h), and expression declines during the 10-h and 18-h stages. In clades B and C, milRNAs exhibit increased expression at the 10-h and 18-h stages. For clades D, milRNAs display exaggerated expression peaks at 24-h. Different expression patterns are found for milRNAs in clade H, which exhibits increased expression from 10-h to 24-h. milRNAs were found to target 631 genes expressed during infection stages in a previous study22 (Fig. 2B; Supplementary Fig. S7). Different expression patterns for target genes are noted for most targets in clades M, O, and P, which exhibit the highest expression at 32-h, 10-h, and 24-h, respectively (Fig. 2B). Five TFs were clustered into clades J and O, including two fungal-specific TFs (AG1IA_05790 and AG1IA_03810) that display expression peaks at the 18-h stage and one bZIP TF (AG1IA_03425) that displays an expression peak at the 10-h stage. AG1IA_05790 is predicted to be regulated by Rhi-milR-54, Rhi-milR-145, and Rhi-milR-163. In contrast, AG1IA_03810 is regulated by Rhi-milR-16. The peak expression of these genes suggests their involvement in regulating other infection-related genes. One secreted protein (AG1IA_00123, GH9) in clade J is regulated by Rhi-milR-165 and is up-regulated at least 2-fold during 10-h and 18-h stages. The expression peak at 18-h for this GH9 enzyme might help to identify its potential role of cellulose digestion. The fungal ABC transporter G family members are connected to pleiotropic drug resistance phenomena and are related to translocation of phospholipid molecules.42–44 During the infection by R. solani, the rice antimicrobials (such as secondary metabolites and small molecules) can be activated and accumulated in response to the pathogen.45 To facilitate the rice infection, R. solani ABC-G transporters may be contributed to drug resistance. One ABC-G transporter (AG1IA_02225; in clade O) shows the abundance of expression, especially for the peak occurred at 10-h (Supplementary Fig. S7), indicating its important role in pathogenesis, which may also be supported by the down-regulated expression of its putative regulator Rhi-milR-57 during 0-h and 10-h stages. In clade M, the expression of GH6 (AG1IA_08328), which is regulated by Rhi-milR-144, is at its highest during the 32-h stage. Previous research reported that biotrophic fungi lack the cellulose of GH6 and hemibiotrophic and necrotrophic fungi employ it to degrade plant cell wall.46–49 AG1IA_08328 was found to use plant cell walls as a substrate22, suggesting its involvement in the infection process and an important role for Rhi-milR-144. AG1IA_05961 is regulated by Rhi-milR-120 and Rhi-milR-145, and an interacting protein (AG1IA_04857, phosphoglucomutase) is regulated by Rhi-milR-77 (Supplementary Fig. S6A), which may facilitate a greater understanding of the heterotrimeric G protein-mediated signalling pathway in R. solani.milRNA may regulate multiple targets that exhibit different expression patterns, whereas milRNAs with similar expression patterns may regulate different expressed targets. Rhi-milR-16 is predicted to specifically regulate six mRNAs that are regulated by Rhi-milR-16 alone (Fig. 2 and Supplementary Table S5). Among these targets, AG1IA_01440 (hypothetical protein) is not expressed during infection, whereas the others differentiate into five different clusters containing AG1IA_02173 (dynactin), AG1IA_03810 (fungal-specific TF), AG1IA_04392 (dehydrogenase E1 and transketolase), AG1IA_05100 (CE10), and AG1IA_07163 (hypothetical protein) in clades P, J, N, K, and O, respectively. The transcriptome expression patterns of these genes in the five clades are distinct. The different expression patterns of the targets of the three milRNAs (Rhi-milR-92, Rhi-milR-94, and Rhi-milR-151) selected for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) confirmation also yielded similar results (Figs 2 and 3A). To investigate the expression patterns of target genes for the milRNAs forming one cluster, targets regulated by four milRNA clusters were selected for analysis and the diversity of expression patterns were represented (Supplementary Fig. S8), supporting that miRNA-dependent regulatory network is very complex.50 However, considering the peak and valley points of expression for target mRNAs, the milRNA expressions seemed to be correlated with the transcript levels of their possible target genes during early infection stages (Supplementary Fig. S8E), such as: occurrence of 0-h peaks for milRNAs in cluster A and 10-h peaks for targets; 10-h and 18-h peaks for milRNAs in cluster B, and 24-h peaks and 10-h valleys for targets; 10-h and 18-h peaks for milRNA in cluster C, and 24-h peaks and 18-h valleys for targets.
Figure 2.
Expression patterns of milRNAs and transcriptome expression of target mRNAs. (A) Eight major clusters (A-H) are presented for expression of the milRNAs. Fourteen conserved milRNAs are denoted with red circles. (B) The transcriptome expression of 631 target mRNAs known to be regulated by milRNAs is presented, revealing seven major clades (J-P). The following examples of milRNA-mRNA interactions are shown: five mRNAs (AG1IA_02173, AG1IA_03810, AG1IA_07163, AG1IA_05100, and AG1IA_04392) regulated by Rhi-milR-16 (red lines), four mRNAs (AG1IA_02819, AG1IA_03572, AG1IA_08376, and AG1IA_09584) regulated by Rhi-milR-92 (blue lines), two mRNAs (AG1IA_02789 and AG1IA_09890) regulated by Rhi-milR-94 (brown lines), and two mRNAs (AG1IA_04083 and AG1IA_07043) regulated by Rhi-milR-151 (black lines).
Figure 3.

Validation of differentially expressed milRNAs obtained in high-throughput sequencing by RT-qPCR/RT-PCR. (A) The expression of selected miRNAs (Rhi-milR-92, Rhi-milR-94, and Rhi-milR-151) was assayed in the first three stages and normalized to the 18S control gene. All analyses were performed with three biological replicates. Each bar represents the mean ± standard deviation (S.D.). (B) RT-PCR for three novel miRNAs (Rhi-milR-92, Rhi-milR-94, and Rhi-milR-151) providing experimental proof that these milRNAs exist in fungi.
3.6. Confirmation of milRNA differential expression during infection
The purpose of this study was to explore the role of milRNAs involved in host–pathogen interactions. During the process of rice infection, the 10-h and 18-h stages are crucial for successful infection by R. solani AG1 IA. Mycelium rapidly develops on the rice surface at 10-h, and infection cushions form at 18-h after infection. We analysed milRNA expression in the initial three samples (0-h, 10-h, and 18-h) by RT-qPCR and found that the expression of randomly selected milRNAs (Rhi-milR-92, Rhi-milR-94, and Rhi-milR-151) declined at 0–10 and 0–18 h stages, suggesting up-regulated of their targets during the key infection stages (Fig. 3A). The Rhi-milR-92 target gene AG1IA_09584 (hydrolase domain-containing protein) was particularly abundant at 10-h and 18-h. After the pathogen infection, special antimicrobials will be produced by rice in response to the pathogen.45 The expression peak at 10-h for AG1IA_09584 that encodes a member of haloacid dehalogenase-like hydrolase superfamily may indicate its involvement in pathogenic progress by degrading the antimicrobial compounds from rice. The trend in milRNA expression level in the initial three samples was similar to that of the normalized tags (Fig. 3A, Supplementary Table S2). RT-PCR results confirm that these three milRNAs exist in R. solani AG1 IA (Fig. 3B).
3.7. Validation of Rhi-milR-16 targets
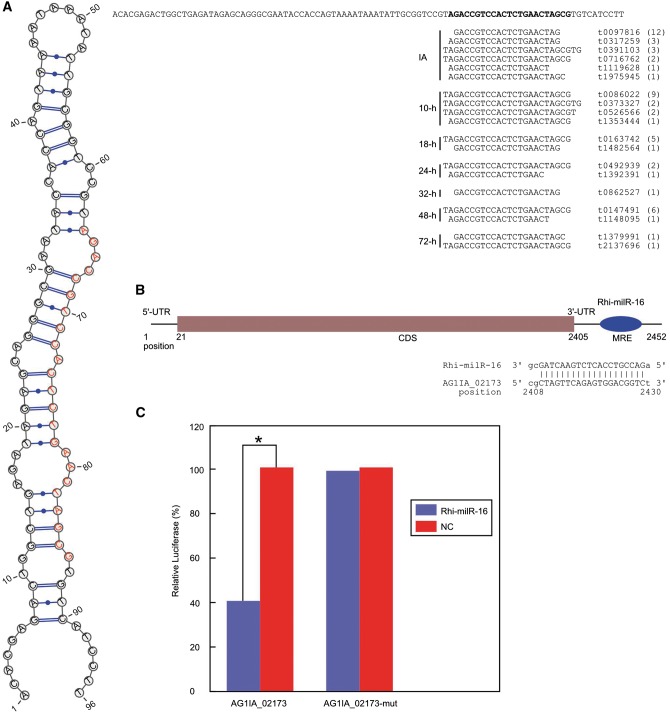
The interactions between a miRNA and its targeted mRNA site(s) are important for understanding miRNA function. The predicted target of Rhi-milR-16 is AG1IA_02173 (Fig. 4A and B). To test the Rhi-milR-16 mimic and its target gene interactions, an exogenous dual-luciferase reporter assay was utilized. HEK293T cells co-transfected with a reporter plasmid (psiCHECK2-AG1IA_02173) and the Rhi-milR-16 mimic decreased the luciferase expression ratio relative to cells transfected with only the psiCHECK-2_AG1IA_02173 reporter. However, the negative control, which contained a mutated Rhi-milR-16 targeting site reporter (psiCHECK2-AG1IA_02173-mut) co-transfected with Rhi-milR-16 mimic (Supplementary Table S10), exhibited the same luciferase expression ratio relative to cells transfected exclusively with the psiCHECK-2_AG1IA_02173-mut reporter (Supplementary Table S11). These results indicate that the Rhi-milR-16 mimic for the psiAG1IA_02173 reporter was inhibitory (P < 0.05) (Fig. 4C, Supplementary Table S12), but the Rhi-milR-16 mimic for the psiAG1IA_02173-mut reporter was not (P > 0.05) (Supplementary Table S12). Therefore, Rhi-milR-16 appears to regulate the predicted target gene AG1IA_02173 (dynactin). Dynactin, an additional multisubunit complex, is required for efficient dynein-mediated transport of vesicles in vitro;51 plays an important role in early endosome motility and apical recycling, which is involved in hyphal tip growth and pathogenicity;52 and results in abnormal hyphal growth and defective in nuclear distribution when mutated.53 This finding suggests that milRNA is associated with the exocytosis, infection, and growth of R. solani AG1 IA.
Figure 4.
Validation of the predicted Rhi-milR-16 target. (A) Rhi-milR-16 secondary structure. (B) The Rhi-milR-16 targeting sequence in the 3'-UTR of AG1IA_02173 mRNA. (C) Rhi-milR-16 targets AG1IA_02173 through a targeting sequence located at its 3'-UTR. Dual-luciferase reporter assays were performed to test the interaction of Rhi-milR-16 and its targeting sequence in the 3'-UTR using constructs containing the predicted targeting sequence and a mutated targeting sequence cloned into the 3'-UTR of the reporter gene. The data represent three independent experiments with three measurements. * indicates P < 0.05.
3.8. Rice miRNAs and their target genes
During the infection, the complex plant defence responses to the broad host-range necrotrophs will be initiated, which may contrast and share mechanisms with biotrophs and is significant to dissect the host–pathogen interactions.54 In rice, several miRNAs are discovered to involve in plant immunity against the hemibiotroph M. oryzae.55 However, no rice miRNA is reported to induce responses to the necrotoph R. solani AG1 IA. We further investigated the sequencing sRNA data and identified 23 rice miRNAs in at least three libraries at more than 2.5 TPM (Supplementary Table S13; Supplementary Fig. S9). Among these miRNAs, two miRNAs (osa-miR164a/b/f and osa-miR166k/l-3p) previously indicated to involve in rice immunity against M. oryzae55 were also identified, and four miRNAs (osa-miR164e, osa-miR396f-5p, osa-miR530-5p, and osa-miR6253) without expression in rice response to M. oryzae (Supplementary Data in study by Li, et al.55) were found in this study, suggesting the multifaceted host resistance mechanisms in rice. For the 59 target genes of these miRNAs, at least of 43 (72.88%) were TFs (such as MYB TFs; Supplementary Table S14) that may regulate responses to necrotrophs. Members of MYB TFs had been reported to mediate responses to necrotrophic fungi with a diversity of virulence strategies through different mechanisms.54 Moreover, other resistance-related genes (such as Os01g02360, a receptor-like kinase) that may contribute to rice defence against R. solani were identified as well. Our analyses provided clues to investigate interactions between rice and R. solani, albeit future research are required to confirm the host resistance mechanisms.
4. Discussion
Over 15,000 miRNAs have been identified in plants, animals, viruses, and some unicellular organisms.2,5,26,56,57 Many studies have reported that fungi encode Dicer-like and Argonaute proteins and that gene expression can be specifically knocked down using RNAi-based methods.58–61 To date, few miRNAs have been reported in plant pathogenic fungi, which could be due to the low accumulation of miRNA or the small number of cells at certain infection stages. Traditional sequencing of sRNAs preferentially identifies abundant miRNAs.62 As a result, less abundant or tissue-specific miRNAs may remain undiscovered in many organisms. Recently, sequencing technologies have allowed the generation of large libraries of sRNAs, which facilitates the identification of less abundant miRNAs.26 Here, a large number of sRNAs from different infection stages of R. solani AG1 were sequenced using high-throughput Illumina sequencing. Our results verified that the plant pathogen R. solani produces miRNAs and identified novel miRNAs in basidiomycetes. Future studies involving the construction of transformants overexpressing or knocking out particular miRNAs in R. solani AG1 will help to confirm their functions. Although the milRNA pathway remains poorly understood, our study provides important evidence for this pathway and broadens our knowledge of the pathogenic mechanism and biology of R. solani at the non-coding RNA level.
The successful identification of miRNAs shows that MIREAP and miRDeep2 are powerful tools33,34 to identify candidate miRNAs based on high-throughput sequencing reads and reference genome sequences. For fungal milRNA discovery,8,9,13–15 fewer than 45 milRNAs were identified in each species from four sRNA sequencing libraries, with the exception of Aspergillus flavus.14 These studies of fungal milRNA8,9,13–15 suggest that the generation of more sequencing data from multiple libraries and the use of suitable prediction workflows should help identify milRNAs. For plant pathogens, the study of milRNAs expressed during infection is lacking. In our work, a method was proposed for fungal milRNA prediction in which the rice miRNA detection workflow was altered, which identified highly credible milRNAs supporting by at least 11 reads in one of the libraries. Sequence comparison indicates that 109 (61.58%) candidate milRNAs share partially conserved sequences with reported miRNAs, suggesting the high accuracy of milRNA prediction by our method as well. Moreover, confirmation of our results by RT-qPCR/RT-PCR supports the accuracy of milRNA identification. The dual-luciferase reporter assay has been used widely to validate human miRNA targets.63 Due to the lack of effective transformation systems and reporter genes for R. solani AG1 IA, HEK293T cells were transfected with a dual-luciferase reporter vector to validate predicted milRNA targets. In future work, we will explore methods to validate the predicted miRNA targets in rice sheath blight pathogen. Moreover, based on expression data obtained during the infection stages and on the pathogenic PPI subnetwork, the expression pattern and character of predicted R. solani miRNAs were revealed. This information could help us to understand pathogenic factors during key infection stages at the systems level. From our analysis, 15 candidate pathogenic milRNAs are identified (Table 2), which is supported by the expression of milRNAs and their target genes, and functions of targets, as well as PHI annotations. Although gene expression was affected by comprehensive factors, such as TFs and alternative splicing, infection-related milRNAs are important pathogenic factors. The results may suggest that the pathogenic mechanisms of milRNAs in the rice sheath blight pathogen are mediated by regulating target ABC-G transporter and hydrolase involved in drug resistance and translocation of various molecules and degrading the antimicrobial compounds from host after penetration respectively, by regulating virulence-associated factors associated with transduction signal pathway, by regulating TFs that may regulate pathogenic factors during infection, and by regulating cell wall-degrading enzymes involving in fungal pathogenicity (Table 2), albeit further research are required to confirm. Our future work will clarify the milRNA pathways and identify critical pathogenic factors. In addition, the diversity of virulence strategies in pathogens corresponds to different host defence response mechanisms,54 which is also supported by the identification of four rice miRNAs that seemed do not involve in rice immunity against M. oryzae55 but expressed during R. solani infection. The target genes of these four miRNAs included TFs that may represent major impacts on rice immunity against the rice sheath blight pathogen. As the mechanisms of interactions between R. solani and rice still remain poor understand, the discovery of rice miRNAs involved in host defence responses to R. solani will facilitate research on pathogen virulence and host resistance.
Table 2.
List of 15 candidate milRNAs involved in pathogenesis
| milRNAs | Clade | Peak | Target Genes | Peak | Description | PHI ID | PHI Description |
|---|---|---|---|---|---|---|---|
| Rhi-milR-95 | A | 0-h | AG1IA_03893 | 10-h | Cell division control | PHI:346 | Reduced virulence |
| Rhi-milR-92 | A | 0-h | AG1IA_09584 | 10-h | Hydrolase domain-containing protein | ||
| Rhi-milR-16 | A | 0-h | AG1IA_03810 | 18-h | Fungal-specific TF | ||
| AG1IA_02173 | 24-h | dynactin | |||||
| Rhi-milR-44 | A | 0-h | AG1IA_00962 | 32-h | G-gamma domain-containing protein | ||
| Rhi-milR-144 | A | 0-h | AG1IA_08328 | 32-h | GH6 | ||
| Rhi-milR-120 | B | 10-h | AG1IA_05961 | 10-h | G-protein beta subunit | PHI:300 | Reduced virulence |
| Rhi-milR-56 | B | 10-h | AG1IA_03425 | 10-h | bZIP TF | ||
| Rhi-milR-54 | B | 10-h | AG1IA_05790 | 18-h | Fungal-specific TF | ||
| Rhi-milR-77 | B | 10-h | AG1IA_04857 | 32-h | Phosphoglucomutase | ||
| Rhi-milR-85 | D | 24-h | AG1IA_06586 | 24-h | g2/mitotic-specific cyclin | PHI:338 | Reduced virulence |
| Rhi-milR-145 | E | 32-h | AG1IA_05961 | 10-h | G-protein beta subunit | PHI:300 | Reduced virulence |
| AG1IA_05790 | 18-h | Fungal-specific TF | |||||
| Rhi-milR-83 | E | 32-h | AG1IA_01341 | 10-h | Ubiquitin ligase | ||
| Rhi-milR-163 | G | 24-h | AG1IA_05790 | 18-h | Fungal-specific TF | ||
| Rhi-milR-165 | G | 24-h | AG1IA_00123 | 18-h | GH9 | ||
| Rhi-milR-57 | F | 0-h | AG1IA_02225 | 10-h | ABC transporter G family | PHI:391 | Unaffected pathogenicity |
The expression peaks of milRNAs and target mRNAs and PHI annotations are shown. Among these targets, AG1IA_00123 is a putative secreted protein.
Authors’ contribution
R.L. and L.H. contributed equally to this work. A.Z. and P.L. managed the project. P.Q. prepared the RNA samples. A.Z. and R.L. designed the analysis. R.L. and A.Z. performed the bioinformatics analysis. Y.W., Z.Y., Q.D., S.L, S.W., W.W., and J.H. participated in milRNA expression and regulation analyses. L.H. performed the experiments. R.L., L.H., A.Z., and P.L. prepared the figures and tables. W.W. and H.L. assisted with preparing the supplementary figures and supplementary tables. R.L. and A.Z. submitted the data to the NCBI. R.L., L.H., and A.Z. wrote the paper. All authors reviewed the manuscript.
Acknowledgements
We thank Erxun Zhou for providing the national standard R. solani isolate AG1 IA from rice, and we acknowledge the Beijing Genomics Institute at Shenzhen for miRNA sequencing of R. solani AG1 IA and Shanghai Ying Biotechnology Co., Ltd for providing the dual-luciferase reporter assay system.
Accession numbers
All data have been deposited at NCBI GEO under the accession code GSE68236.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the National 973 Project (2014CB160304) and the National Natural Science Foundation (31400130).
Supplementary Material
References
- 1.Kim V.N., Han J., Siomi M.C. 2009, Biogenesis of small RNAs in animals, Nat. Rev. Mol. Cell. Biol., 10, 126–39. [DOI] [PubMed] [Google Scholar]
- 2.He L., Hannon G.J. 2004, MicroRNAs: small RNAs with a big role in gene regulation, Nat. Rev. Genet., 5, 522–31. [DOI] [PubMed] [Google Scholar]
- 3.Lee H.C., Chang S.S., Choudhary S., et al. 2009, qiRNA is a new type of small interfering RNA induced by DNA damage, Nature, 459, 274–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghildiyal M., Zamore P.D. 2009, Small silencing RNAs: an expanding universe, Nat. Rev. Genet., 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carthew R.W., Sontheimer E.J. 2009, Origins and mechanisms of miRNAs and siRNAs, Cell, 136, 642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas F.E., Moxon S., de Haro J.P., et al. 2010, Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides, Nucleic Acids Res., 38, 5535–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunes C.C., Gowda M., Sailsbery J., et al. 2011, Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae, BMC Genomics, 12, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R., Jiang N., Jiang Q., et al. 2014, Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum, PLoS One, 9, e104956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q., Wang Z., Zhang J., Meng H., Huang B. 2012, Genome-wide identification and profiling of microRNA-like RNAs from Metarhizium anisopliae during development, Fungal Biol., 116, 1156–62. [DOI] [PubMed] [Google Scholar]
- 10.Jiang N., Yang Y., Janbon G., Pan J., Zhu X. 2012, Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans, PLoS One, 7, e52734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang K., Zhong J., Jiang L., et al. 2013, Identification of microRNA-Like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing, PLoS One, 8, e76288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S.K., Chow W.N., Wong A.Y., et al. 2013, Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei, PLoS Negl. Trop. Dis., 7, e2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J., Fu Y., Xie J., et al. 2012, Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing, Mol. Genet. Genomics, 287, 275–82. [DOI] [PubMed] [Google Scholar]
- 14.Bai Y., Lan F., Yang W., et al. 2015, sRNA profiling in Aspergillus flavus reveals differentially expressed miRNA-like RNAs response to water activity and temperature, Fungal Genet. Biol., 81, 113–9. [DOI] [PubMed] [Google Scholar]
- 15.Yang F. 2015, Genome-wide analysis of small RNAs in the wheat pathogenic fungus Zymoseptoria tritici, Fungal Biol., 119, 631–40. [DOI] [PubMed] [Google Scholar]
- 16.Mueth N.A., Ramachandran S.R., Hulbert S.H. 2015, Small RNAs from the wheat stripe rust fungus (Puccinia striiformis f.sp. tritici), BMC Genomics, 16, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogoshi A. 1987, Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kuhn, Annu. Rev. Phytopathol., 25, 125–43. [Google Scholar]
- 18.Rush M., Lee F. 1992, Sheath blight, In: Gunnell P.S.(ed.) . Compendium of rice diseases, St. Paul, MN: APS Press, pp. 22–33. [Google Scholar]
- 19.Vidhyasekaran P., Ponmalar T.R., Samiyappan R., et al. 1997, Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen, Phytopathology, 87, 1258–63. [DOI] [PubMed] [Google Scholar]
- 20.Yang G., Conner R., Chen Y., Chen J., Wang Y. 2008, Frequency and pathogenicity distribution of Rhizoctonia spp. causing sheath blight on rice and banded leaf disease on maize in Yunnan, China, J. Plant Pathol., 90, 387–92. [Google Scholar]
- 21.Matsuura K. 1986, Scanning electron microscopy of the infection process of Rhizoctonia solarti in leaf sheaths of rice plants, Phytopathology, 76, 811–4. [Google Scholar]
- 22.Zheng A., Lin R., Zhang D., et al. 2013, The evolution and pathogenic mechanisms of the rice sheath blight pathogen, Nat. Commun., 4, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. 2005, Rfam: annotating non-coding RNAs in complete genomes, Nucleic Acids Res., 33, D121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Ai P., Zhang J., et al. 2016, RSIADB, a collective resource for genome and transcriptome analyses in Rhizoctonia solani AG1 IA, Database, 2016, baw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. 2003, MicroRNA targets in Drosophila, Genome Biol., 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data, Nucleic Acids Res., 42, D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q.H., Curaba J., de Lima J.C., Helliwell C. 2012, Functions of miRNAs in rice, In: Sunkar R.(ed.). MicroRNAs in plant development and stress responses. New York, NY: Springer, pp. 149–76. [Google Scholar]
- 28.Lei D., Lin R., Yin C., Li P., Zheng A. 2014, Global protein–protein interaction network of rice sheath blight pathogen, J. Proteome Res., 13, 3277–93. [DOI] [PubMed] [Google Scholar]
- 29.Fiedler S.D., Carletti M.Z., Christenson L.K. 2010, Quantitative RT-PCR methods for mature microRNA expression analysis, In: King N., (ed.) Methods in molecular biology. New York, NY: Springer, pp. 49–64. [DOI] [PubMed] [Google Scholar]
- 30.Livak K.J., Schmittgen T.D. 2001, Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method, Methods, 25, 402–8. [DOI] [PubMed] [Google Scholar]
- 31.Jin Y., Chen Z., Liu X., Zhou X. 2013, Evaluating the microRNA targeting sites by luciferase reporter gene assay, In: Ying S.Y.MicroRNA protocols. New York, NY: Springer, pp. 117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu C., Jeong D.H., Kulkarni K., et al. 2008, Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs), Proc. Natl. Acad. Sci. U. S. A., 105, 4951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedländer M.R., Chen W., Adamidi C., et al. 2008, Discovering microRNAs from deep sequencing data using miRDeep, Nat. Biotechnol., 26, 407–15. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Zhang Z., Liu F., Vongsangnak W., Jing Q., Shen B. 2012, Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis, Nucleic Acids Res., 40, 4298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Q.H., Spriggs A., Matthew L., et al. 2008, A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains, Genome Res., 18, 1456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helwak A., Kudla G., Dudnakova T., Tollervey D. 2013, Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding, Cell, 153, 654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zan H., Tat C., Casali P. 2014, MicroRNAs in lupus, Autoimmunity, 47, 272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban M., Pant R., Raghunath A., Irvine A.G., Pedro H., Hammond-Kosack K.E. 2015, The Pathogen-Host Interactions database (PHI-base): additions and future developments, Nucleic Acids Res., 43, D645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Guo W. 2012, The exocyst complex in exocytosis and cell migration, Protoplasma, 249, 587–97. [DOI] [PubMed] [Google Scholar]
- 40.Gupta A., Chattoo B.B. 2008, Functional analysis of a novel ABC transporter ABC4 from Magnaporthe grisea, FEMS Microbiol. Lett., 278, 22–8. [DOI] [PubMed] [Google Scholar]
- 41.Delgado-Jarana J., Martinez-Rocha A.L., Roldan-Rodriguez R., Roncero M.I., Di Pietro A. 2005, Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways, Fungal Genet. Biol., 42, 61–72. [DOI] [PubMed] [Google Scholar]
- 42.Coleman J.J., Mylonakis E. 2009, Efflux in fungi: la pièce de résistance. PLoS Pathog., 5, e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smriti, Krishnamurthy S., Dixit B.L., Gupta C.M., Milewski S., Prasad R. 2002, ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators, Yeast, 19, 303–18. [DOI] [PubMed] [Google Scholar]
- 44.Kovalchuk A., Driessen A.J. 2010, Phylogenetic analysis of fungal ABC transporters, BMC Genomics, 11, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishi-Kaboshi M., Okada K., Kurimoto L., et al. 2010, A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis, Plant J., 63, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spanu P.D., Abbott J.C., Amselem J., et al. 2010, Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism, Science, 330, 1543–6. [DOI] [PubMed] [Google Scholar]
- 47.Lanver D., Berndt P., Tollot M., et al. 2014, Plant surface cues prime Ustilago maydis for biotrophic development, PLoS Pathog., 10, e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Z., Liu H., Wang C., Xu J.R. 2013, Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics, 14, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Vu B., Itoh K., Nguyen Q.B., Tosa Y., Nakayashiki H. 2012, Cellulases belonging to glycoside hydrolase families 6 and 7 contribute to the virulence of Magnaporthe oryzae, Mol. Plant Microbe Interact., 25, 1135–41. [DOI] [PubMed] [Google Scholar]
- 50.Hausser J., Zavolan M. 2014, Identification and consequences of miRNA-target interactions – beyond repression of gene expression, Nat. Rev. Genetics., 15, 599–612. [DOI] [PubMed] [Google Scholar]
- 51.Schroer T. A. 1996, Structure and function of dynactin, Semin. Cell Dev. Biol., 7, 321–8. [Google Scholar]
- 52.Steinberg G. 2007, On the move: endosomes in fungal growth and pathogenicity, Nat. Rev. Microbiol., 5, 309–16. [DOI] [PubMed] [Google Scholar]
- 53.Bruno K.S., Tinsley J.H., Minke P.F., Plamann M. 1996, Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A., 93, 4775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mengiste T. 2012, Plant immunity to necrotrophs, Annu. Rev. Phytopathol., 50, 267–94. [DOI] [PubMed] [Google Scholar]
- 55.Li Y., Lu Y.G., Shi Y., et al. 2014, Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae, Plant Physiol., 164, 1077–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao T., Li G., Mi S., et al. 2007, A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii, Genes Dev., 21, 1190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu J.Y., Pfuhl T., Motsch N., et al. 2009, Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas, J. Virol., 83, 3333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catalanotto C., Azzalin G., Macino G., Cogoni C. 2002, Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora, Genes Dev., 16, 790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadotani N., Nakayashiki H., Tosa Y., Mayama S. 2004, One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation, J. Biol. Chem., 279, 44467–74. [DOI] [PubMed] [Google Scholar]
- 60.Gong X., Fu Y., Jiang D., Li G., Yi X., Peng Y. 2007, L-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans, Fungal Genet. Biol., 44, 1368–79. [DOI] [PubMed] [Google Scholar]
- 61.Segers G.C., Zhang X., Deng F., Sun Q., Nuss D.L. 2007, Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U. S. A., 104, 12902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartel D.P. 2004, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 116, 281–97. [DOI] [PubMed] [Google Scholar]
- 63.Lee J.Y., Kim S., Hwang do W., et al. 2008, Development of a dual-luciferase reporter system for in vivo visualization of microRNA biogenesis and posttranscriptional regulation, J. Nucl. Med., 49, 285–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.