Abstract
Ehrlichia chaffeensis is an obligate intracellular tick-borne bacterium which causes the disease, human monocytic ehrlichiosis. Ehrlichia chaffeensis contains only two sigma factors, σ32 and σ70. It is difficult to study E. chaffeensis gene regulation due to lack of a transformation system. We developed an Escherichia coli-based transcription system to study E. chaffeensis transcriptional regulation. An E. coli strain with its σ70 repressed with trp promoter is used to express E. chaffeensis σ70. The E. coli system and our previously established in vitro transcription system were used to map transcriptional differences of two Ehrlichia genes encoding p28-outer membrane proteins 14 and 19. We mapped the -10 and -35 motifs and the AT rich spacers located between the two motifs by performing detailed mutational analysis. Mutations within the -35 motif of the genes impacted transcription differently, while -10 motif deletions had no impact. The AT-rich spacers also contributed to transcriptional differences. We further demonstrated that the domain 4.2 of E. chaffeensis σ70 is important for regulating promoter activity and the deletion of region 1.1 of E. chaffeensis σ70 causes enhancement of the promoter activity. This is the first study defining the promoters of two closely related E. chaffeensis genes.
Keywords: gene regulation, intracellular bacteria, Anaplasmataceae
1. Introduction
Human monocytic ehrlichiosis (HME) is caused by the tick-borne pathogen Ehrlichia chaffeensis.1 HME is considered an emerging infectious disease in the USA and is also reported from several other parts of the world.2 HME is an acute flu-like illness with symptoms including fever, headache, myalgia, anorexia and chills and is frequently accompanied by leukopenia, thrombocytopenia, anemia, and upgraded levels of serum hepatic aminotransferases.3 Similarly, several other Anaplasmataceae family pathogens, including the genera Ehrlichia and Anaplasma, have been identified in recent years as the causative agents of important emerging diseases in people and various vertebrate animals.3–5 The limited availability of genetic tools to study obligate intra-phagosomal pathogens impacted our understanding of the molecular mechanisms of pathogenesis and the pathogen’s prolonged persistence in vertebrate and tick hosts.6–8 Host-specific differences in the gene expression of E. chaffeensis are also reported,9,10 but it is entirely unknown how the organism accomplishes such changes in gene expression.
Transcriptional regulation in prokaryotes is accomplished by the action of RNAP holoenzyme. RNAP holoenzyme is a multi-protein complex composed of two alpha (α) subunits, two beta (β) subunits and a sigma (σ) factor.11 Promoter specificity for an RNAP is accomplished by the inclusion of a sigma factor. Ehrlichia chaffeensis genome contains only two sigma factor genes; rpoD (ECH_0760) (the predicted primary housekeeping σ70 gene) and rpoH (ECH_0655) (the predicted alternate σ32 gene) (GenBank # NC_007799.1).1 Both σ32 and σ70 are conserved in most proteobacteria and share extensive similarity at the amino acid level.12 Transcription from a gene promoter by an RNAP typically involves the recognition of and binding to two DNA motifs located upstream from the transcription start site (TSS) of a gene; the motifs -10 and -35, which is a common occurrence for many bacteria.13,14 The -10 motif interacts with the 2.3–2.4 region of a σ70 to bind RNA polymerase,15–19 while the -35 motif is known to interact with the conserved 4.2 region.20–22 Recent studies in Escherichia coli suggest that the spacer sequences located between the -35 and -10 motifs also contribute to transcription initiation and regulation.23–25
We recently mapped the promoters of several E. chaffeensis genes by performing in vitro transcription studies using the RNAP containing recombinant E. chaffeensis sigma factors.26,27 RNA polymerase binding motifs of E. chaffeensis gene promoters are highly homologous for its only two sigma factors, σ32 and σ70. The gene expression in this bacterium can also be accomplished by either of the two factors, but with varying affinities for different gene promoters.27 We reported that the E. chaffeensis outer membrane protein genes encoding for p28-Omp14 and p28-Omp19 proteins (Ech_1136 and Ech_1143, respectively) are transcribed predominantly by σ70. Our initial studies revealed that only the -35 motifs, but not -10 motifs, are required for transcription for these two genes.26 The transcriptional assessment of E. chaffeensis genes requires additional investigations to define the contributions of the pathogen sigma factors for RNAP function, as prior studies were carried out with E. coli RNAP. Such studies are a challenge due to the lack of appropriate molecular tools for this organism.
Most of the current knowledge of bacterial gene regulation comes from studying the gene regulation of E. coli. Such knowledge is severely limited for other Gram-negative bacteria and more importantly, it is unclear how intracellular pathogens, such as E. chaffeensis, regulate gene expression to overcome the host stress. In the current study, we developed an E. coli-based promoter mapping system to study functions of two genes and validated the data using the in vitro transcription system. We took advantage of a previously developed E. coli strain in which the endogenous rpoD gene expression is controlled by the repressible trp promoter.28 In this E. coli, we complemented E. chaffeensis σ70 after suppressing its native σ70. This system was then used to systematically map sequence determinants spanning from the -10 to -35 motifs of two differentially expressed genes recognized primarily by E. chaffeensis σ70. Together, the study allowed us to test the function of E. chaffeensis σ70 and its ability to regulate target genes. In view of the lack of a transformation system in E. chaffeensis and in other related tick-borne intracellular rickettsial pathogens, the assessment of Ehrlichia transcriptional machinery in the surrogate E. coli system along with the validation experiments carried out by in vitro transcription assays offer innovative means in studying gene expression in E. chaffeensis and other important intracellular rickettsial pathogens belonging to the Anaplasmataceae family.
2. Materials and methods
2.1. Escherichia coli strains and plasmids
Escherichia coli strains used in this study were TOP10 (Invitrogen Technologies, Carlsbad, CA), BL21(DE3) (Novagen, San Diego, CA) and CAG20177.28,29 Several plasmid constructs used in this study were obtained from a commercial source or modified from one or more of the existing plasmids. They include pET32a (Novagen) and the derivatives of pSAKT32,30 pQF50K30 and pMT504.31 Genetic makeup of the plasmids described in this study was included in Supplementary Table S1, except those obtained from a commercial source. The plasmid pSAKT32 containing a p15A origin of replication and an ampicillin resistance gene has E. coli rpoH gene under the control of IPTG inducible Plac promoter.30 The E. coli rpoH from this plasmid was replaced with the E. chaffeensis rpoD (Ech_rpoD) gene by digesting the plasmid with Afl II and Sal I, blunt ending the digested fragments with Klenow DNA polymerase (BioLabs, Ipswich, MA), and then ligating with the Ech_rpoD sequence. Ech_rpoD segment was generated by PCR from plasmid pET32-Ech_rpoD32 using Pfu DNA polymerase (Promega, Madison, WI). The modified plasmid is referred to as the pSAKT32-Ech_rpoD. Ehrlichia chaffeensis rpoD variants with substitutions within the 4.2 region of σ70 were constructed by mutagenesis using a QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA). Ehrlichia chaffeensis rpoD variant with deletion of 1.1 region of σ70 was also generated in it by using Q5 Site-Directed Mutagenesis Kit (New England Biolab, Inc, Ipswich, MA). The names of the modified pSAKT32-Ech_rpoD are provided in Supplementary Table S1.
The pQF50K plasmid with a pMB1 origin of replication and with a kanamycin resistance gene contains the β-galactosidase coding sequence (lacZ) driven by E. coli groE promoter.30 The groE promoter in the plasmid was replaced with E. chaffeensis p28-Omp14 or p28-Omp19 gene promoters by employing directional cloning by taking advantage of existing restriction sites with the plasmid surrounding the insertion. The E. chaffeensis promoter segments were generated by PCR using Pfu DNA polymerase (Promega, Madison, WI). The promoter plasmids are referred to as pQF50K-p28-Omp14 and pQF50K-p28-Omp19, respectively. Mutations with deletion of -10 or -35 motifs of the promoters were generated from these plasmids using Q5 Site-Directed Mutagenesis Kit (New England Biolab, Inc, Ipswich, MA). Site directed mutagenesis at every nucleotide of the -35 motif of the promoters was also generated from the plasmids using the Quick-change Multisite Mutagenesis Kit (Agilent Technologies, La Jolla, CA). Mutations to modify the AT rich spacer sequence of the p28-Omp14 promoter were generated by modifying the pQF50K-p28-Omp14 plasmid using Q5 Site-Directed Mutagenesis Kit. The names of all engineered plasmids are listed in Supplementary Table S1. Mutagenic oligonucleotides were described in the Supplementary Table S2.
The expression plasmids of E. chaffeensis wild-type (WT) σ70 or its variants were constructed for preparing purified recombinant proteins using the E. chaffeensis σ70 plasmid reported earlier.32 Ehrlichia chaffeensis σ70 variants within the 4.2 region of E. chaffeensis σ70 were constructed by modifying the plasmid pET32a-Ech_rpoD by using a QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA) and the modified expression constructs were then used to prepare modified recombinant proteins. The names of the modified pET32-Ech_rpoD are provided in Supplementary Table S1.
For in vitro transcription analysis, E. chaffeensis promoter segments of p28-Omp14 and p28-Omp19 or their mutants were cloned in front of the G-less casette of pMT504 plasmid at the EcoR V site to serve as transcription templates.31 The constructs with various mutations at -35 motif for the p28-Omp14 and p28-Omp19 promoters were generated by PCR using the -35 motif mutant-specific plasmids in pQF50K as the templates from the respective gene promoter plasmids. The lengths of transcripts for the various promoter segments of p28-Omp14 and p28-Omp19 genes are 162 nucleotides. Integrity of all cloned segments in the plasmid constructs was confirmed by automated DNA sequence analysis using CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA).
2.2. Escherichia coli growth conditions and β-galactosidase assays
The CAG20177 E. coli strain alone or with the recombinant plasmids was grown as described earlier.28 Briefly, cultures were grown at 37 °C in Luria–Bertani medium with chloramphenicol (30 μg/ml) plus indole-3-acrylic acid (IAA) (0.2 mM) to maintain expression of endogenous σ70. To express E. chaffeensis σ70 from plasmid pSKAT32-Ech_rpoD or its derivatives, E. coli CAG20177 strain containing the plasmid were grown with ampicillin overnight along with the IAA and chloramphenicol then diluted 1:100 into a fresh medium containing the same antibiotics, but without IAA to suppress the E. coli σ70 and to induce the expression of WT E. chaffeensis σ70 or its derivatives. Due to the leaky expression from the lac promoter, E. chaffeensis σ70 expression was adequate to sustain the bacterial growth in the absence of IPTG. Accordingly, all experiments were carried out without adding IPTG. To assess the functions and impact of various mutations within the promoter regions of genes encoding p28-Omp14 and 19, pQF50K plasmid containing the promoter segments were maintained by growing the E. coli cultures with the addition of kanamycin. The β-galactosidase assays were performed on the lysates prepared from the cultures grown until the OD at 600 nm reached to ∼0.6 using a β-gal assay kit (Invitrogen Technologies, Carlsbad, CA). The experiments were performed thrice with independently grown cultures; specific activity of β-galactosidase was calculated as outlined in the kit protocol.
2.3. In vitro transcription assays
In vitro transcription reactions were performed in 10 μl reaction mixture containing 0.13 pmol each of the supercoiled plasmid DNA as the template and using RNAP holoenzyme containing either recombinant E. chaffeensis σ70 or its derivatives.32 The holoenzyme was prepared by mixing 0.5 μl of 1:10 diluted stock of E. coli core enzyme (Epicentre, Madison, WI) mixed with 10-fold molar excess of purified recombinant E. chaffeensis σ70 or its derivatives and kept in ice for 30 min prior to using for the reactions. The transcription reactions were performed at 37 °C for 20 min, and the reactions were terminated by adding 7 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol). Six microliters each of the samples were resolved on a 6% polyacrylamide sequencing gel with 7 M urea, then gels were transferred to a Whatman paper, dried and 162 nucleotide transcripts were visualized by exposing an X-ray film to the gels. The transcripts were quantified using ImageJ software (http://rsb.info.nih.gov/ij).
2.4. Native PAGE analysis
The DNA promoter segments of p28-Omp14 gene (222 bp) which included the AT-rich spacer (WT) or the modified spacers were generated by PCR from the p28-Omp14 gene. The modified derivatives of the spacer containing complementary sequence (SP1), GC-rich spacer (SP2), or the p28-Omp19 gene spacer inserted in place of the p28-Omp14 spacer (SP3) cloned in plasmid pQF50K were used as templates for amplification using the primers, Gene14-up and Gene14-down (Supplementary Table S2). The PCR products were separated by electrophoresis at 4 °C in 0.5× TBE buffer on a non-denaturing 8% polyacrylamide gel. The DNA in the gel was stained with ethidium bromide and visualized by UV illumination and images captured using KODAK 1D Image Analysis system.
2.5. Modelling of DNA fragments in silico
The spacer sequence DNA segments (WT, SP1, SP2 and SP3) described above were assessed computationally using the online software, ‘model.it’ (http://hydra.icgeb.trieste.it/dna/model_it.html) using the parameter ‘Electrophoresis (dinucleotide)’ to predict the DNA structure. The resulting predicted structures were downloaded to pdb format and PyMOL was used to prepare figures.
2.6. Statistical analysis
Statistical analyses were performed using Student’s t-test, and a P-value <0.05 was considered significant. P-values between 0.05 and 0.01 are identified with a single asterisk and P-value <0.01 are identified with double asterisks.
3. Results
3.1. Ehrlichia chaffeensis genes encoding for the p28-Omp14 and 19 proteins recognized by σ70 require the -35 motif, but not -10 motif, for transcription
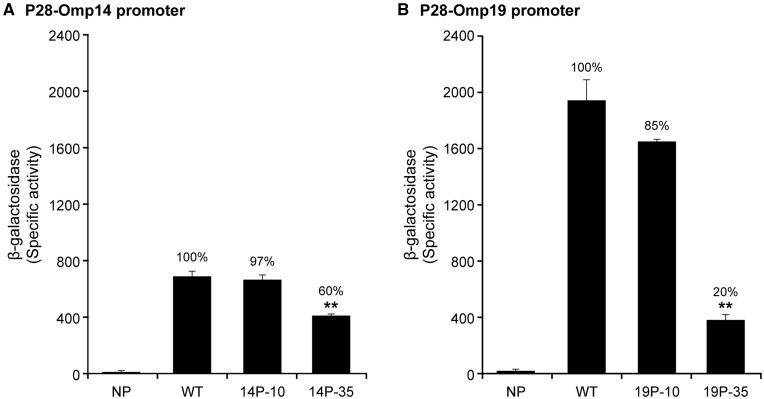
Our prior studies demonstrated that the -35 motif, but not the -10 motif, is critical to E. chaffeensis promoter activity.26 That study was carried out using the E. coli σ70 containing RNAP holoenzyme and thus the data may not be a true reflection of the outcome from the RNAP of the pathogen. To validate the data, we developed an E. coli surrogate system expressing E. chaffeensis σ70 by taking advantage of a previously described E. coli strain (CAG20177) in which the endogenous σ70 gene (rpoD) expression is controlled by the repressible trp promoter.28 In particular, the expression of chromosomally encoded E. coli σ70 requires IAA for optimal growth, as it relieves the tryptophan repression (Supplementary Fig. S1). In the absence of IAA, the E. coli growth is significantly inhibited (e.g. ∼6-fold difference between the cultures with IAA or without IAA at 3 h). The inhibition was also significantly releaved when complemented by another related σ70, as we observed with the introduction of the plasmid expressing the E. chaffeensis σ70 gene (Ech-rpoD) from the lac promoter in the presence of IPTG (Supplementary Fig. S1). This modified E. coli expressing Ech-rpoD is then used for studying the pathogen gene promoters. We used this E. coli system to map promoters of two E. chaffeensis genes; Ech_1136 and Ech_1143 encoding for the proteins p28-Omp14 and p28-Omp19, respectively. These genes were previously identified as transcribed by the E. chaffeensis σ70.32 These gene promoter segments, cloned in front of a reporter gene for β-galactosidase in a plasmid, were used to transform the modified CAG20177 strain of E. coli (Supplementary Fig. S2). Our initial experiments in media lacking IAA tested to assess differences in transcription with or without the induction of E. chaffeensis σ70 by IPTG (Supplementary Fig. S3). As the β-galactosidase activity is also observed for the induced bacteria that is not significantly different from the non-induced, due to the leaky expression from lac promoter,28 all subsequent assessments were carried out without adding IPTG to the culture media. The p28-Omp19 gene promoter induced ∼3-fold more β-galactosidase compared with that found for the promoter of p28-Omp14 (Fig. 1). The complete deletion of -35 motifs from promoters of the genes encoding p28-Omp14 and p28-Omp19 caused a 40% and 80% reduction of the promoter activity, respectively (P ≤0.005), while deletion of -10 motifs from these two promoters resulted in non-signifant change (Fig. 1).
Figure 1.
Importance of -10 and -35 motifs of two Ehrlichia chaffeensis gene promoters assessed in the surrogate system of E. coli strain CAG20177. The β-galactosidase expression, driven by E. chaffeensis p28-Omp14 (A) or p28-Omp19 (B) gene promoters or from the promoters containing deletion mutations at -10 and -35, was assessed relative to no promoter controls. (Constructs; NP, no promoter; WT, wild-type promoter; 14P-10 and 19P-10 represent deletions at -10 motifs and 14P-35 and 19P-35 refer to deletion constructs with -35 motif deletions.) Significant changes in the β-galactosidase activity were identified compared with the data observed for the WT constructs.
3.2. Identifying the critical sequence determinants of -35 motifs in E. chaffeensis genes recognized by σ70
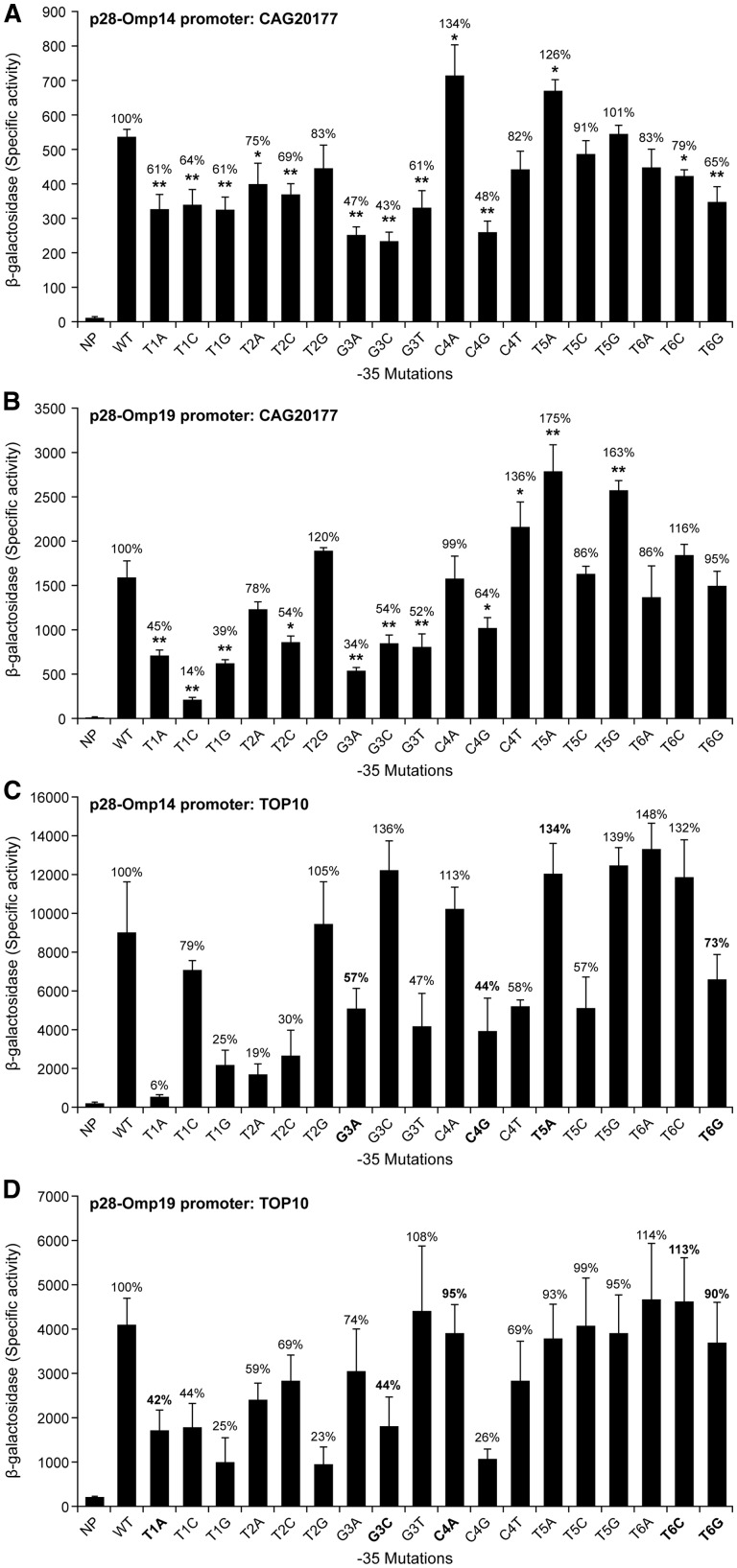
The -35 motifs are extensively conserved for E. chaffeensis genes; its consensus sequence is TTGWNW.27 Further, this motif is identical for p28-Omp14 and p28-Omp19 genes (TTGCTT) (Supplementary Fig. S2A). To define the critical sequence determinants for the promoter activity, substitutions at each base of the six nucleotide motifs were made in the p28-Omp14 and 19 gene promoters and the impact of mutations was assessed by changes in the β-galactosidase expression with E. chaffeensis σ70 in the E. coli surrogate system (Fig. 2A and B). For each base pair substitution, the combination of letters and numbers indicates a specific substitution mutation. For example, T1A indicates a change from T to A transversion at the first position in the -35 motif. The mutations in the first three nucleotides (TTG) had a significant impact in reducing the promoter activities of both the genes. The impact of mutations was also gene-specific. For p28-Omp14 gene promoter, substitution at the first position T to any other nucleotide resulted in ∼40% decline in the promoter activity. Mutations in p28-Omp19 gene at this position also caused a decline in the promoter activity, however, the nucleotide changes caused a greater decline which ranged from 55% to 86%. The promoter activity for this gene is also different for different substitutions; T1C had the greatest impact. Mutations in the second T for both genes had lesser impact compared with the first position mutations. T2A mutation in both genes had an approximately equal amount of decline in the promoter activities (22–25% decline), whereas the T2C mutation caused slightly variable declines in the promoter activities (31% for p28-Omp14 and 46% for p28-Omp19) and T2G had an opposite effect trend for the two gene promoters; this mutation resulted in decline in promoter activity for gene 14 and enhancement for gene 19 promoter. The G3 position resulted in the strongest reduction of promoter activity of both the genes; 39–57% for p28-Omp14 and 46–66% for p28-Omp19. Substitutions in the fourth position also caused significant variations in the promoter activities; C4A mutation in p28-Omp14 and C4T in p28-Omp19 caused increases in the respective promoter activites by ∼35%, whereas no significant change was observed for the C4A mutation for p28-Omp19 and for the C4T mutation for p28-Omp14. C4G transversions for both genes resulted in the promoter activities decline to 52% and 36%, respectively. Substitutions in the fifth position T to A caused a substantial enhancement of the promoter activities for both the genes (26% and 75%, respectively). T5C mutation had no significant effect for both the gene promoters, whereas T5G caused about a 63% increase for p28-Omp19 promoter and had no significant for p28-Omp14 promoter. Mutations in the sixth position had no significant impact for p28-Omp19 promoter, but notable declines in the promoter activities were observed for the T6C and T6G mutations for the p28-Omp14 promoter (21% and 35% declines, respectively). The extensive mutational analysis spanning all six positions of the -35 were also assessed for both the gene promoters in an E. coli strain (TOP10) with its native σ70 (Fig. 2C and D). The data revealed that the E. chaffeensis σ70 differed considerably compared with the E. coli σ70 in responding to various point mutations assessed. In particular, only four substititions in p28-Omp14 gene promoter and five substitions in p28-Omp19 gene promoter correlated well in altering the promoter activities when using σ70 of E. chaffeensis and E. coli (within ∼10% variations). Mutations that correlated well in altering the promoter activity with σ70 of E. coli and E. chaffeensis were identified with bold text in Fig. 2C and D. These data suggest that, while the E. coli σ70 may complement the function of E. chaffeensis σ70, the promoter specificities the two sigma factors are distinct in recognizing the Ehrlichia promoters.
Figure 2.
Mapping the sequence determinants of -35 motifs in Ehrlichia chaffeensis genes. The β-galactosidase expression driven by E. chaffeensis promoters constructs containing point mutations at each of the six nucleotide positions of the -35 motifs of genes encoding p28-Omp14 (A) and p28-Omp19 (B) were measured in the CAG20177 strain of E. coli expressing E. chaffeensis σ70. The experiment included the no promoter (NP) and wild-type promoter (WT) controls. Each mutation is identified with a change of the nucleotide at each position to the modified nucleotide. β-galactosidase expression was presented relative to the respective wild-type promoters. The β-galactosidase expression driven by E. chaffeensis promoters constructs containing point mutations at each of the six nucleotide positions of the -35 motifs of genes encoding p28-Omp14 (C) and p28-Omp19 (D) also were measured in the TOP10 strain of E. coli expressing its native chromosomally expressed σ70 with only the promoter plasmid pQF50K-p28-Omp14 or pQF50K-p28-Omp19 . The experiment also included the no promoter (NP) and wild-type promoter (WT) controls. Only four substititions in p28-Omp14 gene promoter and five substitions in p28-Omp19 gene promoter correlated well in altering the promoter activities when using σ70 of E. chaffeensis and E. coli (within ∼10% variations); these mutations were identified in this figure with bold text.
3.3. In vitro transcription for the sequence determinants of -35 motif by recombinant E. chaffeensis σ70
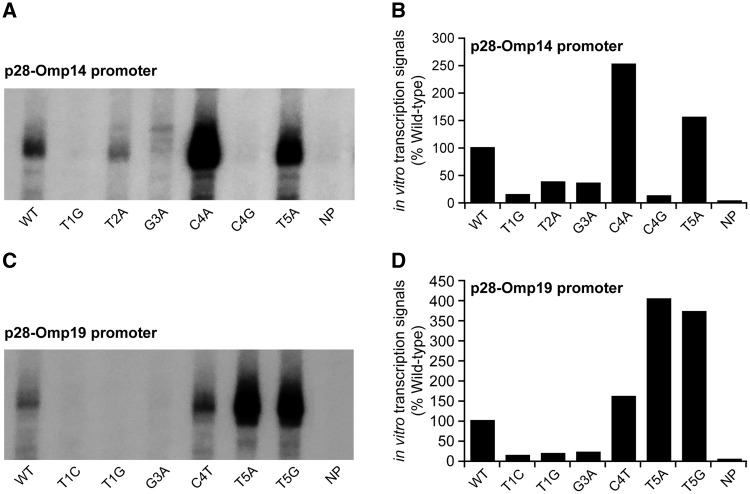
To validate the results of -35 motif mutational analyses in CAG20177, we tested several promoter mutations by performing in vitro transcription assays with the holoenzyme reconstituted with recombinant E. chaffeensis σ70.32 We randomly selected five mutants of genes encoding p28-Omp14 and p28-Omp19 and the mutant promoters were re-cloned into the G-less cassette and used as the transcription templates of in vitro transcription assays (Fig. 3). The mutants causing decline in the transcriptional activity in E. coli surrogate system also yielded reduced levels of in vitro transcripts and likewise the mutants which caused an enhancement of transcription also resulted in the increased synthesis of in vitro transcripts. We also noted minor bands migrating slightly larger than the predicted transcripts in two mutnats (T2A and G3A); it is possible that these products may have generated by the RNAP binding to other sites near the promoter in the absence of specific binding. As we previously described,32 the recombination σ70 alone or E. coli core enzyme without the sigma factor did not generate in vitro transcripts (data not shown).
Figure 3.
In vitro transcription analysis validating the Ehrlichia chaffeensis gene promoter mutants spanning the -35 motifs. Six each of the randomly selected mutations at -35 motifs of p28-Omp14 (A and B) and p28-Omp19 (C and D) were exmined by in vitro transcription assays using RNAP holoenzyme containing E. chaffeensis recombinant σ70. The abundance of transcripts for each gene was captured from the 32P incorporation. The intensity of a band signals in a gel for in vitro transcriptions made for the wild-type and mutant promoters was determined using the software ImageJ. Panels A and C have the image data, and panels B and D included the quantitative data collected from the image signals. The bars show the relative transcription products of mutant promoters as the percentage of transcripts compared with the wild-type promoter for σ70. (NP is a construct without a promoter; WT refers to a wild-type promoter, and various mutant promoter constructs are identified as in Fig. 2).
3.4. Substitutions in region 4.2 of E. chaffeensis σ70
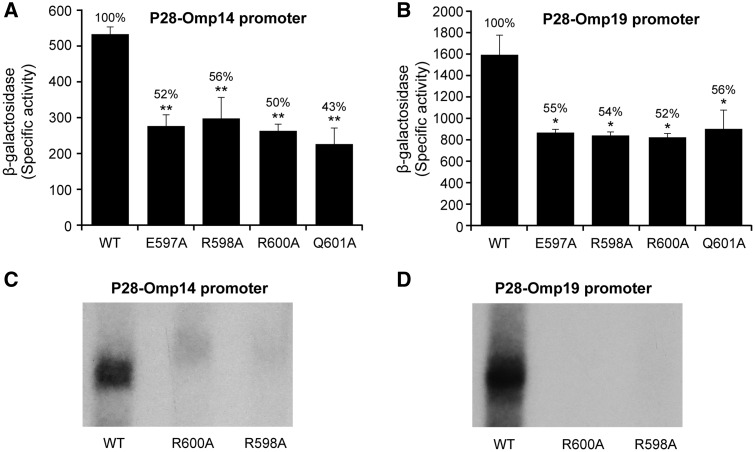
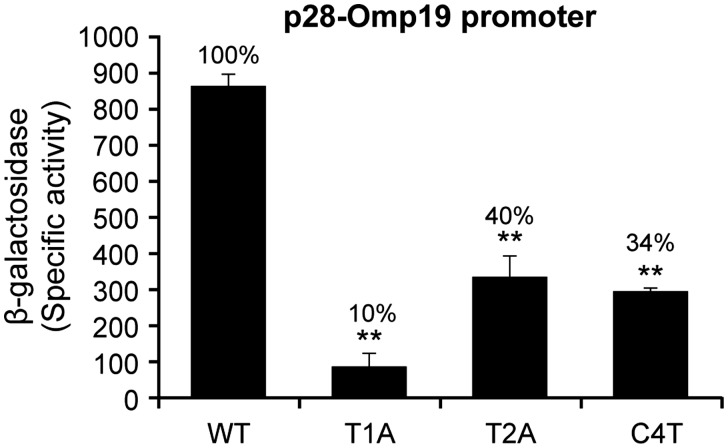
A conserved region near the C-terminus of the E. coli sigma factor is identified as essential for DNA binding and promoter activity, specifically to the -35 motif,22,28,34 which includes four amino acids in the 4.2 regions of E. coli σ70 and are also conserved in E. chaffeensis σ70.27 In E. chaffeensis σ70, the conserved amino acids are; glutamic acid at 597, two arginines at 598 and 600, and glutamine at 601. To evaluate if mutations in these four amino acids in E. chaffeensis σ70 would affect the promoter activity, individual substitution mutation were created to modify these four amino acids in the E. chaffeensis σ70 gene coding sequence in the expression plasmid to alanine. Transcriptional activities of the modified sigma factors were assessed with the WT promoters of p28-Omp14 and p28-Omp19 in the E. coli surrogate system (CAG20177). Mutations at all four locations for both the gene promoters resulted in significant reduction of the promoter activity (∼48–57% decline) (Fig. 4A and B). To verify these data, we also performed in vitro transcription assays with the E. chaffeensis σ70 mutants where arginine at position 598 and 600 was modified to alanine using the WT promoters of both p28-Omp14 and p28-Omp19 genes (Fig. 4C and D). The in vitro transcription also was reduced for the mutants. We also tested the ability of one of the mutant forms (E579A mutation) of E. chaffeensis σ70 in driving transcription from three mutant promoters of p28-Omp19 with substitutions T1A, T2A and C4T in E. coli surrogate system (Fig. 5). The promoter activities for all three mutations in the of p28-Omp19 promoter caused a further decline of 90%, 60% and 66% relative to the WT promoter, respectively.
Figure 4.
Substitutions in region 4.2 of Ehrlichia chaffeensis σ70 influence the promoter activity of the wild-type E. chaffeensis promoters (A, p28-Omp14 and B, p28-Omp19). Mutations to change amino acids to alanine at four conserved residues (E597, R598, R600 and Q601) of E. chaffeensis σ70 were assessed with the wild-type promoters; p28-Omp14 (A) and p28-Omp19 (B). β-galactosidase expression was measured for the mutant proteins relative to the wild-type (WT) E. chaffeensis σ70. Mutations in the conserved amino acids of E. chaffeensis σ70 4.2 region also cause reduction in the in vitro transcript synthesis from the wild-type promoters (C and D). In vitro transcription analysis was performed using RNAP holoenzyme containing E. chaffeensis recombinant wild-type σ70, or with its mutants R598A or R600A and with wild-type p28-Omp14 (C) and p28-Omp19 (D) promoters.
Figure 5.
A change in a conserved amino acid of Ehrlichia chaffeensis σ70 4.2 region further reduced the promoter activity in -35 motif mutants. The activities for the -35 motif mutants T1A, T2A and C4T of p28-Omp19 promoter were assessed with E. chaffeensis σ70 mutant (E597A) by measuring changes in the β-galactosidase expression in the E. coli strain CAG20177. The reduction of the enzyme activity was expressed relative to the wild-type promoter.
3.5. The spacer sequences affect promoter activity
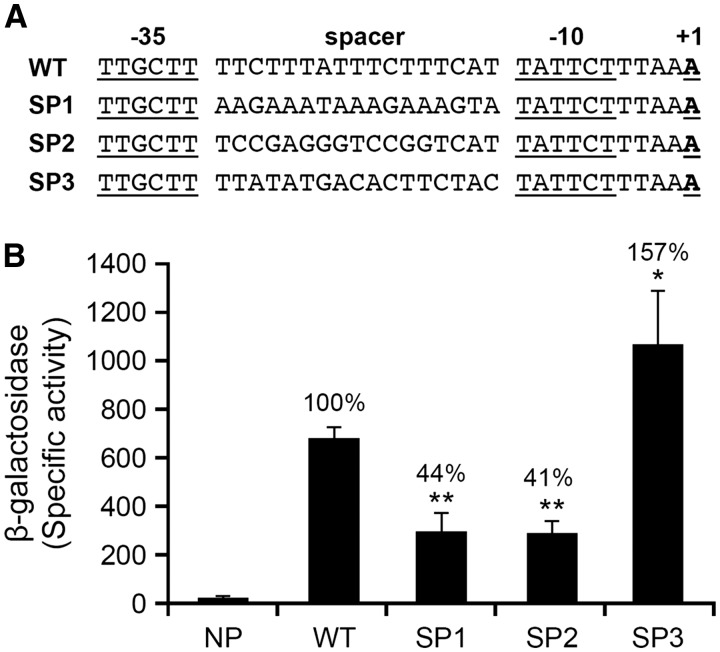
The extensive experimental analysis described above revealed that the -35 motif, but not -10, impacted the E. chaffeensis transcription driven by its σ70 for the two genes assessed. The deletion of -35 motifs caused significant decline of the promoter activities, but did not completely abolish the transcription, suggesting that the promoter function requires the contributions of additional sequences. In particular, we hypothesized that the sequences other than the -35 motifs also contribute to the differences in the promoter activities of the two genes. In E. coli, the length of a spacer sequence between the -10 and -35 motifs influences promoter activity.33,35 Previous studies in E. coli also demonstrated that the nucleotide differences within the spacer sequences also influence the promoter activity.23–25 We investigated if spacer sequences also similarly influence the promoter activity of E. chaffeensis genes by modifying the spacer sequences. We constructed three mutants to modify the spacer region of the p28-Omp14 gene promoter: (i) the AT-rich spacer sequence of the gene is replaced with complementary nucleotides at each position of the spacer (SP1) to keep the AT and GC content constant, (ii) the sequence is replaced with a sequence having GC-rich spacer sequence (SP2), or (iii) the spacer sequence of p28-Omp14 gene promoter is replaced with the p28-Omp19 gene promoter spacer sequence (SP3) (Fig. 6A). The SP3 construct is included to test if replacing the spacer sequence of p28-Omp14 gene with p28-Omp19 gene is sufficient in enhancing the promoter activity to that observed for the p28-Omp19 gene promoter, as both the promoters have identical -35 motifs. The WT and the modified constructs were tested in the E. coli system by measuring the β-galactosidase activity; the SP1 and SP2 caused the reduction of the enzyme activity by 56% and 59%, respectively (Fig. 6B). The substitution with the p28-Omp19 gene spacer (SP3) caused enhancement of the promoter activity by ∼1.6-fold (Fig. 6B). These results suggested that the spacer sequences of the E. chaffeensis gene promoters play an important role in transcriptional variations and may also account for some of the differences in the activities of p28-Omp14 and p28-Omp19 gene WT promoters.
Figure 6.
AT-rich spacer sequence located between -10 and -35 motif contributes to altering the promoter activity of Ehrlichia chaffeensis genes. Promoter fragments used in the assays are as in Supplementary Fig. S2 for wild-type p28-Omp14 gene. (A) sequence spanning from +1 to -35 motif and the AT-rich spacer sequence is presented for the wild-type construct (WT) and for the constructs with modified spacer sequences which included replacing the AT-rich spacer with complementary sequence (SP1), with GC-rich spacer sequence (SP2) or with the p28-Omp19 gene spacer sequence (SP3). (B) The β-galactosidase expression driven by promoters of WT, SP1, SP2 and SP3 in E. coli (CAG20177) expressing E. chaffeensis σ70 was measured and the data were presented. The assay also included the data generated from the promoterless construct control (NP).
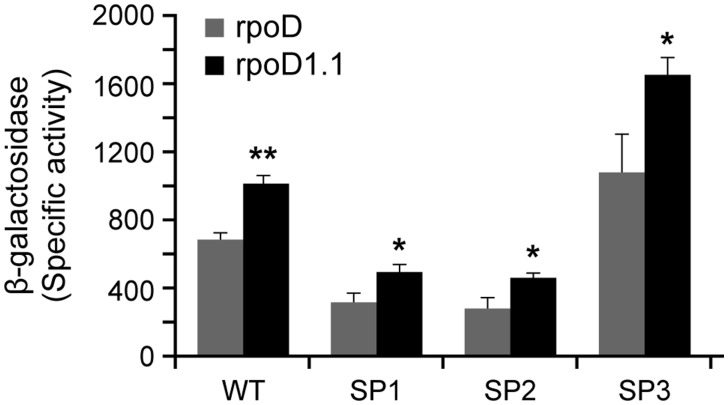
Previous studies in E. coli revealed that the 1.1 region of its σ70 contributes to the promoter activity by modulating the formation of stable polymerase and promoter complexes.24,36,37 Deletion of this region causes enhanced or decreased promoter activity depending on a promoter.24,36 The impact of 1.1 deletion is also variable for different spacer sequences for the promoters containing the same -35 and -10 motifs, as evidenced for Ptac and PuvsX/sigma.36 As E. chaffeensi p28-Omp14 and p28-Omp19 promoters have the identical -35, and that -10 was found to be less important for the transcription from these two gene promoters, we investigated if the 1.1 deletion E. chaffeensis σ70 also cause variations in the promoter activities if we modify the spacer sequences. The deletion of 1.1 region in E. chaffeensis σ70 led to significant enhancement of the promoter activity when assessed for the WT p28-Omp14 promoter segment. Modified spacer sequences containing the complementary sequence (SP1) or GC sequence (SP2) or the replacement of the spacer sequence with WT p28-Omp19 promoter spacer sequence (SP3) also resulted in the enhancement of promoter activities (Fig. 7).
Figure 7.
WT and SP1, SP2 and SP3 constructs were assessed for their promoter activities in E. coli expressing wild-type Ehrlichia chaffeensis σ70 from rpoD gene or from its mutant having deletion at 1.1 region (rpoD 1.1). The β-galactosidase expression was significantly higher for all four promoters when assessed with rpoD 1.1 compared with the wild-type rpoD.
The changes of base sequence for the spacer DNA fragments with identical length possibly render different conformations or curvatures to a DNA molecule24,25 and may aid in altering the affinities of RNAP binding and transcription. DNA conformational changes can also impact migration patterns in a polyacrylamide gel (PAG).24,25 To test this, we compared the mobility of DNA fragments of p28-Omp14 promoter segments, WT, SP1, SP2 and SP3, by subjecting to electrophoresis in a non-denaturing PAG (Fig. 8A). The gel migration patterns are different for all four DNAs. Consistent with these results, the predicted DNA structures have different topologies as judged by the ‘model it’ program (Fig. 8B).38
Figure 8.
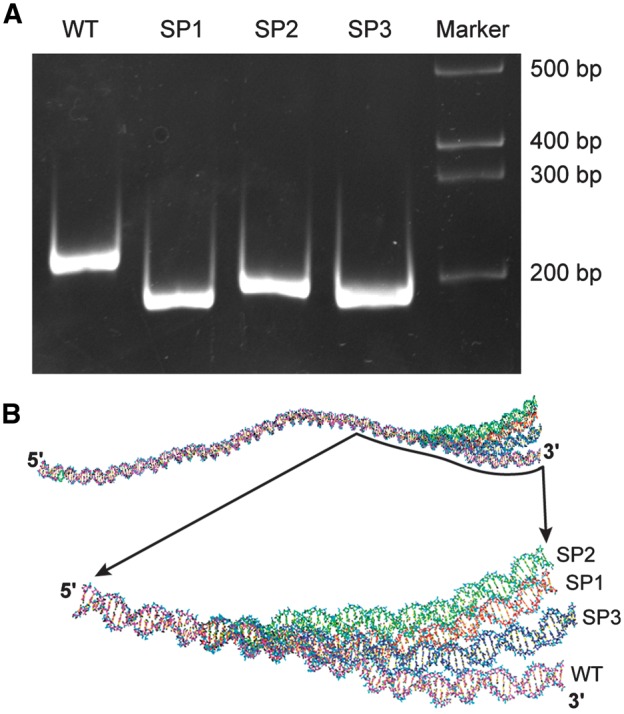
Changes in the AT-rich spacer sequence impact DNA gel migration and conformation. (A) Wild-type p28-Omp14 gene promoter and with modified spacers; SP1, SP2 and SP3 (described in Fig. 6) had variable migration patterns when resolved in a non-denaturing PAGE. (B) Topology of the wild-type and the three spacer modified promoter segments revealed conformational changes when assessed by the prediction program;38 WT, pink; SP1, red; SP2, green and SP3, blue.
4. Discussion
It is unclear how E. chaffeensis and the related Anaplasmataceae family pathogens transmitted from ticks regulate their gene expression in vertebrate and tick hosts. Ehrlichia chaffeensis genome contains genes only for two sigma factors (σ32 and σ70) and for very few predicted transcriptional regulators (GenBank # NC_007799.1).1 To understand how the Anaplasmataceae family pathogens adapt to their vertebrate and tick hosts and sense nutrient and starving environments within an infected host cell requires a detailed knowledge about the pathogens’ gene regulation. Studying the regulation of gene expression is also important in defining the molecular basis for the conversions to the pathogens’ infectious form (dense core cells) and replicating form (reticulate cells) within a phagosome of an infected host cell. Our recent data support the hypothesis that the E. chaffeensis sigma factors, σ32 and σ70, function cooperatively in transcribing pathogen genes.27 The current study is the first to undertake a detailed investigation at the gene level to map differences in gene expression accomplished by two distinct and closely related genes; Ech_1136 and Ech_1143, of the pathogen encoding for the proteins; p28-Omp14 and p28-Omp19, respectively.
In the absence of a genetic transformation system, researchers investigating the gene expression of intracellular Chlamydia species pathogens relied on the use of in vitro transcription method to study the bacterial gene regulation and to define the transcriptional mechanisms.31,39–41 In vitro transcription assays are proven the most valuable in defining the transcriptional machinery of several Chlamydia genes.40–46 Ehrlichia species research is also challenging due to lack of a well-established genetic transformation system and the lack of natural plasmids in them further complicates the research focused on studying gene regulation. In the current study, we developed and utilized the E. coli surrogate system to map the DNA binding domains involved in regulating the gene expression in E. chaffeensis. Further, we used the in vitro transcription system to validate the data. The approaches also aided in determining the molecular basis for differences in gene expression from two closely related genes.
Escherichia coli transcriptions for housekeeping genes are driven by RNAP holoenzyme containing σ70 which recognizes two highly conserved motifs; referred as -10 and -35 motifs.13 The consensus motif sequences are TATAAT and TTGACA, respectively. The σ70 homologs are also extensively conserved in several other Gram-negative bacteria.12 We recently reported that the consensus -10 and -35 motifs for E. chaffeensis σ70 are TATTNT and TTGNTT, respectively.27 We also reported that the -10 and -35 motifs for the alternative sigma factor, σ32, in E. chaffeensis (TATATN and TTGAAA, respectively) are very similar to σ70 consensus sequences for the genes we assessed.27 The -10 motif of E. chaffeensis, however, differs considerably from the E. coli σ32 consensus -10 motif (CCCCATNT), while the consensus -35 motif is identical (TTGAAA).47,48 Consistent with the extensive homology of σ32 and σ70 consensus -10 and -35 motifs, E. chaffeensis genes can also be transcribed by both the sigma factors, but with varying affinities.27 Ehrlichia chaffeensis has two morphological forms; dense core and reticulate cells49,50 and it is entirely unknown how the organism and the related rickettsial organisms having two distinct morphological forms and also having the ability to adapt to dual hosts regulate their gene expression. Considering the lack of genetic tools and transformation system, the methods described in the current study will be valuable in defining the gene regulation in this organism, the related Anaplasmataceae family organisms, and in extending studies to other intracellular Gram-negative pathogens having two distinct morphological forms, such as Chlamydia species and Coxiella burnetii.51,52
The consensus -35 motifs in all mapped E. chaffeensis genes, independent of a gene primarily transcribed by σ32 or σ70, contain the extensively conserved first three nucleotides at the 5′ end.27 In this study, we presented data demonstrating that the -10 motifs are not critical for the gene activities of two closely related outer membrane protein genes (p28-Omp14 and p28-Omp19) driven by its primary sigma factor, σ70. At this time, it is not clear if -10 motif is similarly less important for other pathogen genes. This hypothesis needs further investigation. We also presented evidence that the -35 motifs are critical for the σ70 function for the two genes assessed. Further, we reported that any changes to the first three nucleotides of the -35 motif, TTG, result in significant decline in the promoter activities, despite different degrees of variations observed for the two gene promoters. The TTG in -35 motifs is conserved in most of the E. chaffeensis genes suggesting that its interactions with σ70 may be vital for its function, although the remaining three nucleotides on the -35 motif may also play a critical role for the gene-specific transcription. The TTG is also found to be important for σ70 gene promoters of E. coli.53 It is well known that sigma factors possess variable numbers of DNA binding regions.12,54 Each region holds a specific role in promoter recognition. For example, the region 4 located in the C-terminus contains a helix-turn-helix (HTH) motif of known DNA-binding protein.55 Previous studies revealed that the 4.2 region in E. coli σ70 is involved in the base-specific recognition with the -35 motif.15,20 Moreover, the substitutions in four conserved charged amino acids at E265, R266, R268 and Q269 in E. coli σ32 to a non-polar amino acid, alanine, cause reduction of the promoter activity.22 Sequence allignment revealed that the E. chaffeensis σ32 has the same four amino acids as conserved and mutating these amino acids to alanine also resulted in the reduction of its function in driving the promoter activities of the genes recognized by it.27 These four amino acids are also conserved in E. chaffeensis σ70 and that the mutations in these amino acids to alanine also negatively impacted the promoter activity. In Fig. 5, when combined E579A substitution in σ70 and T1A, T2A and C4T substitution in the p28-Omp19 promoter, respectively, the lower activity of promoter was observed compared with WT promoter. The results suggest that the E579 may not interact with these bases of the -35 motif for the p28-Omp19 promoter, as reported previously for E. coli.22 Additional experimental analysis is necessary to test this hypothesis and to evaluate if this domain in E. chaffeensis is also involved in base-specific recognition.
It is well demonstrated in E. coli that the length of a spacer sequence between the -10 and -35 motifs influences promoter activity.33,35 Recent studies also suggest that the kind of specific nucleotides present within a spacer region also influence the promoter activity.23–25 In the current study, we investigated the role of spacer sequences for E. chaffeensis RNAP function and their contributions to differences in transcription levels of two closely related genes, as both the genes have different nucleotide sequences in the spacers while the lengths remain the same. Indeed, our data demonstrated that modifying the spacer sequence with complementary sequence in the p28-Omp14 gene promoter or by replacing with a randomly selected GC-rich spacer sequence caused significant reduction in the promoter activity. Interestingly, replacing the WT p28-Omp14 spacer with the spacer from the p28-Omp19 gene promoter enhanced the promoter activity by ∼1.6-fold. The p28-Omp19 gene promoter is ∼3-fold stronger than the p28-Omp14 gene promoter, as evidenced by the 3-fold higher β-galactosidase expression observed in the E. coli surrogate system. The 1.6-fold enhancement of the p28-Omp14 gene promoter activity when replaced with the p28-Omp19 spacer suggests that the spacer sequence is a major contributer for the differences in the promoter activities of the two colesely related outer membrane protein genes. As reported earlier for an E. coli gene,24 the data for E. chaffeensis genes also demonstrate that the variations in spacer sequences influence in altering the promoter activity of a gene, possibly due to differing conformations or curvatures. In particular, we present the evidence that the nucleotide differences within a spacer sequence are important contributors in influencing the promoter strengths, possibly due to altering the curvature of a promoter leading to altered interactions with RNAP. Previous studies in E. coli demonstrated that the spacer sequences affect the RNA polymerase binding affinity.56 This hypothesis remains to be tested for E. chaffeensis.
Depending on the promoter assessed, the loss of region 1.1 within the E. coli σ70 protein can influence the promoter activity of a gene positively or negatively or can cause no impact.36 For example, Hook-Barnard24 reported that the deletion of region 1.1 domain within the E. coli σ70 protein increases the transcription by ∼2-fold from Pmin7 gene promoter. On the contrary, its deletion has no significant effect on the amount of mRNA made from the Pmin7/GC or Pmin/comp promoters when assessed with modified spacer sequences (GC-rich spacer or complementary spacer).24 It is reported that region 1.1 at the N-terminal of σ70 of E. coli affect spacer-mediated changes in transcriptional initiation via converting the trajectory of the spacer of promoter.24 In the current study, we presented evidence that E. chaffeensis σ70 with a mutation of region 1.1 significantly enhances the activity of WT p28-Omp14 gene promoter and the promoter with mutant spacer sequences (SP1, SP2 and SP3).
This work is the first to utilize various molecular approaches in defining the -10 and -35 motifs and the AT-rich spacer sequences located between the two motifs of two closely related E. chaffeensis genes encoding for differentially expressed proteins; p28-Omp14 and p28-Omp19. The differences in the spacer sequences alone are sufficient in altering the gene transcription by 1.6-fold. In particular, we presented the first evidence demonstrating that the difference in transcription by ∼50% from two closely related genes can be accounted due to differences in their AT-rich spacer sequences. DNA binding proteins may be additional contributors in influencing the gene expression. Previous studies by Cheng et al.57 using the E. coli RNAP holoenzyme demonstrated that an E. chaffeensis DNA regulator, EcxR, serves as an activator in promoting the gene expression of several type IV secretion system genes of the pathogen. The role of DNA transcription regulators remain to be investigated for their contributions to differential expression from p28-Omp genes. The E. coli surrogate system described in the current study can facilitate greatly in evaluating the DNA transcription regulators of E. chaffeensis. We believe that the current study will also be valuable for furthering our understanding of the regulation of gene expression in E. chaffeensis and in defining the detailed molecular basis of differential gene expression and its contributions to the pathogen adaptations to dual hosts and in sensing the distinct host cell environments. The molecular methods described here are also valuable for studies focused on understanding the gene regulation in other related rickettsial pathogens.
In summary, we developed an E. coli surrogate system and used it to extensively map E. chaffeensis two σ70 gene promoters. The E. coli system was also used to present evidence that the loss of -10 motifs has no role for the gene expression for the two genes assessed in the current study. We also mapped the critical determinants of the -35 motif by performing mutational analysis. Further, we demonstrated that the AT-rich sequences are involved in contributing to promoter-specific variations in the gene transcriptions.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by PHS grant number AI070908 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. This manuscript is contribution number 16-109-J from the Kansas Agricultural Experiment Station. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
Acknowledgements
We thank Dr Pieter L. deHaseth of Case Western Reserve University, Cleveland, OH for providing pQF50K_groE and pSAKT32 plasmids and Dr Carol A. Gross of University of California, San Francisco, CA for providing the E. coli CAG20177 strain and Dr Ming Tan of the University of California, Irvine, CA for providing the G-less cassette plasmid, pMT504. We also thank Ms Mal Rooks Hoover for her help in preparing the figures.
References
- 1.Dunning Hotopp J.C., Lin M., Madupu R., et al. 2006, Comparative genomics of emerging human ehrlichiosis agents, PLoS Genet., 2, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yabsley M.J. 2010, Natural history of Ehrlichia chaffeensis: vertebrate hosts and tick vectors from the United States and evidence for endemic transmission in other countries, Vet. Parasitol., 167, 136–48. [DOI] [PubMed] [Google Scholar]
- 3.Walker D.H., Paddock C.D., Dumler J.S. 2008, Emerging and re-emerging tick-transmitted rickettsial and ehrlichial infections, Med. Clin. North Am., 92, 1345–61. [DOI] [PubMed] [Google Scholar]
- 4.Rikihisa Y. 2010, Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells, Nat. Rev. Microbiol., 8, 328–39. [DOI] [PubMed] [Google Scholar]
- 5.Walker D.H., Dumler J.S. 1996, Emergence of the ehrlichioses as human health problems, Emerg. Infect. Dis., 2, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson W.R., Lockhart J.M., Stallknecht D.E., Howerth E.W., Dawson J.E., Rechav Y. 2001, Persistent Ehrlichia chaffeensis infection in white-tailed deer, J. Wildl. Dis., 37, 538–46. [DOI] [PubMed] [Google Scholar]
- 7.Dumler J.S., Sutker W.L., Walker D.H. 1993, Persistent infection with Ehrlichia chaffeensis, Clin. Infect. Dis., 17, 903–5. [DOI] [PubMed] [Google Scholar]
- 8.Unver A., Rikihisa Y., Stich R.W., Ohashi N., Felek S. 2002, The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts, Infect. Immun., 70, 4701–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo G.M., Cheng C., Tomich J., Ganta R.R. 2008, Total, membrane, and immunogenic proteomes of macrophage- and tick cell-derived Ehrlichia chaffeensis evaluated by liquid chromatography-tandem mass spectrometry and MALDI-TOF methods, Infect. Immun., 76, 4823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuriakose J.A., Miyashiro S., Luo T., Zhu B., McBride J.W. 2011, Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation, PLoS One, 6, e24136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlin M., Kingston R., Gilman M., Wiggs J., deVera A. 1983, Isolation of bacterial and bacteriophage RNA polymerases and their use in synthesis of RNA in vitro, Methods Enzymol., 101, 540–68. [DOI] [PubMed] [Google Scholar]
- 12.Lonetto M., Gribskov M., Gross C.A. 1992, The sigma 70 family: sequence conservation and evolutionary relationships, J. Bacteriol., 174, 3843–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross C.A., Chan C., Dombroski A., et al. 1998, The functional and regulatory roles of sigma factors in transcription, Cold Spring Harb. Symp. Quant. Biol., 63, 141–55. [DOI] [PubMed] [Google Scholar]
- 14.Paget M.S., Helmann J.D. 2003, The sigma70 family of sigma factors, Genome Biol., 4, 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegele D.A., Hu J.C., Walter W.A., Gross C.A. 1989, Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase, J. Mol. Biol., 206, 591–603. [DOI] [PubMed] [Google Scholar]
- 16.Roberts C.W., Roberts J.W. 1996, Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase, Cell, 86, 495–501. [DOI] [PubMed] [Google Scholar]
- 17.Panaghie G., Aiyar S.E., Bobb K.L., Hayward R.S., de Haseth P.L. 2000, Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex, J. Mol. Biol., 299, 1217–30. [DOI] [PubMed] [Google Scholar]
- 18.Koo B.M., Rhodius V.A., Campbell E.A., Gross C.A. 2009, Dissection of recognition determinants of Escherichia coli sigma 32 suggests a composite -10 region with an ‘extended -10’ motif and a core -10 element, Mol. Microbiol., 72, 815–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juang Y.L., Helmann J.D. 1994, A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids, J. Mol. Biol., 235, 1470–88. [DOI] [PubMed] [Google Scholar]
- 20.Gardella T., Moyle H., Susskind M.M. 1989, A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity, J. Mol. Biol., 206, 579–90. [DOI] [PubMed] [Google Scholar]
- 21.Young B.A., Gruber T.M., Gross C.A. 2002, Views of transcription initiation, Cell, 109, 417–20. [DOI] [PubMed] [Google Scholar]
- 22.Kourennaia O.V., Tsujikawa L., Dehaseth P.L. 2005, Mutational analysis of Escherichia coli heat shock transcription factor sigma 32 reveals similarities with sigma 70 in recognition of the -35 promoter element and differences in promoter DNA melting and -10 recognition, J. Bacteriol., 187, 6762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hook-Barnard IG H.D. 2007, Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters, Gene Regul. Syst. Biol., 1, 275–93. [PMC free article] [PubMed] [Google Scholar]
- 24.Hook-Barnard I.G., Hinton D.M. 2009, The promoter spacer influences transcription initiation via σ70 region 1.1 of Escherichia coli RNA polymerase, Proc. Natl. Acad. Sci. U. S. A., 106, 737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S.S., Typas A., Hengge R., Grainger D.C. 2011, Escherichia coli σ70 senses sequence and conformation of the promoter spacer region, Nucleic Acids Res., 39, 5109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peddireddi L., Cheng C., Ganta R.R. 2009, Promoter analysis of macrophage- and tick cell-specific differentially expressed Ehrlichia chaffeensis p28-Omp genes, BMC Microbiol., 9, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Von Ohlen T., Cheng C., Faburay B., Ganta R.R. 2013, Transcription of Ehrlichia chaffeensis genes is accomplished by RNA polymerase holoenzyme containing either sigma 32 or sigma 70, PLoS One, 8, e81780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonetto M.A., Rhodius V., Lamberg K., Kiley P., Busby S., Gross C. 1998, Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit, J. Mol. Biol., 284, 1353–65. [DOI] [PubMed] [Google Scholar]
- 29.Sanderson A., Mitchell J.E., Minchin S.D., Busby S.J.W. 2003, Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended −10 elements at promoters, FEBS Lett., 544, 199–205. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., deHaseth P.L. 2003, Sigma 32-dependent promoter activity in vivo: sequence determinants of the groE promoter, J. Bacteriol., 185, 5800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan M., Engel J.N. 1996, Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter, J. Bacteriol., 178, 6975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faburay B., Liu H., Peddireddi L., Ganta R.R. 2011, Isolation and characterization of Ehrlichia chaffeensis RNA polymerase and its use in evaluating p28 outer membrane protein gene promoters, BMC Microbiol., 11, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulligan M.E., Brosius J., McClure W.R. 1985, Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter, J. Biol. Chem., 260, 3529–38. [PubMed] [Google Scholar]
- 34.Gregory B.D., Nickels B.E., Darst S.A., Hochschild A. 2005, An altered-specificity DNA-binding mutant of Escherichia coli σ70 facilitates the analysis of σ70 function in vivo, Mol. Microbiol., 56, 1208–19. [DOI] [PubMed] [Google Scholar]
- 35.Aoyama T., Takanami M., Ohtsuka E., et al. 1983, Essential structure of E. coli promoter effect of spacer length between the two consensus sequences on promoter function, Nucleic Acids Res., 11, 5855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuthoori S., Bowers C.W., McCracken A., Dombroski A.J., Hinton D.M. 2001, Domain 1.1 of the σ70 subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes, J. Mol. Biol., 309, 561–72. [DOI] [PubMed] [Google Scholar]
- 37.Wilson Bowers C., Dombroski A.J. 1999, A mutation in region 1.1 of σ70 affects promoter DNA binding by Escherichia coli RNA polymerase holoenzyme, EMBO J., 18, 709–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlahoviček K., Kaján L., Pongor S. 2003, DNA analysis servers: plot.it, bend.it, model.it and IS, Nucleic Acids Res., 31, 3686–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathews S.A., Douglas A., Sriprakash K.S., Hatch T.P. 1993, In vitro transcription in Chlamydia psittaci and Chlamydia trachomatis, Mol. Microbiol., 7, 937–46. [DOI] [PubMed] [Google Scholar]
- 40.Bao X., Pachikara N.D., Oey C.B., et al. 2011, Non-coding nucleotides and amino acids near the active site regulate peptide deformylase expression and inhibitor susceptibility in Chlamydia trachomatis, Microbiology, 157, 2569–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao X., Deighan P., Hua Z., et al. 2009, A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the β subunit and the primary σ subunit, Genes Dev., 23, 1818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan M., Gaal T., Gourse R.L., Engel J.N. 1998, Mutational analysis of the Chlamydia trachomatis rRNA P1 Promoter defines four regions important for transcription in vitro, J. Bacteriol., 180, 2359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akers J.C., Tan M. 2006, Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis, J. Bacteriol., 188, 4236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiu Yin Yu H., Tan M. 2003, σ28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia, Mol. Microbiol., 50, 577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bao X., Nickels B.E., Fan H. 2012, Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of σ66, Proc. Natl. Acad. Sci. U. S. A., 109, 16870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L., Li M., Zhang Y. 2004, Chlamydia trachomatis σ28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae, Microbiology, 150, 205–15. [DOI] [PubMed] [Google Scholar]
- 47.Nonaka G., Blankschien M., Herman C., Gross C.A., Rhodius V.A. 2006, Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress, Genes Dev., 20, 1776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koo B.-M., Rhodius V.A., Nonaka G., deHaseth P.L., Gross C.A. 2009, Reduced capacity of alternative σs to melt promoters ensures stringent promoter recognition, Genes Dev., 23, 2426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J.-Z., Popov V.L., Gao S., Walker D.H., Yu X.-J. 2007, The developmental cycle of Ehrlichia chaffeensis in vertebrate cells, Cell Microbiol., 9, 610–8. [DOI] [PubMed] [Google Scholar]
- 50.Dedonder S.E., Cheng C., Willard L.H., Boyle D.L., Ganta R.R. 2012, Transmission electron microscopy reveals distinct macrophage- and tick cell-specific morphological stages of Ehrlichia chaffeensis, PLoS One, 7, e36749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hogan R.J., Mathews S.A., Mukhopadhyay S., Summersgill J.T., Timms P. 2004, Chlamydial persistence: beyond the biphasic paradigm, Infect. Immun., 72, 1843–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coleman S.A., Fischer E.R., Howe D., Mead D.J., Heinzen R.A. 2004, Temporal analysis of Coxiella burnetii morphological differentiation, J. Bacteriol., 186, 7344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djordjevic M. 2011, Redefining Escherichia coli σ70 promoter elements: -15 motif as a complement of the −10 motif, J. Bacteriol., 193, 6305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruber T.M., Gross C.A. 2003, Multiple sigma subunits and the partitioning of bacterial transcription space, Annu. Rev. Microbiol., 57, 441–66. [DOI] [PubMed] [Google Scholar]
- 55.Pabo C.O., Sauer R.T. 1984, Protein-DNA recognition, Annu. Rev. Biochem., 53, 293–321. [DOI] [PubMed] [Google Scholar]
- 56.Liu M., Tolstorukov M., Zhurkin V., Garges S., Adhya S. 2004, A mutant spacer sequence between -35 and -10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent, Proc. Natl. Acad. Sci. U. S. A., 101, 6911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Z., Wang X., Rikihisa Y. 2008, Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein, J. Bacteriol., 190, 2096–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.