Abstract
Metabolic and physiological potential evaluator (MAPLE) is an automatic system that can perform a series of steps used in the evaluation of potential comprehensive functions (functionome) harboured in the genome and metagenome. MAPLE first assigns KEGG Orthology (KO) to the query gene, maps the KO-assigned genes to the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional modules, and then calculates the module completion ratio (MCR) of each functional module to characterize the potential functionome in the user’s own genomic and metagenomic data. In this study, we added two more useful functions to calculate module abundance and Q-value, which indicate the functional abundance and statistical significance of the MCR results, respectively, to the new version of MAPLE for more detailed comparative genomic and metagenomic analyses. Consequently, MAPLE version 2.1.0 reported significant differences in the potential functionome, functional abundance, and diversity of contributors to each function among four metagenomic datasets generated by the global ocean sampling expedition, one of the most popular environmental samples to use with this system. MAPLE version 2.1.0 is now available through the web interface (http://www.genome.jp/tools/maple/) 17 June 2016, date last accessed.
Keywords: metagenome, MAPLE, metabolic pathway, global ocean sampling (GOS)
1. Introduction
A primary objective of genomic and metagenomic analyses is to deduce potential comprehensive functions (the functionome) harbored by an individual organism or an entire community in diverse environments. However, evaluation of the potential functionome still remains difficult in comparison with the functional annotation of individual genes or proteins, principally because no standard methodology to extract functional category information, such as metabolism, energy generation, and membrane transport systems, has yet been established. In recent years, detailed and comprehensive functional categories present in the Kyoto Encyclopedia of Genes and Genomes (KEGG)1 and SEED2 databases have been used to identify functional features in comparative genomics and metagenomics represented by the KEGG Automatic Annotation Server (KAAS),3 Metagenomics Rapid Annotation using Subsystem Technology,4 and Metagenome Analyzer5 systems. Each system employs a similarity-based method for functional annotations but utilizes different databases for protein sequences, default threshold values, and ortholog identification (ID) numbers for mapping annotated sequences to functional categories depending on the desired outputs, specifically pathways in KEGG or subsystems in SEED. However, these functional categories still remain too broad to distinguish metabolic or physiological features. Thus, the evaluation of existing tools and development of new tools are required to characterize potential physiological and metabolic pathways in actual ecosystems.6
To resolve this problem, we previously developed a new method to evaluate the potential functionome based on calculating the completion ratio of four types of KEGG modules, i.e., pathways, molecular complexes, functional sets, and signatures,7 represented as the percentage of a module component filled with the input KEGG Orthology (KO)-assigned genes by KAAS. In conjunction with the development of this method, a stand-alone calculation system of the module completion ratio (MCR) has been developed.8 However, it is difficult for general scientists who are not familiar with bioinformatic tools to handle the massive sequence data required for evaluation of potential functionomes in their own data. In addition, it is not easy to visualize the mapping patterns of KO-assigned query genes to the KEGG modules in detail. Thus, in December of 2013, we launched a prototype system, the metabolic and physiological potential evaluator (MAPLE), to automate our newly developed method.9 MCR is an easy-to-understand measure to evaluate the functional potential, and generally, there is a correlation between the completeness of a KEGG module and the likelihood that an organism can perform the physiological function corresponding to the module; however, its interpretation may become difficult when the KOs used for a module are shared with other independent modules, (e.g. the modules such as the reductive TCA cycle and TCA cycle, glycolysis and gluconeogenesis), and the MCR does not necessarily reflect the working probability of each functional module. Moreover, when the module is complete, its abundance becomes a more important measure for comparing the functional robustness among different genomes or metagenomes from diverse environments. Thus, we aimed to improve the MAPLE system to increase the usefulness of our evaluation method and to facilitate the interpretation of the results of functional analyses using this system.
In this study, we present MAPLE version 2.1.0, an improvement of the original MAPLE system that permits the estimation of functional abundance and indicates the working probability of the MCR results by statistical testing. We also highlight the significant differences in abundance and biodiversity of the physiological functions among four metagenomic datasets generated by the global ocean sampling (GOS) expedition with this system.
2. Materials and methods
2.1. Overview
MAPLE is an automatic system for mapping genes in an individual genome and metagenome to the functional modules defined by KEGG and for calculating the MCR and module abundance. Overview of MAPLE system is described in Supplementary text and Figure S1.
2.2. Module completion ratio
The completion ratio of all KEGG functional modules in each sample was calculated based on a Boolean algebra-like equation as previously described.8 The calculation program was slightly modified according to the changes in the Boolean algebra-like equation defined for some modules by the KEGG.
2.3. Calculation of module abundance
The total number of sequence reads assigned to each KO constructing a module was divided by the average length of each KO group for normalization of KO abundance. This normalized KO abundance is described at the lower right corner of each KO box when the mapping pattern of the query genes to the module is displayed. The module abundance is calculated based on the normalized KO abundance. For example, the module M00529, defined as a reaction of denitrification, is composed of four reaction steps (Supplementary Fig. S2). In each K number set, vertically connected K numbers indicate a complex whereas horizontally located K numbers indicate alternatives.8,9 Because the enzyme responsible for the first reaction (nitrate reductase) is composed of four (left side) or two KO complexes (right side), the abundance of the first reaction step becomes 0 unless all KOs vertically connected are filled with the KO assigned genes. When all vertically connected KOs are filled, the minimal value of KO abundance in the vertically connected boxes becomes the abundance at the first step. Thus, the abundance of the first step in module M00529 is 63. As the second step, when two horizontally located KOs are filled with the KO assigned genes, the abundance at the second step becomes 1,129, which is sum of both KOs. The abundance at the third step becomes 56 in a similar manner as the first step, and that of the last step is 46 (Supplementary Fig. S2). Because the module abundance becomes the minimal value among all steps, the abundance of module M00529 is calculated to be 46. The method for comparison of the module abundances among different metagenomic samples is described in Supplementary text.
2.4. Analysis of the Q-value for determining the significance of module completeness
Generally, it is expected that the MCR is linked to the likelihood that the organisms perform the physiological function corresponding to the module. However, when the KOs used for a module are shared with the other modules, the MCR does not necessarily reflect the working probability of each functional module (Supplementary Fig. S3-F). Thus, the relationships among module completion of the targeted module, module completion of other modules to which the same KOs are assigned, and the contribution of specific KOs of each module to module completion should be considered when a module is not complete.8 Namely, even if the same MCR was observed in different modules, the working probability of the physiological function is not necessarily equal among these MCRs. To avoid these problems, we proposed the use of the Q-value for determining the significance of module completion (see Supplementary text). This measure, which represents the probability that a reaction module is identified by chance, is calculated based on the statistics of sequence similarity scores (e.g. E-values) using the concept of multiple testing corrections, according to the definition of the KEGG reaction module (i.e. Boolean algebra-like equation).
2.5. Characterization of the microbial community structure based on ribosomal proteins
Ribosomal proteins are well conserved among all organisms and possess sequences specific to each individual organism; therefore, ribosomal proteins can be used for identification of organisms. Accordingly, we examined how accurately KAAS, which is used in the MAPLE system, can annotate the ribosomal proteins taxonomically. All prokaryotic ribosomal proteins available from the NCBI database (189,020 proteins) were subjected to annotation using MAPLE, and the results were then compared with the original taxonomic annotation. Consequently, since there was no significant difference between the original annotation at the phylum level and those by KAAS, despite the fact that the MAPLE system uses the KEGG database, which contains only completed genome data (Supplementary Fig. S4), we concluded that MAPLE could be effectively applied for identification of organisms by construction of a metagenome based on ribosomal proteins. For application of MAPLE to taxonomic analysis, we calculated the ratio of bacteria and archaea in the metagenome based on the mapping pattern to the module M90000 for prokaryotic ribosomes and the taxonomic annotation of each ribosomal protein. As mentioned above, because the archaeal ribosome, which contains 58 ribosomal proteins, has six more proteins than the bacterial ribosome, we normalized the total number of archaeal ribosomal proteins to the number of bacterial ribosomal proteins by multiplying by 52/58. We summed up the number of bacterial ribosomes and normalized archaeal ribosomes and then used this sum as a denominator to calculate the ratio of archaea and bacteria. We also calculate the ratio of each taxonomic level defined by KEGG, such as phylum and class, in the metagenome using the same method.
2.6. Metagenomic analysis of environmental samples using the MAPLE system
We downloaded the metagenomic sequences generated by GOS expedition10,11 and the environmental data for each sampling site from the iMicrobe database (http://data.imicrobe.us/project/view/26 17 June 2016, date last accessed). Among 83 samples, we selected four metagenomic samples including over 1 million protein-coding genes, two from Sargasso Sea (GS000c and GS000d) and two from near the Galápagos Islands (GS030 and GS031). Detail information about these sites is provided in Supplementary Table S1. Approximately 1.21 million to 1.43 million amino acid sequences in the multi-FASTA format were submitted to MAPLE through the web interface (Supplementary Fig. S1-1).
3. Results
3.1. Comparison of complete modules
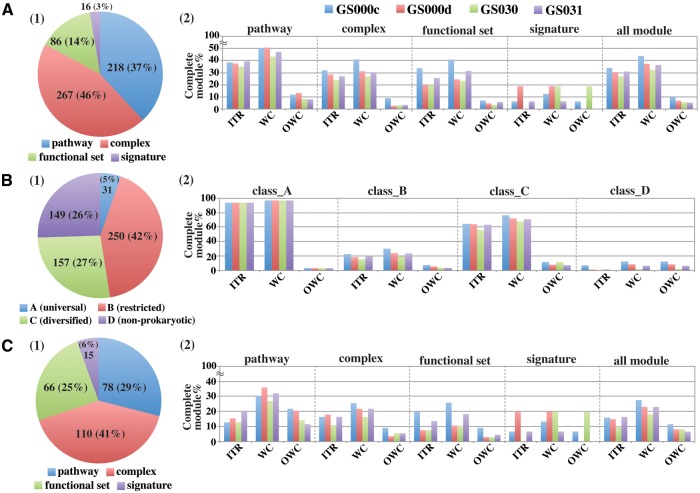
KEGG modules (587 modules as of December 2014) are modular functional units based on the KEGG pathways and are categorized into pathway modules, structural complexes, functional sets, and genetic signatures. We applied metagenomic sequence data from four GOS sites to the MAPLE system based on the KEGG modules and summarized all MAPLE results (Supplementary Table S2). Using the MAPLE results, we first investigated the ratio of complete modules to all KEGG modules to characterize the differences in potential functionomes among the four GOS sites. According to individual taxonomic rank (ITR), the highest ratio of the complete module was found in the GS000c site (33.9%), whereas the lowest ratio (26.8%) was found in the GS030 site (Fig. 1A–2 and Supplementary Table S3). ITR is a second taxonomic subcategory, containing mostly phylum-level information, such as that for Firmicutes, Actinobacteria, and Alphaproteobacteria, as well as class and order information, as defined in the KEGG Organisms database. These values for ITR were 5.3% lower than those for the whole community (WC) containing various taxonomic ranks appearing in GS031 and 9.7% lower than those for the WC in GS000c. Namely, some of incomplete modules by ITR were completed by WC, which is composed of a combination of various taxonomic ranks. We defined the module that was completed only by the WC as the OWC module (Fig. 1 and Supplementary Table S3). A similar trend was observed for the other two sites (GS000d and GS031), and there were approximate differences of 5–7% between the ratios of complete modules by ITR and WC. The GS000c site tended to have a high completion module rate for pathway modules, structural complexes, and functional sets, but not for signature modules, with a particularly remarkable completion rate in the functional set module, which contained functions such as aminoacyl-tRNA biosynthesis, nucleotide sugar biosynthesis, and various two-component regulatory systems (Fig. 1A–2).
Figure 1.
Comparison of the complete module patterns among the four GOS sites. A: Classification of complete modules based on the module type defined by the KEGG database. 1: Ratio of each module type in the KEGG module. 2: Comparison of the ratio of complete modules in each type of KEGG module among the four GOS sites. B: Classification of complete modules by module class based on the MCR patterns of 1,488 prokaryotes. 1: Ratio of each module class in the KEGG module. 2: Comparison of ratios of complete modules in each module class among the four GOS sites. ITR: individual taxonomic rank. WC: whole community, OWC: only whole community.
The KEGG modules primarily fall into four classes (A, B, C, and D) based on distribution patterns of MCRs in all species registered in the KEGG Organisms database.8 Of the 587 KEGG functional modules, only 31 (5%) of modules in prokaryotic species were grouped into class A defined as ‘universal’. All modules grouped into class A were completed by ITR except for module M00526 (lysine biosynthesis, DAP dehydrogenase pathway) but M00526 was not completed even by WC for all GOS sites (Fig. 1B–2). The ratio of complete modules by ITR in class B defined as ‘restricted’ was <20%, except for GS000c, and even those by WC did not exceed 25% for sites GS000d, GS030, and GS031, although the GS000c site had a ratio of more than 30%. Modules over 63% in class C defined as ‘diversified’ were completed by the ITR in GS000c, GS000d, and GS031, and the ratio of complete modules by the WC ranged from 71% to 76% for all GOS sites except GS030. As an overall trend, there were no significant differences in the ratios of complete modules in four categorized classes based on the distribution patterns of MCRs in all classes from A to D (Fig. 1B–2).
Modules completed by <10% of the prokaryotic or eukaryotic species were defined as ‘rare’ in the modules grouped into classes B and C; 269 modules (pathway: 78, structural complex: 110, functional set: 66, and signature: 15) were identified as rare modules in the prokaryotic species (Fig. 1C–1). Generally, few rare modules were completed by the ITR, and the ratio of complete modules was <20% for all GOS sites. However, some incomplete rare modules became complete by the WC, and the highest ratio (∼36%) of complete modules was observed in the pathway modules for all GOS sites (Fig. 1C–2). Class D, which accounted for 26% (149 modules) of all modules, comprised nonprokaryotic modules that were not completed by any single prokaryotic species. Unlike the case of all pathway modules shown in Fig. 1A–2, the GS000d and GS031 sites tended to have higher completion rations for rare pathway modules than the GS000c site.
In the natural environment, the microbial community is mainly composed of various uncultivable microbes, including those that are phylogenetically distant from the registered microbes in the KEGG Organisms database. Thus, the genes derived even from the same microbial genome in the environmental sample are not necessarily assigned to those from the microbes within the same taxonomic rank. This is one of major explanations for the differences in the ratios of complete modules between the WC and ITR, which were observed mainly for the pathway module. Indeed, almost all modules that were completed only by the WC were categorized into class D, with rare modules belonging to classes B and C. Two class D modules (M00013: malonate semialdehyde pathway and M00077: chondroichin sulphate degradation) and two other rare modules (M00358: coenzyme M biosynthesis and M00088: ketone body biosynthesis) were common modules completed only by the WC for all four GOS sites. Module M00013 was completed by organisms within several eukaryotic taxonomic ranks; however, M00077 was completed by microbes within several types of bacterial taxonomic ranks, such as Planctomyces, Firmicutes, and Bacteroidetes, with the exception of GS0030, despite the fact that M00077 was a nonprokaryotic module. Chondroichin sulphate, a sulphated glycosaminoglycan composed of a chain of alternating sugars, is an important structural component of cartilage.12 Thus, several bacterial species may cooperatively degrade chondroichin sulphate derived from the cartilage of marine creatures or some unknown single microbes that are phylogenetically distant from known prokaryotic species.
3.2. Biodiversity and abundance of the complete modules
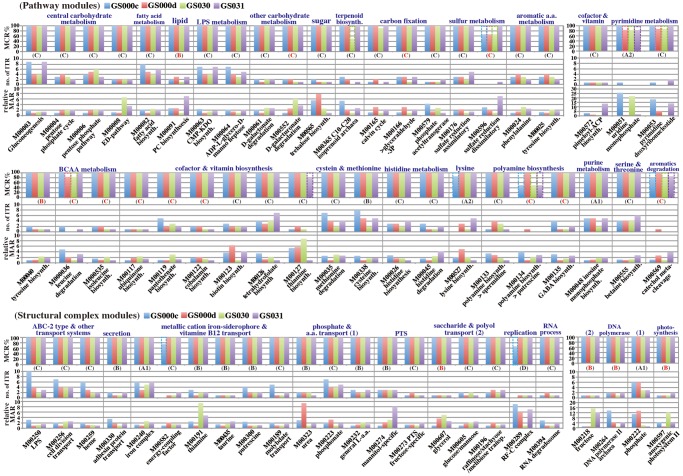
MAPLE highlights the differences in the potential functions of organisms and environmental samples if the MCRs of various modules differ. However, in the first version of MAPLE, we could not clarify the differences in the functional potential of modules commonly completed in all samples. Accordingly, we improved the MAPLE system in order to determine the diversity of the ITRs that completed the module and the module abundance for every ITR (Supplementary Fig. S5). We then characterized the common complete modules among the four GOS sites based on these two measures. Out of 444 KEGG modules (excluding the nonprokaryotic modules), 155 modules were completed by the WC, whereas 136 modules were completed by ITRs. Among 155 complete modules (88 pathway, 55 complex, and 12 functional set modules), there were 42 pathway modules and 24 structural complex modules showing more than 2-fold differences in module abundance ratios among the four GOS sites; these data were normalized according to the average length of each KO and number of ribosomal proteins identified at each GOS site (see Materials and methods; Fig. 2). For example, module M00572, responsible for pimeloyl-ACP biosynthesis related to biotin metabolism, showed a difference of more than 10-fold in the module abundance ratio although the module was completed by only one ITR, Gammaproteobacteria-others, i.e. Gammaproteobacteria excluding the order Enterobacteriales (γ-others) for the four GOS sites. This module, which included six reaction steps, contained two module-specific KOs, K02169 and K02170, and there were substantial differences in the abundances of K02170 between GS031 and the other three sites. Indeed, most of the sequences assigned to K02170 were derived from γ-others, whereas some sequences from Deltaproteobacteria, having MCRs of 83.3%, also contributed to the increase in the total module abundance. To compare the abundance of this module by γ-others among GS000c, GS000d, GS030, and GS031 sites, the module abundance was normalized by the number of ribosomal proteins appearing in each metagenome (see Supplementary Text). The normalized abundances of each site were 0.009 (GS000c), 0.004 (GS000d), 0.003 (GS030), and 0.025 (GS031), respectively, with a ratio of 3:1.3:1:8.3. In contrast, for the structural complex module responsible for the fructose transport system (M00218), there was a 12-fold greater difference in the module abundance ratio between Sargasso Sea sites (GS000c and GS000d) and the other two sites near the Galápagos Islands (GS030 and GS031), despite the fact that the module was completed by only one ITR of Alphaproteobacteria for all sites (Fig. 2). The population of Alphaproteobacteria for the GS000c site was 36%, whereas that of GS000d was almost the same as those from the Galápagos Islands (57–59%). Thus, the abundance of this module did not necessarily depend on the total bacterial population within Alphaproteobacteria, and only limited species within this class presumably contribute to the abundance of this function. In contrast to module M00218, the two Sargasso Sea sites exhibited abundance ratios more than 10-fold higher than those of the other two sites near the Galápagos Islands in module M00222 (phosphate transport system), which was categorized into the universal module (class A). Although most major ITRs contributing to the abundance of module M00222 were from Alphaproteobacteria, this module was also completed by two other common ITRs, γ-others, and Deltaproteobacteria for all sites. However, the diversity of ITRs contributing to the module abundance for the Sargasso Sea sites was greater than that of the sites near the Galápagos Islands, and other ITRs, such as Thaumarchaeota, Firmicutes-Clostridia (Firmicutes-C), Chloroflexi, and Cyanobacteria, also contributed to the total module abundance. Besides these modules, four other modules classified in functional categories, such as pyrimidine metabolism (M00051 and M00053), photosynthesis (M00597), and DNA polymerase (M00264), showed 10-fold greater differences in module abundance ratios among the four GOS sites.
Figure 2.
Comparison of biodiversity contributing to the complete module and relative module abundance in the pathway and structural complex modules among the four GOS sites. The upper histogram shows MCR patterns of the KEGG module. The bars outlined by dashed lines indicate the MCRs of the WC. Uppercase characters A to D under the upper histogram show the module class used in Fig. 1, and ‘R’ indicates rare modules. The middle histogram shows the number of ITRs that completed the functional module. The lower histogram shows the relative module abundance ratio (MAR). The MAR was calculated by dividing abundance of each module by the minimum abundance among the four GOS sites. The modules showing 2-fold greater differences in the MAR among the four GOS sites are represented.
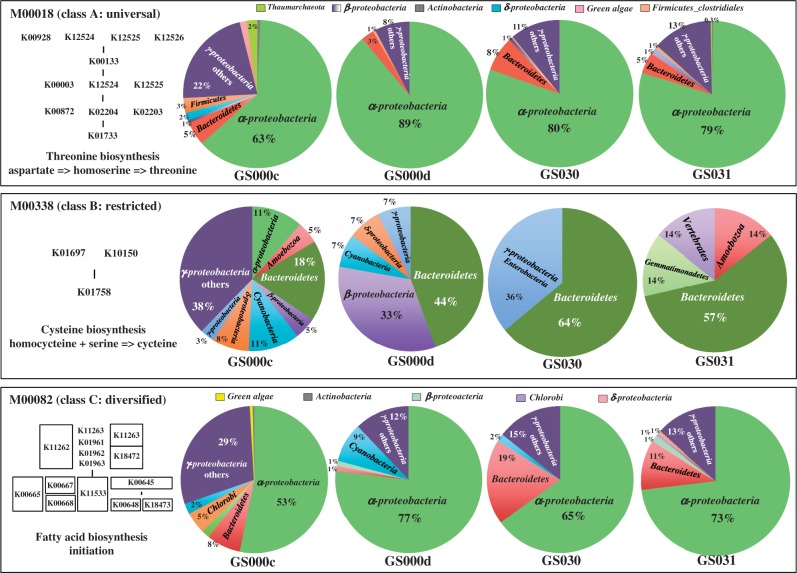
M00018 (threonine biosynthesis), which was categorized into the universal module (class A), was completed by various ITRs; however, there were no substantial differences in module abundance ratios among the four GOS sites. As shown in Fig. 3, the population patterns of ITRs contributing to the module abundance were similar for the four GOS sites, with the exception of GS000c. Alphaproteobacteria accounted for 80% or more of the module abundance in GS000d, GS030, and GS031, and after adding the second and third major ITRs, i.e. Gammaproteobacteria and Bacteroidetes, more than 98% of the modules were accounted for. With respect to GS000c, the ratio of Alphaproteobacteria was low (63%), whereas that of Gammaproteobacteria for the second major ITR was more than two times higher than that of the other three sites. The remaining 15% was nearly evenly occupied by various ITRs, such as Bacteroidetes, Firmicutes-C, Deltaproteobacteria, and Thaumarchaeota. Modules M00338 and M00082, which were categorized as restricted (class B) and diversified (class C) modules, respectively, did not show significant differences in module abundance ratios, except for GS000d, which had a lower module abundance than the other sites (Fig. 2). Similar to the case of module M00018, the patterns of a variety of ITRs, including those that completed the module M00082 (fatty acid biosynthesis-initiation), and the module abundance ratio of each ITR were similar for GS030 and GS031 but obviously different for GS000c and GS000d, despite the fact that Alphaproteobacteria was the major ITR contributing to module abundance for both sites (Fig. 3). In contrast, with respect to M00338 (cysteine biosynthesis), various ITRs and the module abundance ratios differed completely from each other. However, Bacteroidetes, one of the major contributors to the module abundance, was shared among the four GOS sites. For example, this module was completed by eight types of ITRs in GS000c but only by two types of ITRs in GS031, and two eukaryotic ITRs (Vertebrate and Amoebozoa) accounted for 14% of the total module abundance (28% in total) in GS030. In GS000c, two ITRs (γ-others and Alphaproteobacteria) that never appeared at the other three sites accounted for 38% and 11% of the total module abundance, respectively.
Figure 3.
Comparison of abundance ratios of the modules completed by ITRs. Three typical pathway modules categorized into classes A (universal), B (restricted), and C (diversified). There were no substantial differences in relative module abundances among the four GOS sites selected for comparison of the contributions of ITR patterns to the module abundance.
3.3. Evaluation of the MCR according to the Q-value
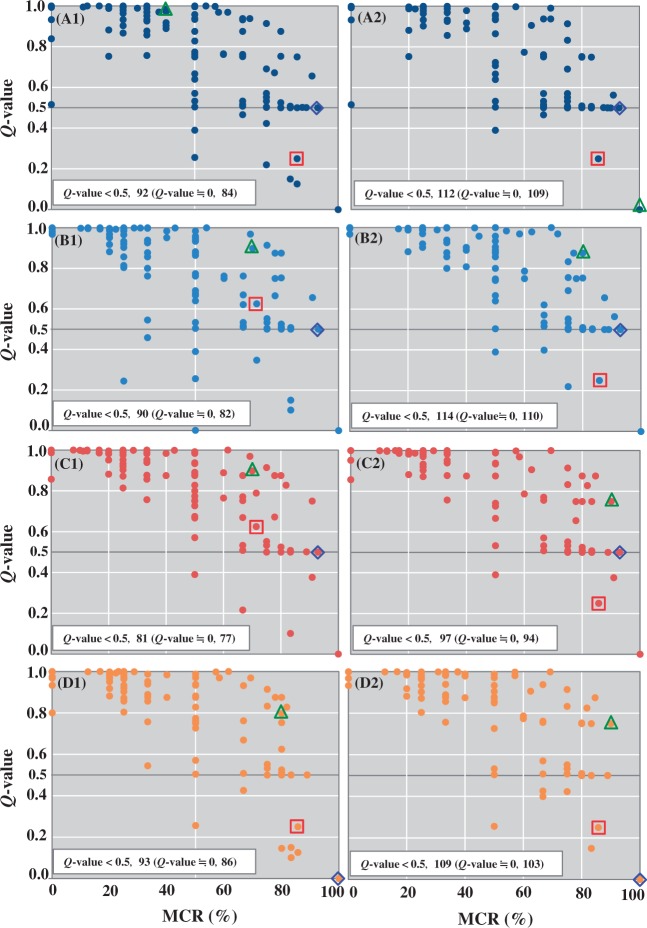
As mentioned in the section Analysis of the Q-value for determining the significance of module completeness, the Q-value is useful because the MCR has limitations. We here investigate the relationship between MCR and Q-value in each module for the four GOS sites, in order to show the advantage of Q-value.
We found higher MCRs were generally associated with lower Q-values, and the Q-value was almost 0 when the MCR was 100%. However, the Q-value is not necessarily negatively correlated with the MCR. For example, we considered the module M00532 (photorespiration). This module was not completed by the WC for the GS000d, GS030, and GS031 sites; however, the MCR was higher than 80% (Fig. 4). Notably, the Q-value of this module was high for each site (0.875 for GS000d and 0.75 for both GS030 and GS031; Supplementary Table S2). In this module, which comprised 10 reaction steps, the KOs assigned to seven steps (steps 2–5 and 8–10) were module-specific, sites GS030 and GS031 lacked K14272 in step 5, and GS000d lacked K03781 and K14272 in step 4. Moreover, module M00373 (ethylmalonyl pathway related to glyoxylate and dicarboxylate metabolism) was not completed by the WC at sites GS000c, GS000d, and GS030, although the MCR was 92.9%. Half of the 14 reaction steps included in this module were module-specific, and these three sites lacked only one KO, K14451, which was assigned to the last module-specific step in this module. According to the definition, the Q-value emphasizes the completion of module-specific KOs for evaluating the working probability; thus, even though a high MCR value is shown, the Q-value is high when the module-specific KO is missing.
Figure 4.
Comparison of MCR and Q-value patterns of pathway modules by ITRs and WCs among the four GOS sites. (A) GS000c, (B) GS000d, (C) GS030, and (D) GS031. 1: ITR, 2: WC. Triangle: photorespiration (M00532), diamond: ethylmalonyl pathway (M00373), square: C5-isoprenoid biosynthesis (M00095). Number of modules with the Q-value <0.5 in ITR and WC is shown in each sample. Among those with the Q-value <0.5, the number of modules with that of nearly zero is shown in the parentheses. The number of modules with the Q-value <0.5 in WC is somewhat larger than that in ITR in all GOS sites.
Although module M00095 (C5 isoprenoid biosynthesis in the mevalonate pathway) was not completed in the WC, the MCR was high (85.7%) for the four GOS sites, similar to those of the modules M00532; however, the Q-value of this module was low (0.25). There are four module-specific reaction steps (steps 3–6) in the module comprising seven reaction steps, and the KO assigned to step 5 was missing for all GOS sites, similar to the results observed for the module M00532. However, there was a notable difference in the module structure of the missing reaction step; that is, reaction step 5 consisted of alternative genes (KOs), i.e. isozymes. According to the definition, the Q-value becomes lower when more isozymes are available; thus, it is low in such a case, even if module-specific KOs are missing.
3.4. Microbial community structure based on ribosomal proteins
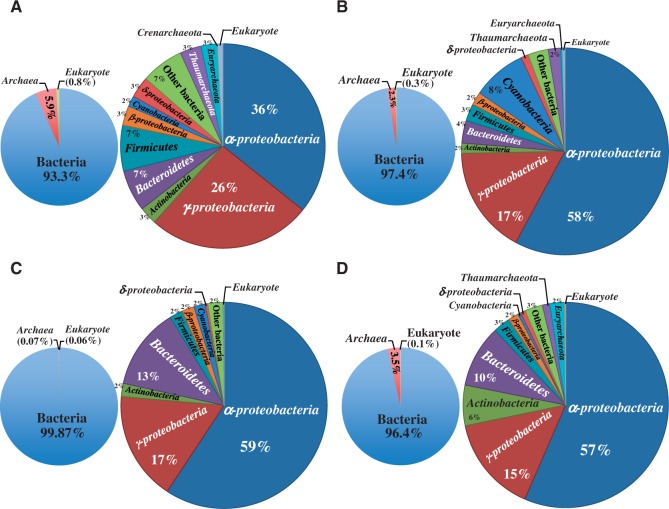
To compare the microbial community structure among the four GOS sites, we analysed the taxonomic composition at the phylum level based on the ribosomal proteins comprising prokaryotic and eukaryotic ribosomes (see Materials and methods). As shown in Supplementary Table S1, because filters with a pore size of 0.1–0.22 µm were used to recover the cells from seawater collected at the four GOS sites in the previous studies,10,11 after prefiltration with filters having a pore size of 0.8 µm, most of the recovered cells were expected to be prokaryotic. As expected, more than 99% of the microbial community was composed of prokaryotes for all GOS sites, particularly the GS030 site; bacteria accounted for 99.9%, whereas archaea accounted for ∼2–6% at the other three sites (Fig. 5). Although the ratio of archaea at site GS000c (5.9%) was higher than that at other sites (GS000d: 2.3%, GS030: 0.07%, and GS031: 3.5%), the archaeal population in the microbial community was generally very low.
Figure 5.
Microbial community structure for the four GOS sites based on ribosomal proteins. (A) GS000c, (B) GS000d, (C) GS030, and (D) GS031.
The major bacterial taxon at all GOS sites was Alphaproteobacteria; the proportion of Alphaproteobacteria within the WC was ∼60%, except for at site GS000c, where the proportion was only 36%. Conversely, site GS000c exhibited the highest proportion of Gammaproteobacteria (26%), which was the second most common taxon for all sites. Another major difference between sites GS000c and GS000d was the proportions of Firmicuctes and Bacteroidetes; for both organisms, the proportion tended to be higher at site GS000c than at site GS000d. In contrast, the population of Cyanobacteria was four times higher in GS000d (8%) than in GS000c, despite the similarities in environmental and sampling conditions of these two sites.10 With respect to the other two sites near the Galápagos Islands, there were no substantial differences in bacterial proportions, with the exception that the proportion of Actinobacteria (6%) was three times higher at site GS031 than at site GS0030, whereas almost no archaea were detected at site GS030.
4. Discussion
The original MAPLE system has been used for metagenomic analysis of the human gut microbiome13 and for comparative functional analysis of individual microbes, even within candidate phyla.14,15 There is generally a correlation between the completeness of a KEGG module and the likelihood that an organism or microbial community performs the physiological function. However, the MCR does not necessarily reflect the working probability of each functional module because the KOs used for each module are shared between other independent modules (e.g. between TCA cycle [M00009] and the reductive TCA cycle [M00173], and between glycolysis [M00001] and gluconeogenesis [M00003]). Thus, we introduced the Q-value parameter to the MAPLE system to statistically re-evaluate the working probability based on the MCR, and several modules, which were deduced to have a higher working probability, were suggested based on the Q-values of the incomplete modules in this study. The Q-value is also helpful for evaluating the working probability of the functional module in the individual genome, and modules with lower Q-values, such as glycolysis (MCR: 90%, Q-value: 0.281) and gluconeogenesis (MCR: 85.7%, Q-value: 0.375), could be completed by manual curation in the genome of Ca. C. subterraneum.15 Thus, the Q-value is thought to be a good suggestive index to determine the working probability when the functional modules are not completed in the metagenome or in individual genomes.
In addition to the Q-value, we also introduced the concepts of biodiversity and the abundance of complete modules for more detailed analyses of the complete modules because we could not distinguish the properties of complete modules shared between the different environmental samples using the original MAPLE system. To demonstrate the usefulness of the new MAPLE system, we used four metagenomic sequence datasets generated by the GOS expedition. When we compared the abundance of each functional module among the four GOS sites, we used the module abundance normalized by the sum of ribosomal proteins constructing the prokaryotic ribosome module (M90000). Generally, most environmental metagenomic analyses target prokaryotes, and very few eukaryotic sequences (<1%) were detected in the metagenomic sample in this study. In such cases, it is easy to normalize the module abundance according to the total number of prokaryotic ribosomal proteins; however, when eukaryotic sequences are frequently detected, eukaryotic ribosomal proteins also should be considered for normalization of the module abundance. The eukaryotic ribosome is composed of 79 ribosomal proteins, including two accessory proteins; importantly, 33 KOs corresponding to the ribosomal proteins are shared between archaea and eukaryotes, similar to the case of bacteria and archaea. Thus, when the number of eukaryotic ribosomal proteins is calculated, we have to consider this point.
In many metagenomic analyses, 16S rRNA gene sequences obtained by polymerase chain reaction (PCR) amplification are used to compare microbial community structures among different environments.16 In recent studies, this PCR-based amplicon approach has targeted the V4 region because different regions of the 16S rRNA gene have been shown to yield varying degrees of accuracy in taxonomic assignments.17 However, prokaryotic species exhibit variations in the copy number of the 16S rRNA gene, and it is impossible to determine the copy numbers of individual uncultivable, unknown microbes present in actual microbial communities. Thus, because taxonomic compositions based on amplicon sequences are strongly influenced by copy numbers in addition to basic PCR bias, this approach is not useful for the analysis of microbial community structure. On the other hand, a new method based on universal single-copy genes, which provide prokaryotic species boundaries at higher resolution than 16S rRNA gene, has been used to estimate relative abundances of known and currently unknown microbial community members with metagenomic data at species-level resolution.18 However, community structure analysis at such high resolution is not necessarily required in metagenomic analyses of natural environments, unlike that of the human gut microbiome, because many community members have not yet been cultivated or identified at the species level. Thus, community structure analyses at the phylum or class level based on ribosomal proteins using the new version (version 2.1.0) of the MAPLE system are thought to be feasible for analysis of metagenomes from the natural environments. Although the sequence lengths generated by Illumina HiSeq and MiSeq sequencers (at most 400–500 bp as an assembled contig of the paired-end sequences) are shorter than the Sanger sequences used in this study, we have already confirmed that there are no discernible differences in KO ID assignment and mapping ratios of KO-assigned coding sequences for the KEGG modules in the metagenomic sequences of the human gut microbiome when comparing Sanger and Illumina reads.8,19 Finally, user’s guide of MAPLE is described in the Supplementary Data.
Supplementary Material
Acknowledgements
We wish to thank H. Uehara of SGI Japan Ltd. for assistance with optimization of the computer environment. We also thank Dr H. Huang of Chuo University for technical assistance.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported in part by grants from the collaborative research program of the Institute for Chemical Research, Kyoto University to H.T. and S.G. (grants #2013-23 and #2014-24). This work was also supported in part by KAKENHI to H.T. and K.T. (No. 26550053) and by a grant from the Cross-ministerial Strategic Innovation Promotion Program to H.T. and W.A.
References
- 1.Kanehisa M., Goto S. 2000, KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overbeek R., Begley T., Butler R.M., et al. 2005, The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res., 33, 5691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriya Y., Itoh M., Okuda S., Yoshizawa A., Kanehisa M. 2007, KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res., 35, W182–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer F., Paarmann D., D’Souza M., et al. 2008, The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics, 9, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. 2011, Integrative analysis of environmental sequences using MEGAN4. Genome Res., 21, 1552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filippo C.D., Ramazzotti M., Fontana R., Cavalieri D. 2012, Bioinformatic approaches for functional annotation and pathway inference in metagenomic data. Brief Bioinform., 13, 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M., Araki M., Goto S., et al. 2008, KEGG for linking genomes to life and environment. Nucleic Acids Res., 36, D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takami H., Taniguchi T., Moriya Y., Kuwahara T., Kanehisa M., Goto S. 2012, Evaluation method for the potential functionome harbored in the genome and metagenome. BMC Genomics, 13, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takami H. 2014, New method for comparative functional genomics and metagenomics using KEGG modules In: K. Nelson. ed., Encyclopedia of Metagenomics. Springer-Verlag, Berlin, Heidelberg, Germany, pp. 525–39. [Google Scholar]
- 10.Venter J.C, Remington K., Heidelberg J.F., et al. 2004, Environmental genome shotgun sequencing of the Sargasso Sea. Science, 304, 66–74. [DOI] [PubMed] [Google Scholar]
- 11.Yutin N., Suzuki M.T., Teeling H., et al. 2007, Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol., 9, 1464–75. [DOI] [PubMed] [Google Scholar]
- 12.Baeurle S.A., Kiselev M.G., Makorava E.S., Nogovitsin E.A. 2009, Effect of counterion behavior on the frictional-compressive properties of chondroitin sulfate solutions. Polymer, 50, 1805–13. [Google Scholar]
- 13.Obregon-Tito A.J., Tito R.Y., Metcalf J., et al. 2015, Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun., 6, 6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoemaker W.R., Muscarella M.E., Lennon J.T. 2015, Genome sequence of the soil bacterium Janthinobacterium sp. KBS0711. Genome Announc., 3, e00689–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takami H., Arai W., Takemoto K., Uchiyama I., Taniguchi T. 2015, Functional classification of uncultured “Candidatus Caldiarchaeum subterraneum” using the MAPLE system. PLoS One, 10, e0132994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tringe S.G., Rubin E.M. 2005, Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet., 6, 805–14. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Lozupone C., Hamady M., Bushman F.D., Knight R. 2007, Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res., 35, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mende D.R., Sunagawa S., Aeller G., Bork P. 2013, Accurate and universal delineation of prokaryotic species. Nat. Methods, 10, 881–4. [DOI] [PubMed] [Google Scholar]
- 19.Qin J., Li R., Raes J., et al. 2010, A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.